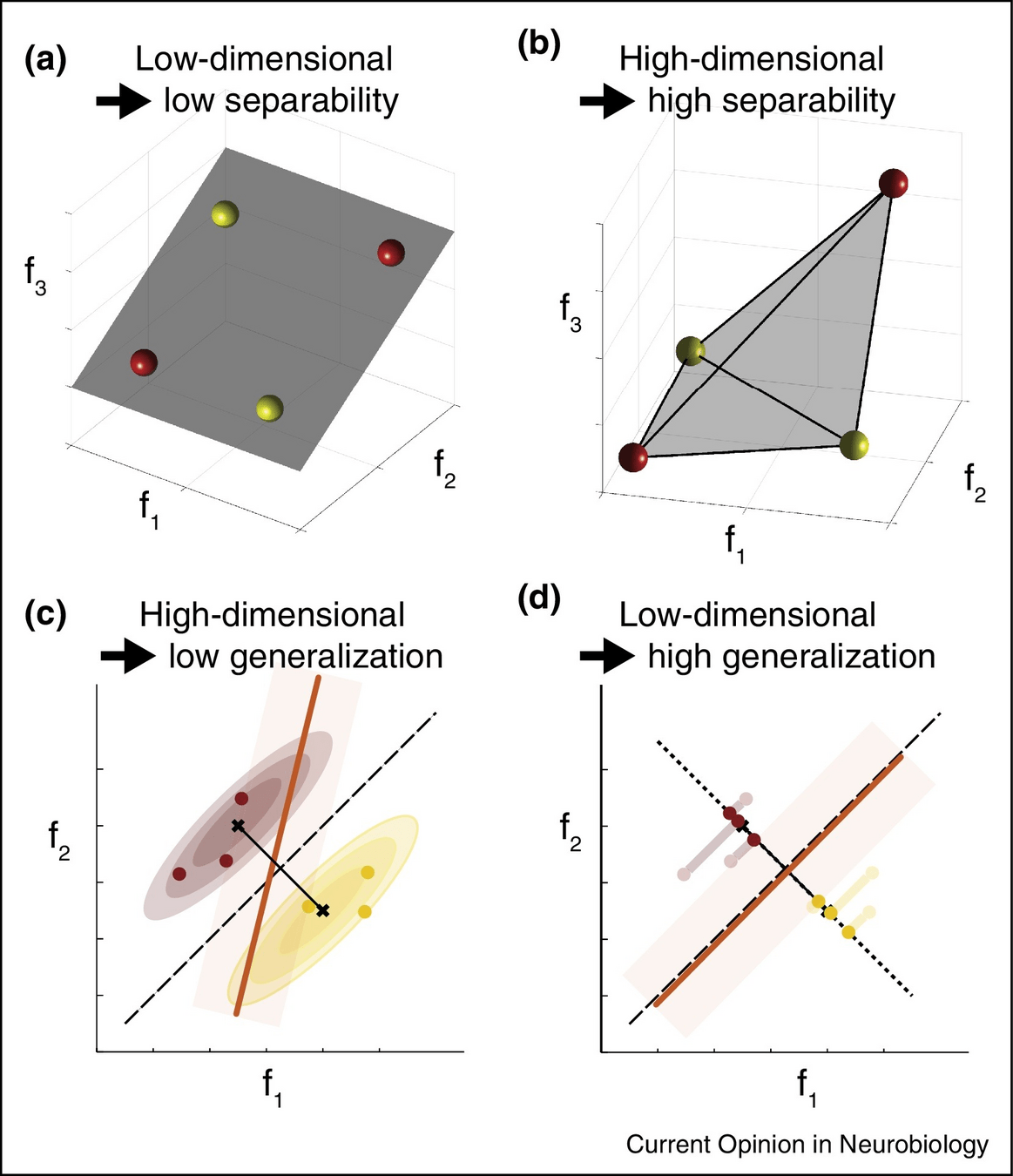
High-dimensional latent structure in visual cortex responses to natural images
Theories should be as simple as possible ...
Theories should be as simple as possible ...
... but no simpler!



We often simplify the primate visual system ...
dimensionality reduction
PCA
NMF
ICA

... yielding valuable insights ...
faces
words
bodies
scenes
food
social interactions


yeeteigendecompositionkayfabe








... but perhaps we oversimplify it.
large-scale datasets
high-SNR neuroimaging
modern analysis techniques
Implications for visual processing
high-dimensional code
low-dimensional code
explains many invariances observed in visual cortex
readily interpretable!
high representational capacity, expressive
makes learning new tasks easier











Is the visual code low-dimensional?
" [...] the topographies in VT cortex that support a wide range of stimulus distinctions, including distinctions among responses to complex video segments, can be modeled with 35 basis functions"

Computational goal: compressing dimensionality?
~87 dimensions of object representations in monkey IT
"A progressive shrinkage in the intrinsic dimensionality of object response manifolds at higher cortical levels might simplify the task of discriminating different objects or object categories."

But high-dimensional coding has benefits!
detecting high-dimensional codes requires large datasets!
high representational capacity, expressive

makes learning new tasks easier

Does human vision use a
low- or high-dimensional code?

8 subjects

7 T fMRI

"Have you seen this image before?"
continuous recognition

Ideal dataset to characterize dimensionality
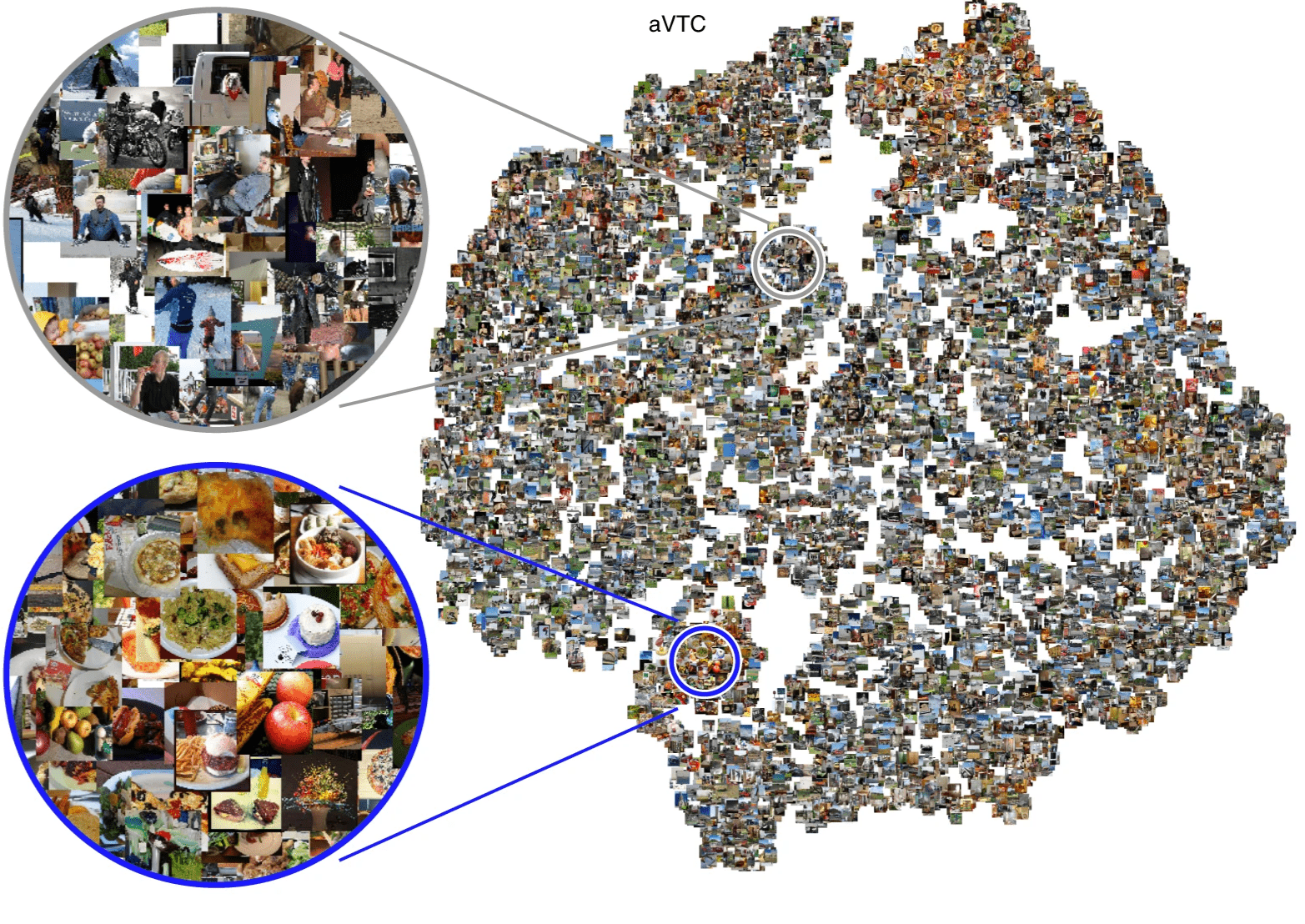
- very large-scale
- complex naturalistic stimuli
How can we estimate dimensionality?
same geometry,
new perspective!
ambient
dimensions

voxel 1
voxel 2

rotation

latent
dimensions
Key statistic: the covariance spectrum


latent dimensions sorted by variance

variance along the dimension
low-dimensional code
high-dimensional code




variance along each dimension
stimulus-related signal
trial-specific noise



Should we just apply PCA?
Cross-decomposition: a better PCA
generalizes
... across stimulus repetitions
... to novel images
trial 1
trial 2


neurons (or) voxels
stimuli
Learn latent dimensions
Step 1
Step 2
Evaluate reliable variance


If there is no stimulus-related signal, expected value = 0

PCA vs cross-decomposition
logarithmic binning + 8-fold cross-validation

Our covariance spectra
latent dimensions sorted by variance on the training set

reliable variance on the test set





- no small "core subset"
- thousands of dimensions!
- limited by dataset size

A power-law covariance spectrum
more data
more dimensions?

i love sci-fi and am willing to put up with a lot. Sci-fi movies/tv are usually underfunded, under-appreciated and misunderstood. I tried to like this, I really did, but it is to good tv sci-fi as babylon 5 is to star trek (the original). Silly prosthetics, cheap cardboard sets, stilted dialogues, cg that doesn't match the background, and painfully one-dimensional characters cannot be overcome with a'sci-fi'setting. (I'm sure there are those of you out there who think babylon 5 is good sci-fi tv. It's not. It's clichéd and uninspiring.) while us viewers might like emotion and character development, sci-fi is a genre that does not take itself seriously (cf. Star trek). [...]
i love and to up with a lot. Are, and. I to like this, I really did, but it is to good as is to (the).,,, that doesn't the, and characters be with a''. (I'm there are those of you out there who think is good. It's not. It's and.) while us like and character, is a that does not take [...]

What about smaller visual cortex regions?


Is this high-dimensional structure shared across people?


vs


Cross-decomposition still works!
generalizes
... across stimulus repetitions
... to novel images
... across participants
subject 1
subject 2



neurons (or) voxels
stimuli
Learn latent dimensions
Step 1
Step 2
Evaluate reliable variance



Cross-individual covariance spectra


Cross-individual covariance spectra



Shared high-dimensional structure across individuals











limited by 1,000 shared images

Shared high-dimensional structure across individuals
Consistent with power laws in mouse V1
similar neural population statistics
- in different species
- across imaging methods
- at very different resolutions



also shared across individuals!




10,000 neurons
2,800 images
calcium imaging


Spatial topography of latent dimensions


low-rank
high-rank
1
10
100
1,000
4
8
20
1,000
large spatial scale
small spatial scale
Coarse-to-fine spatial structure





1
1,000

30 mm
80 μm

Spatial scale: a potential organizing principle?






Spatial scale: a potential organizing principle?




number of neurons in human V1

spacing between neurons
a code that leverages all latent dimensions
Conclusions
high-dimensional
- power-law covariance
- unbounded




or

striking universality
- across individuals
- across species
New approaches!

- impossible to interpret every single latent dimension
- generative mechanisms that induce this high-dimensional latent structure
Thank you!




The Bonner Lab!







The Isik Lab!