Stuff I've been doing
1. Reading papers
2. Trying to come up with project ideas
3. Some preliminary analysis ...
4. Logistics + settling in
Some short-term goals
1. Re-analyze some existing fMRI data
2. Play with some existing sEEG data
while I
3. Design a theory/computation project
4. Get some practical experience with EEG

primary sensory cortices
- auditory
- somatosensory
- visual
simple stimuli
- pure tones
- air puffs
- moving dot array
collect fMRI responses
measure variability across trials
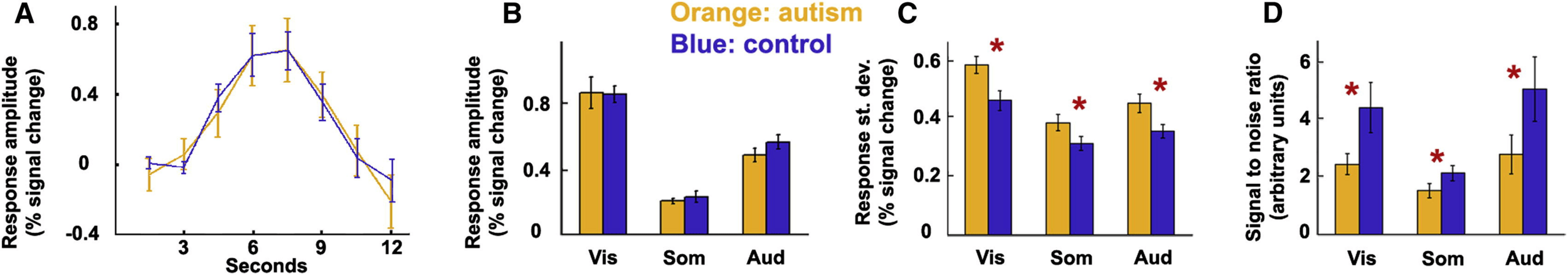
Are the earliest neural representations of basic sensory stimuli as stable as neurotypicals or noisier?
neurotypical
ASD
In ASD, the earliest neural representations of basic sensory stimuli are noisier than those of neurotypicals
true in all primary sensory cortices (!)



2016
Replication + autism-specificity
Some limitations
- ROI analysis: coarse resolution
- Univariate analysis: ignores population coding
Some mixed findings in the literature
(negative results with EEG, electrophysiology)
Things I'm trying
- Single-trial betas at single-voxel resolution
- Multivariate analysis
Are there spatial patterns of reliability?
e.g. posterior-anterior gradient
Is the unreliability confined to a subspace of the population response?
e.g. is the noise variance orthogonal to the signal dimensions
Where I am
[x] Got access to the data
[x] BIDS-ified it
[x] Preprocessed (part of) it
[ ] Missing/conflicting metadata :(
[ ] Estimate single-trial betas
[ ] The rest of the analysis
last collected fMRI data in 2020; analyzed it in MATLAB
Software (recs? shared code?)
- heudiconv (DICOMs to BIDS)
- pydeface (via bidsonym)
- fMRIprep
- GLMsingle
mind:/lab_data/behrmannlab (17 TB, 100% full)
Can parts be archived? Where?
I/O sometimes super slow
HDDs vs SSDs?
- {missing, extra, truncated} runs
- some runs explicitly labelled "bad" for some reason
- one subject has a single DICOM missing (!) in their anatomical scan
- some DICOMs appear corrupted/incomplete; can't create NII
missing/conflicting metadata :(

Got ~event onset times... but don't know stimulus identity
- Which tones?
- Which direction were the dots moving?
- Which hand was the air puff delivered to?
after all the constraints*
37 out of 72 subjects
13 Autism
13 Schizophrenia
11 Control
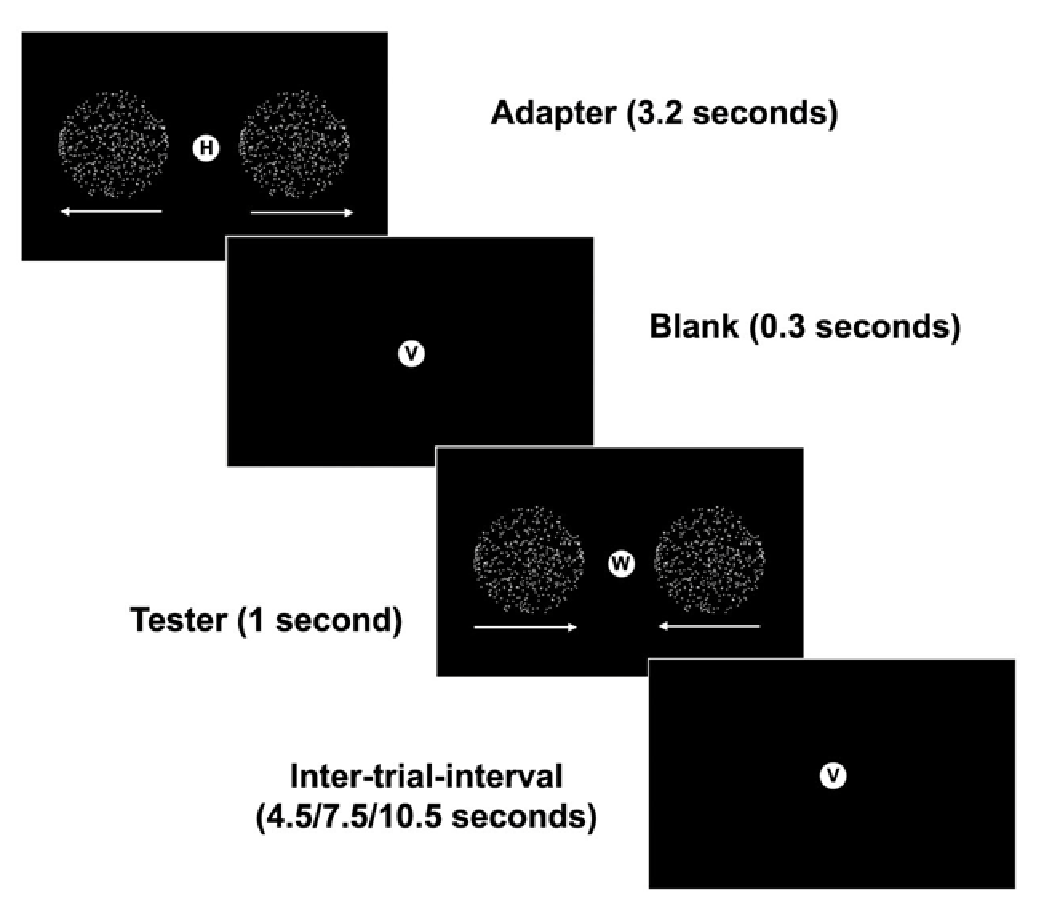
Orthogonal 1-back task on letters at fixation,
but I have no idea what the metadata means

range: [76, 90]
ASCII codes? L to Z
H
@Marlene: Why adaptation?
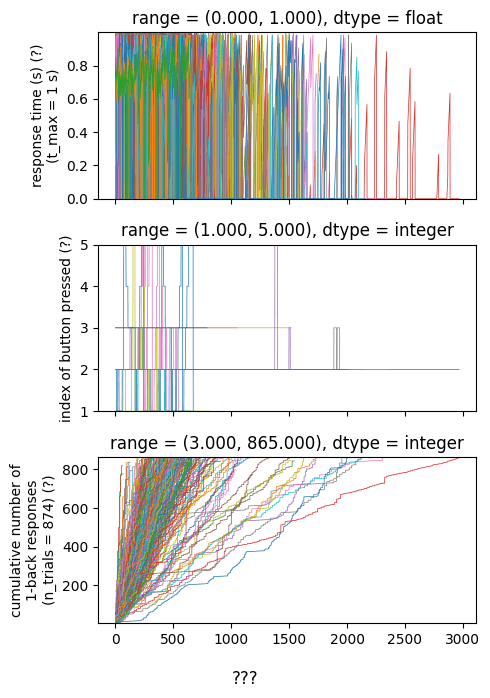
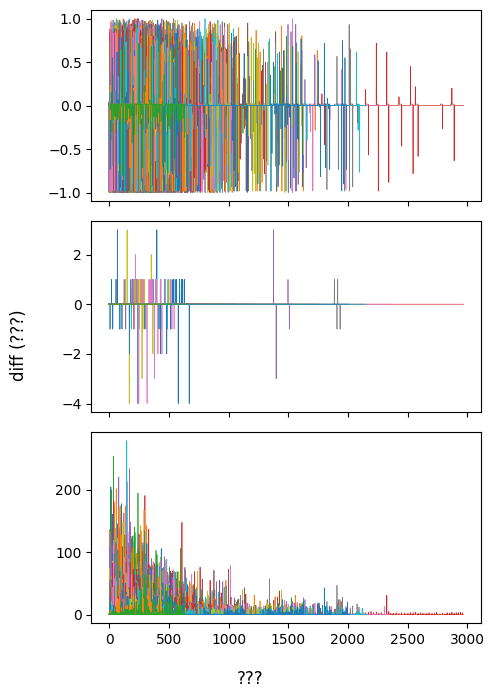
For each run, I have a file called "responses" with 3 numerical columns
But I don't know what the columns are yet
And the lengths of the files are all different (!!!) and don't correspond to any known quantity
Anyway... I also want to play with some sEEG data
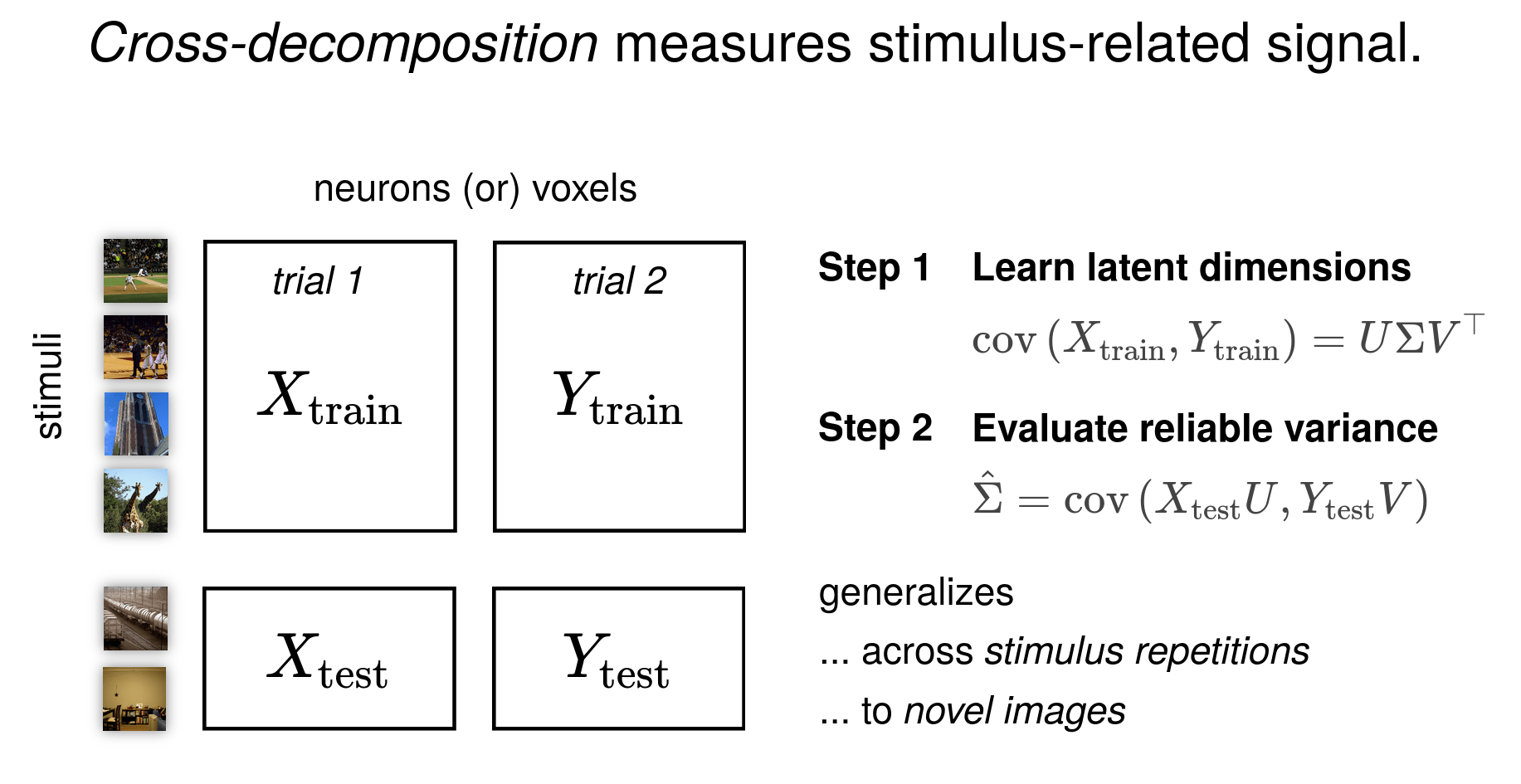
(from my PhD work)
- need single-trial responses
- TODO: split the localizer blocks of the sEEG data (0.8 s ON + 0.2 s OFF) into single-trial epochs for each stimulus
Haven't really done much time-/spectral- domain analyses before; gonna be fun
Goal: to estimate dimensionality of sEEG responses
Some other ideas I'm thinking about
Multi-(image spatial scale) visual processing:
- Relationship to multiple spatial scales of organization of cortex?
- Incremental learning of visual features?
- Constraining model-brain comparisons with local-only connections in linear fits
- Only allow "local"/constrained feature weighting in downstream layers (i.e., later features can only mix input features in particular ways)
Causal manipulation of model representations
- Re-weight their latent dimensions; measure output performance
- See if weight structure is similar across networks; alter it