PROJECT CARDIOWAVE
CS460: COMPUTER GRAPHICS
TEAM 23
ROHINI DESHMUKH
The aim of this project is to develop an interactive web-based 3D heart model that will visualize the pumping action of the atriums and ventricles in synchronization with ECG data.
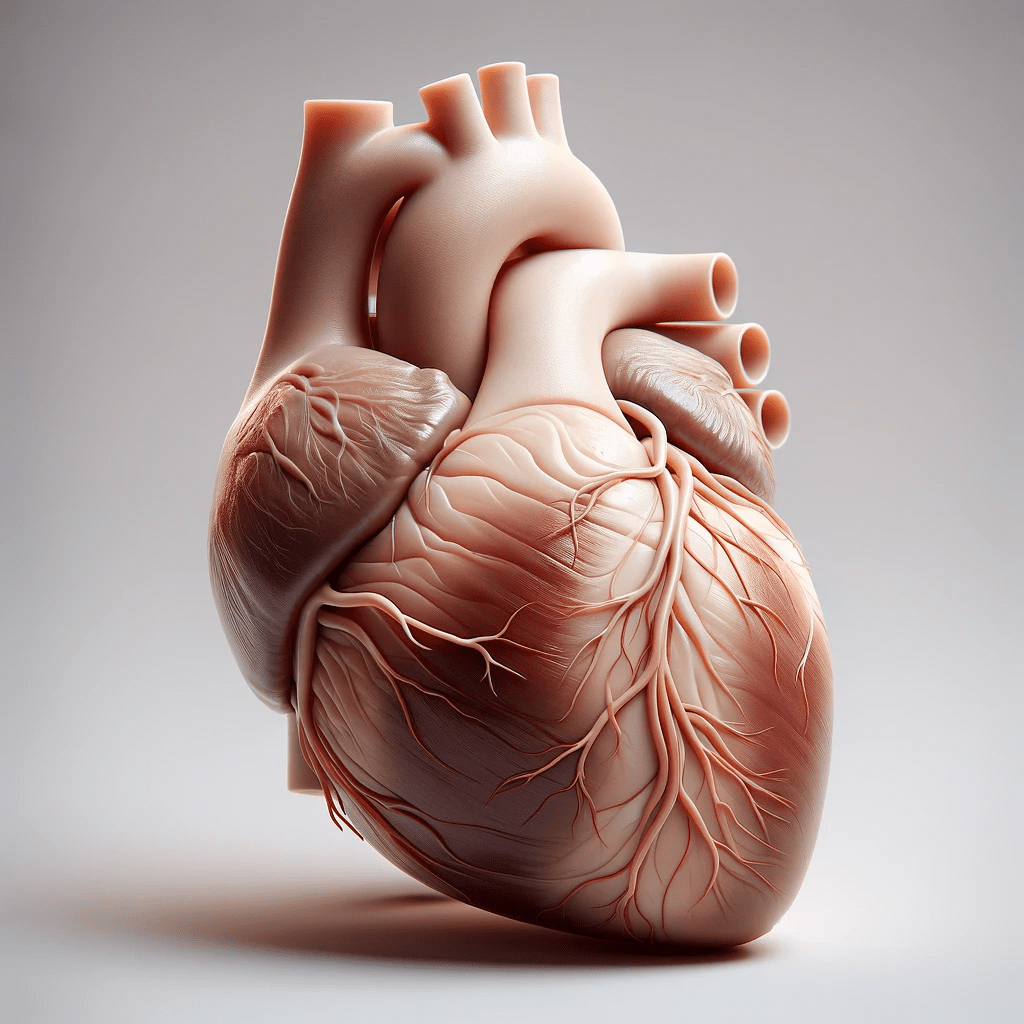
Aim:
Step 1: Render 3D model on web
Downloaded the mesh from web
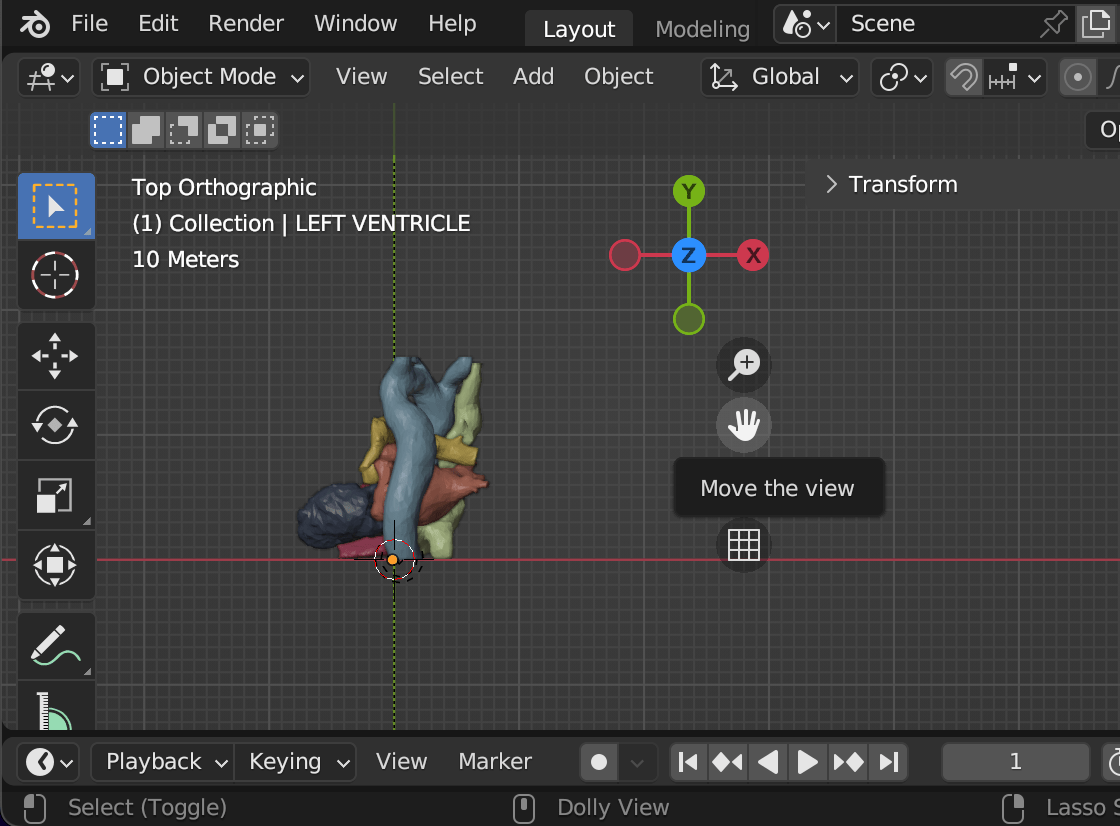
Edited using Blender
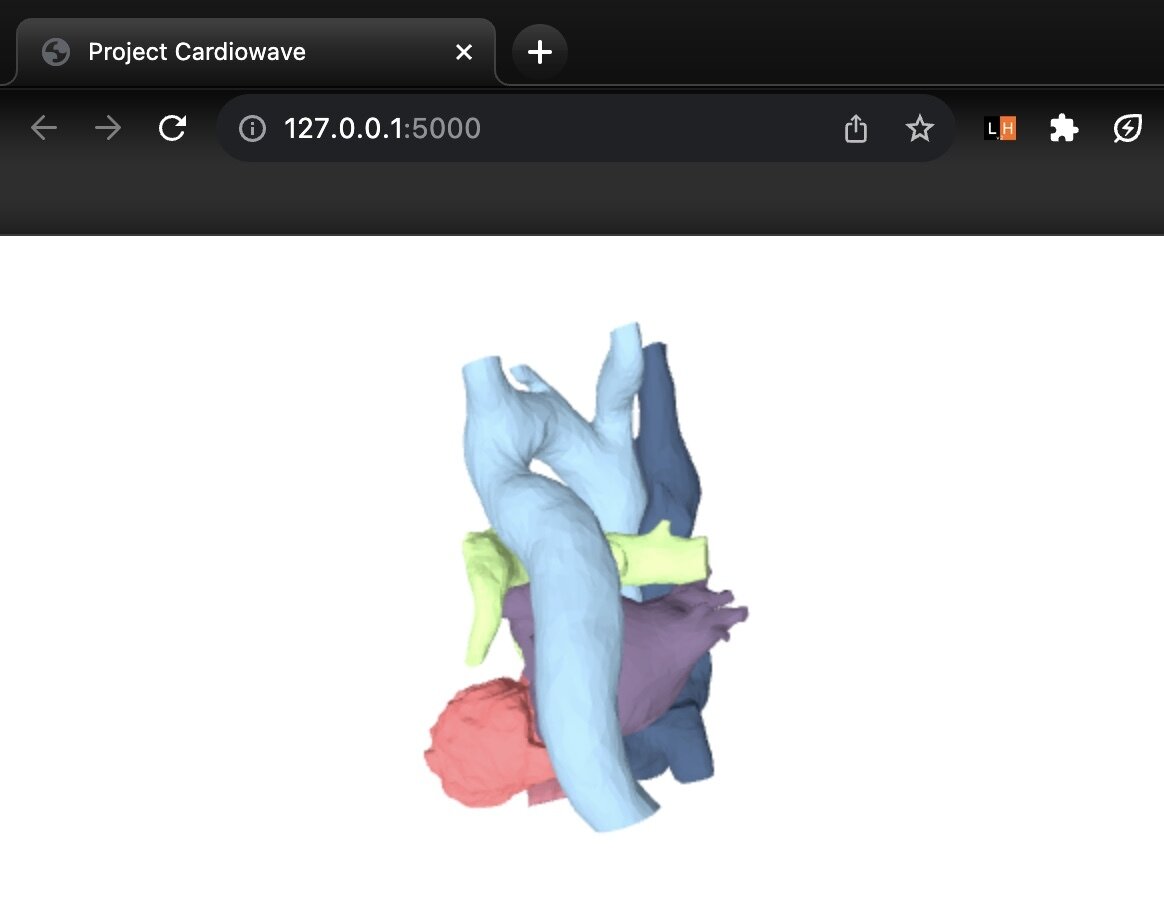
//display the 6 meshes atrium
var loader = new GLTFLoader();
loader.load('static/heart.glb', function (gltf) {
const objectsInfo = [
{ name: 'RIGHT_ATRIUM', color: 0x355070 },
{ name: 'LEFT_ATRIUM', color: 0x6D597A },
{ name: 'RIGHT_VENTRICLE', color: 0xB56576 },
{ name: 'LEFT_VENTRICLE', color: 0xE56B6F },
{ name: 'AORTA', color: 0xEAAC8B },
{ name: 'PULMONARY_ARTERY', color: 0xB5E48C },
];
for (const objInfo of objectsInfo) {
const object = gltf.scene.getObjectByName(objInfo.name);
object.scale.set(0.5, 0.5, 0.5);
object.material = new THREE.MeshStandardMaterial({ color: objInfo.color });
object.visible = true;
scene.add(object);
}
});Loaded the gltf file using Three.js
3D model on web
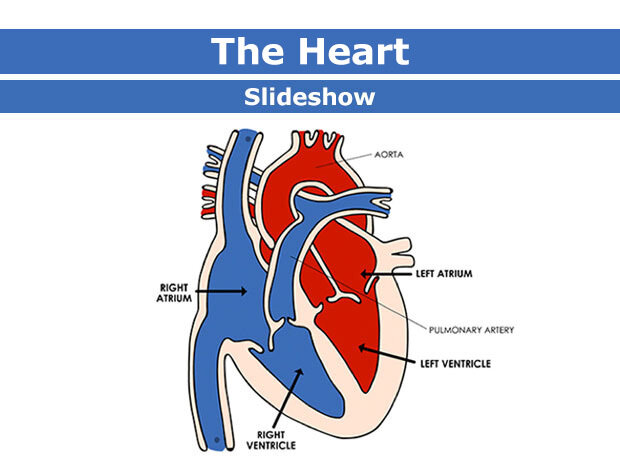
Step 2: Study related to Heart and ECG
6 primary meshes of the heart model
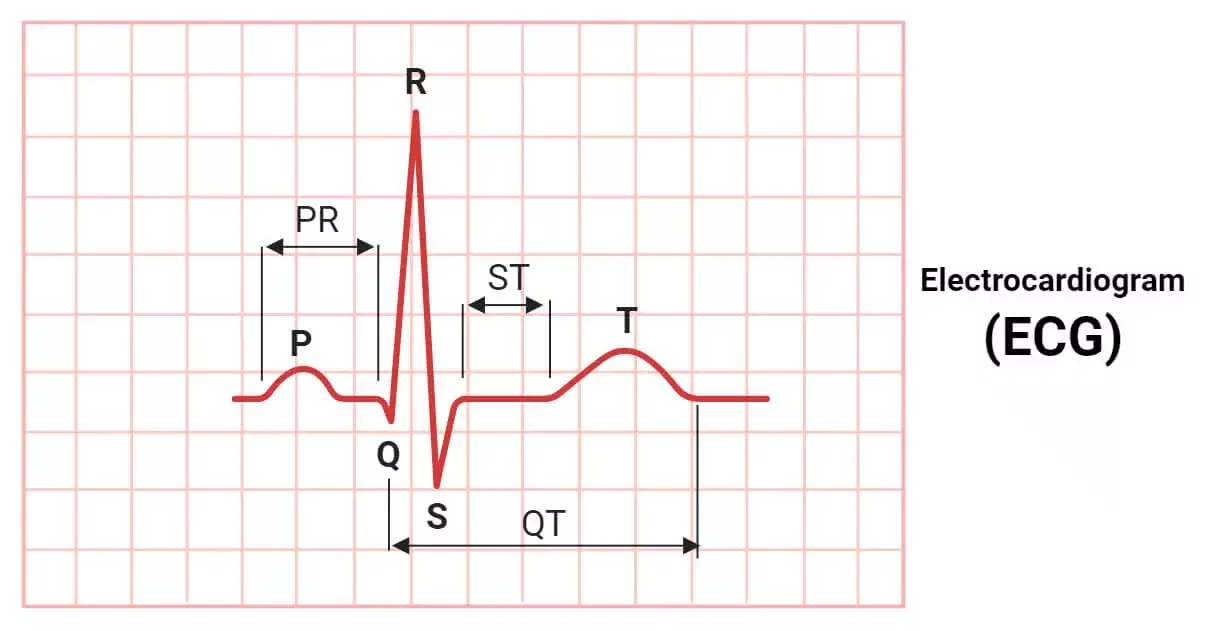
ECG signal
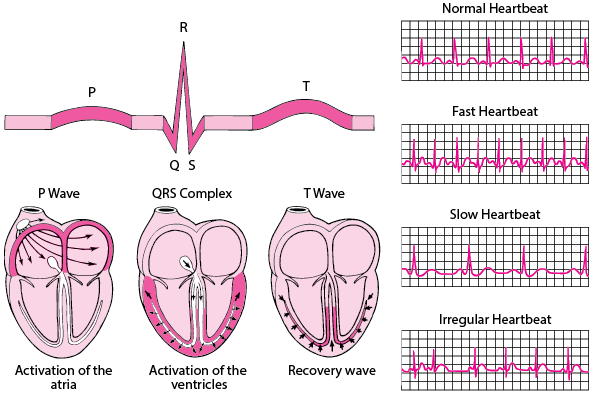
Activation of atria and ventricles depending upon the ECG signal
Step 3: Medical libraries used for data processing using python


import numpy as np
from biosppy.signals import ecg
# load raw ECG signal
signal = np.loadtxt('./examples/ecg.txt')
# process it and plot
out = ecg.ecg(signal=signal, sampling_rate=1000., show=True)import neurokit2 as nk
# Download example data
data = nk.data("bio_eventrelated_100hz")
# Preprocess the data (filter, find peaks, etc.)
processed_data, info = nk.bio_process(ecg=data["ECG"], sampling_rate=100)
# Compute relevant features
results = nk.bio_analyze(processed_data, sampling_rate=100)
Step 4: Python Dict to JSON format
'jsonify()'

# Download the ECG data file
url = "https://cs666.org/data/ecg.txt"
r = requests.get(url, allow_redirects=True)
open("ecg.txt", "wb").write(r.content) # Save the content in binary mode
# Load the dataset using biosppy library
from biosppy.signals import ecg
signal = np.loadtxt('ecg.txt')
out = ecg.ecg(signal=signal, sampling_rate=1000., show=False)
#jsonify
@app.route('/ecg_data')
def ecg_data_route():
ecg_data = {
"signal": out['filtered'].tolist(), # Filtered ECG signal
"rpeaks": out['rpeaks'].tolist(), # R-peak
}
return jsonify(ecg_data)
Output
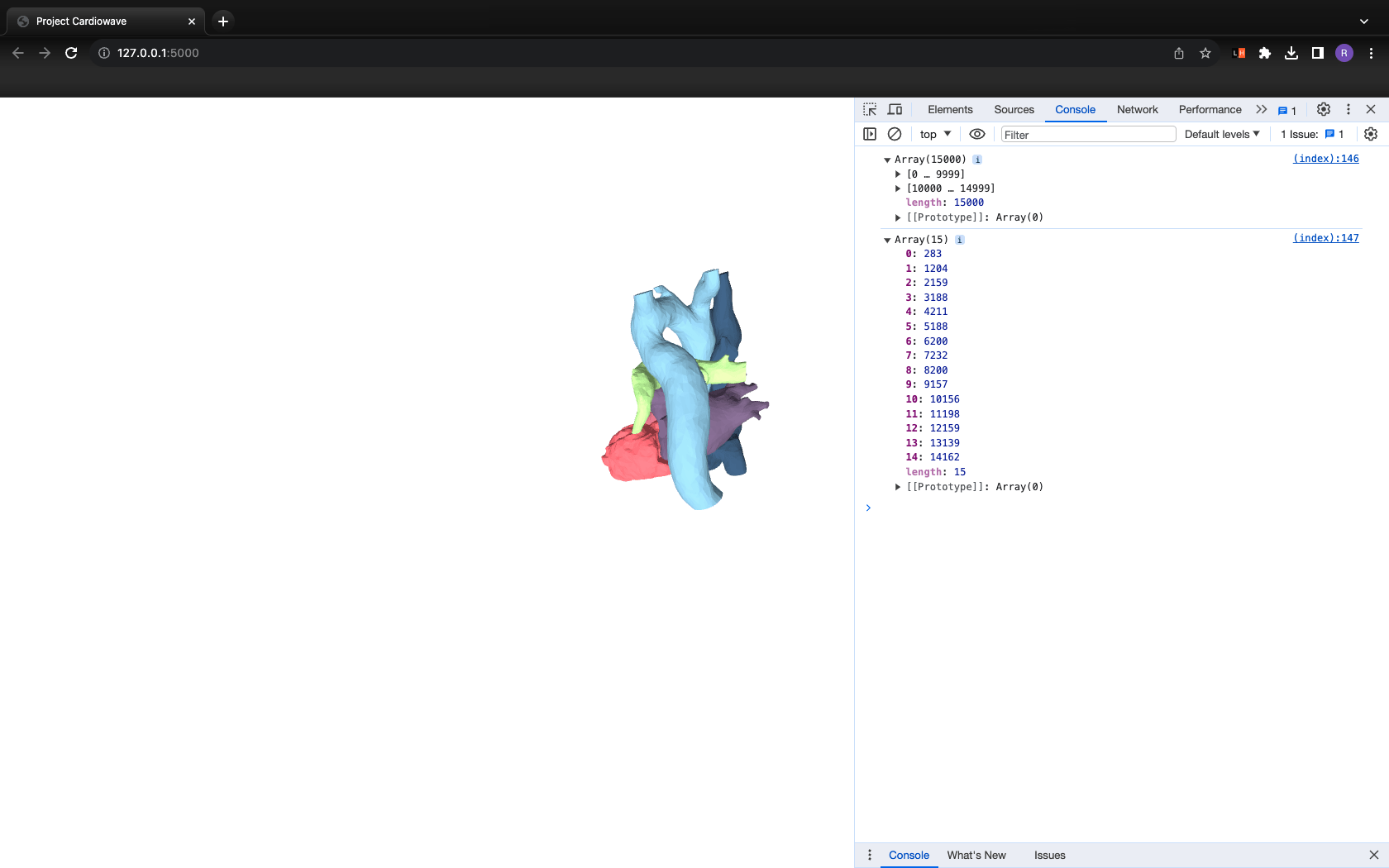
Step 5: Work in progress
- Parsing data using Neurokit for p, q, s, t wave
- Synchronizing heart frequency with the ECG data
Thank you!

PROJECT CARDIOWAVE
Interactive web-based 3D heart model that will visualize the pumping action of the atriums and ventricles in synchronization with ECG data.