Basic Graph Theory
UMD CP Club Summer CP Week 6


What is a Graph?

Graph Terminology
A graph \(G = (V,E)\) is a structure consisting of a vertex set \(V\) and edge set \(E\)
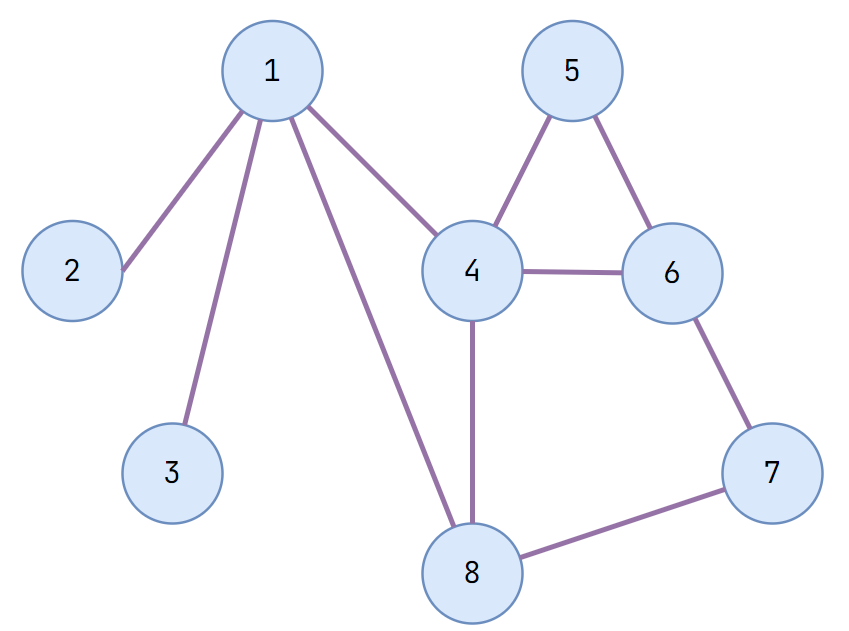

Graph Terminology
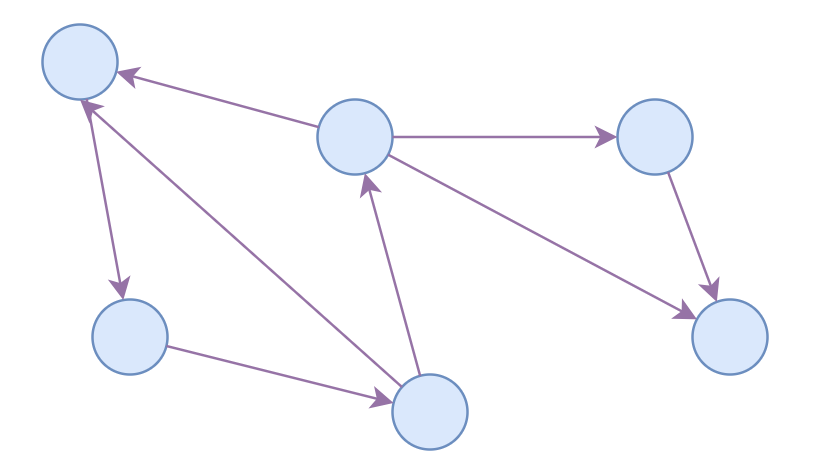
Directed Graph
Undirected Graph

Graph Terminology
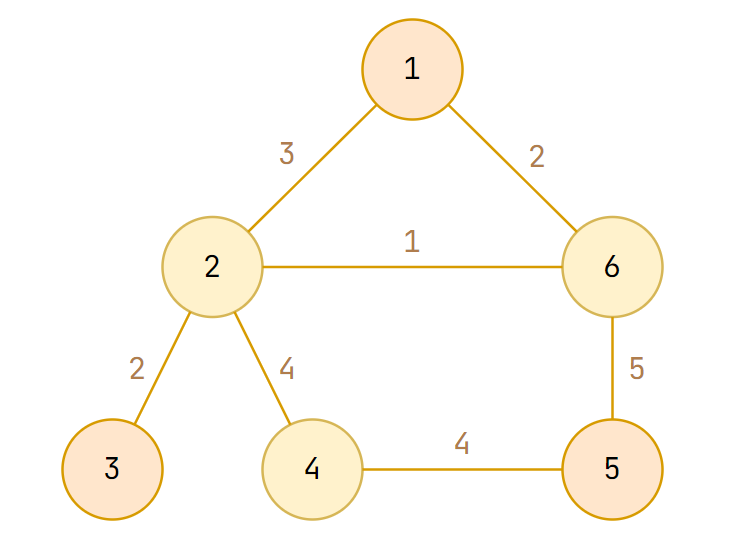
There can be weight on edges, and we call the graph weighted

Graph Terminology
An outdegree of a vertex \(u\) is the number of edge in a form of \((u,v)\)
An indegree of a vertex \(u\) is the number of edge in a form of \((v,u)\)
An degree of a vertex \(u\) is the number of edges incident to \(u\)
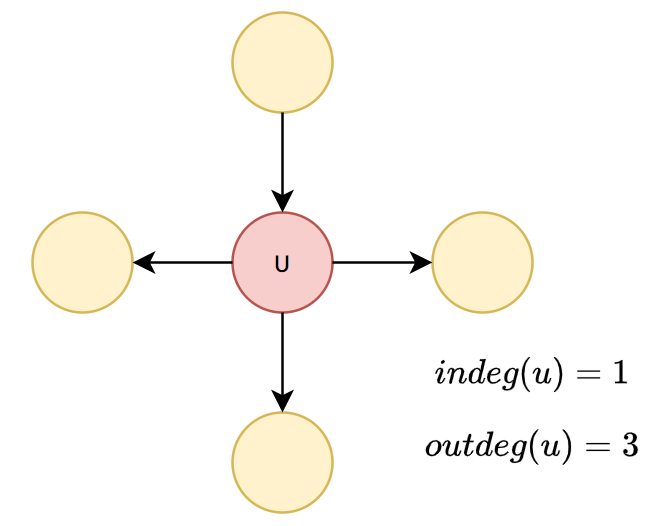

Graph Terminology
- A path is a sequence of vertices \(v_1, v_2, \cdots, v_n\)
- A simple path does not include repeating vertices
- A cycle is a path that starts and ends with same vertex
- A circuit is a cycle that goes through all vertices

Graph Terminology
Two vertices \(u,v\) are connected if you can reach \(v\) from \(u\) and \(u\) from \(v\)
The connected components are the set of vertices that are connected
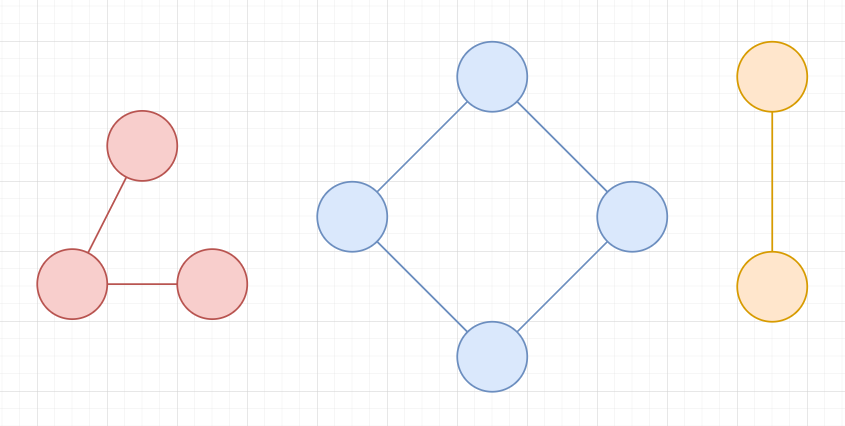

Graph Terminology
Two vertices \(u,v\) are connected if you can reach \(v\) from \(u\) and \(u\) from \(v\)
The connected components are the set of vertices that are connected


Graph Terminology
A tree is a connected graph without cycles, it can be rooted or not
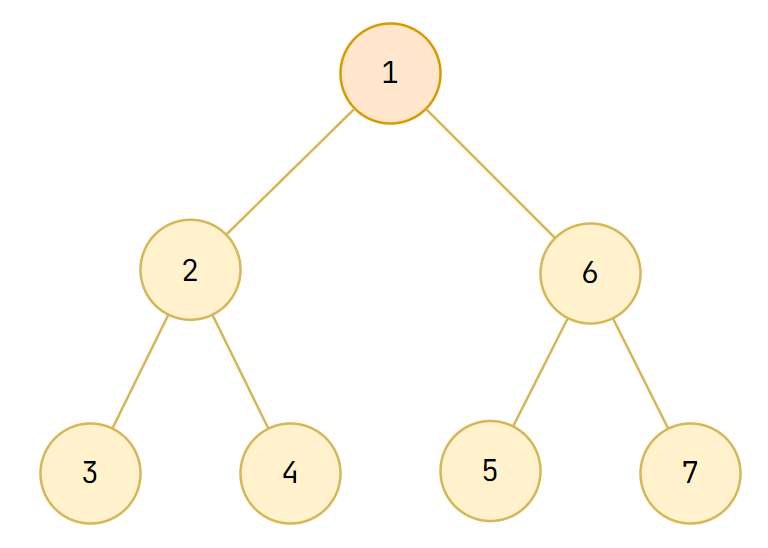

Graph Terminology
A directed acyclic graph (DAG) is a directed graph without cycles
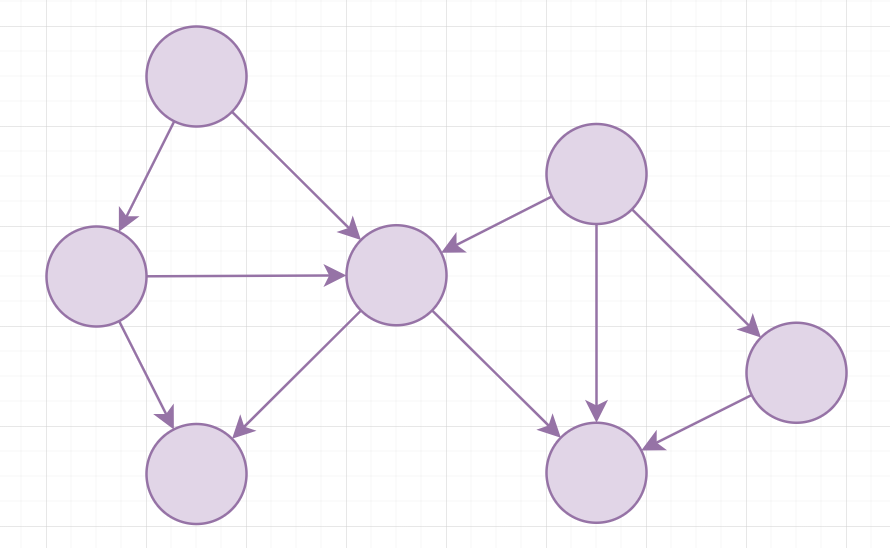
It is possible to find a topological order

Graph Terminology
A functional graph is a directed graph where all vertices has outdegree exactly 1
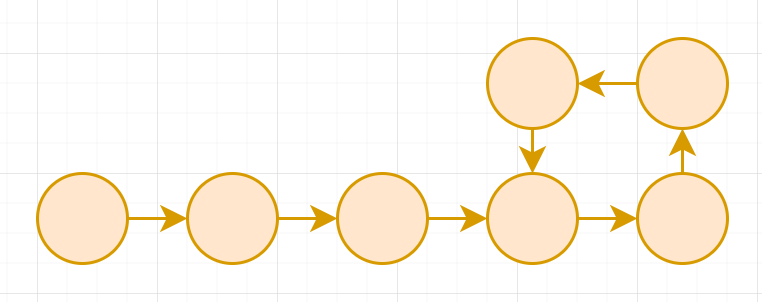
A permutation can be drawn into a functional graph

Graph Terminology
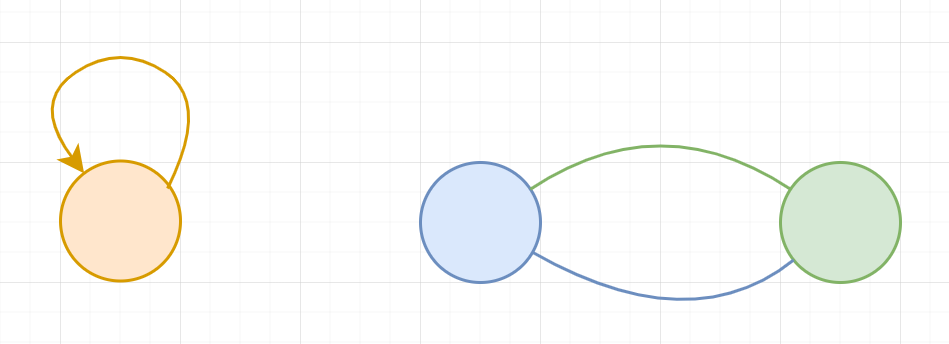
A simple graph is a graph that does not contain self loops or multi-edge

How to store Graphs

What do we want?
Suppose you are given a graph, and you want to know whether two vertices are adjacent, how will you do it?

A Simple Solution
Let's start an 2D Array \(A\), and we call it the adjacency matrix

A Simple Solution
Let's start an 2D Array \(A\), and we call it the adjacency matrix
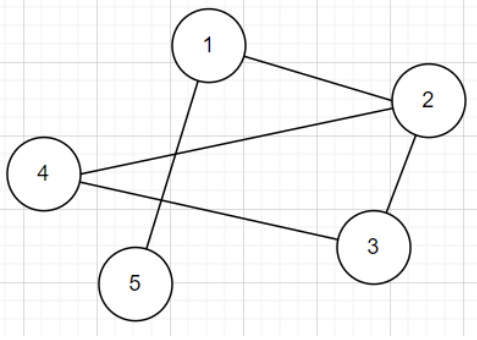
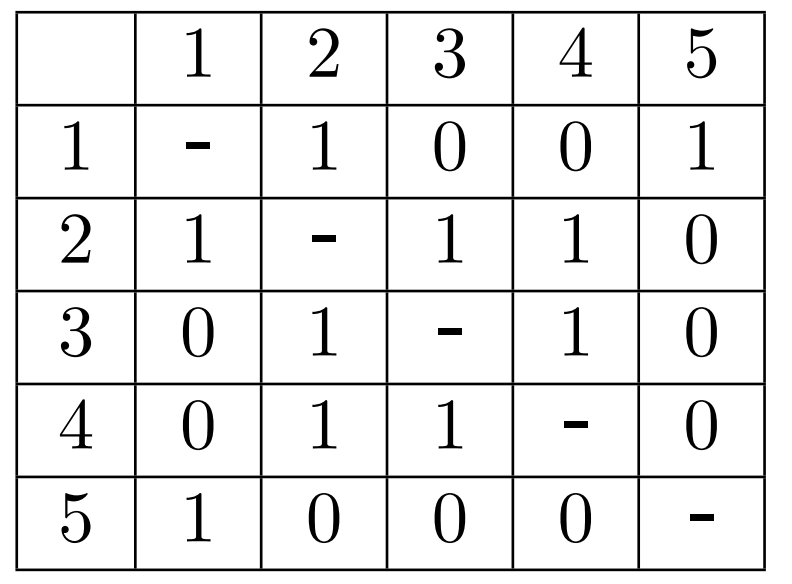

A Simple Solution
This enables us to know whether \(u,v\) are adjacent in \(O(1)\), but it requires \(O(|V|^2)\) space

An Arising Problem
While the previous way makes it easy to know whether two vertices are adjacent
It is actually not too useful for us later on

An Arising Problem
Is there a way to reduce memory usage?

Adjacency List
Consider storing an array of lists \(A\), and \(A[u]\) stores all \(v\) connected to \(u\)
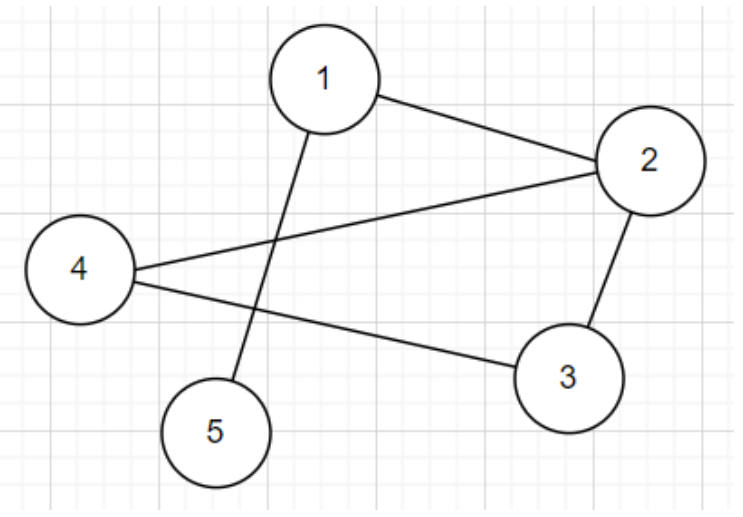
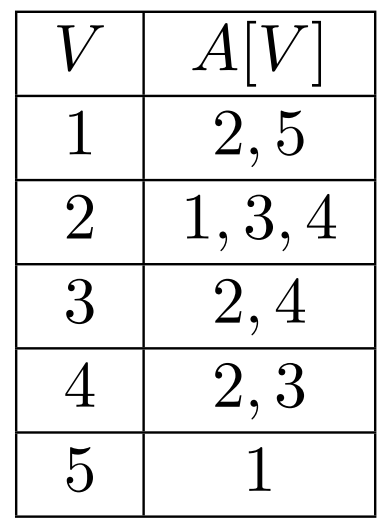

Adjacency List
This makes finding whether \(u, v\) are connected into \(O(|V|)\)
But it will make traversing the graph easier

Code
const int N = 5005;
int adj[N][N];
int main(){
int n,m;
cin >> n >> m;
for(int i = 0; i < m; i++){
int u,v;
cin >> u >> v;
adj[u][v] = 1;
adj[v][u] = 1; //if undirected
}
}Adjacency Matrix
const int N = 1e6+5;
vector<int> adj[N];
int main(){
int n,m;
cin >> n >> m;
for(int i = 0; i < m; i++){
int u,v;
cin >> u >> v;
adj[u].push_back(v);
adj[v].push_back(u); //if undirected
}
}Adjacency List

Graph Traversal

Graph Traversal
There are two ways of doing graph traversal
DFS (Depth First Search)
BFS (Breath First Search)

Depth First Search
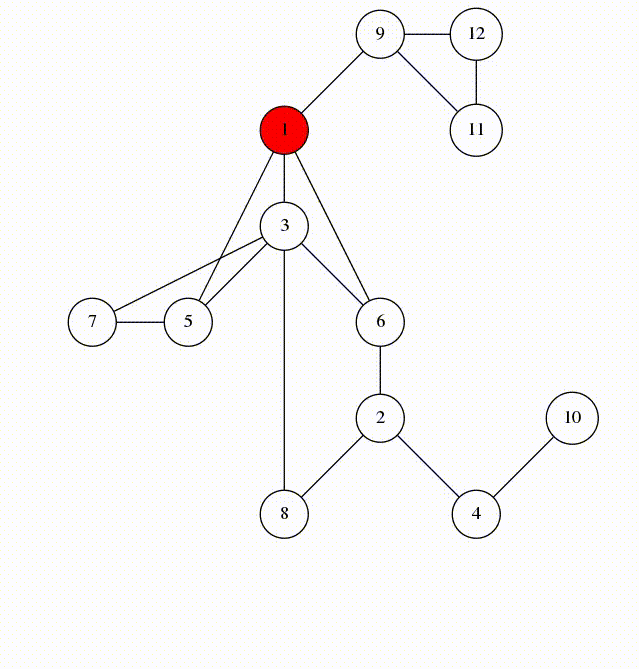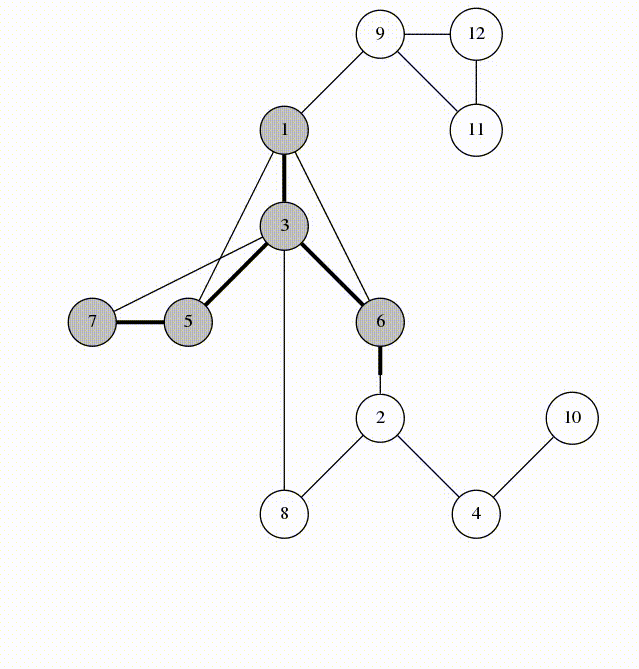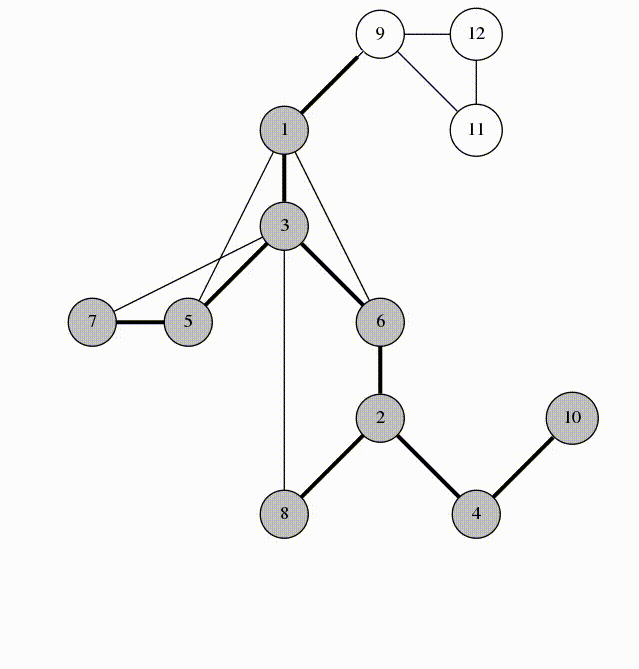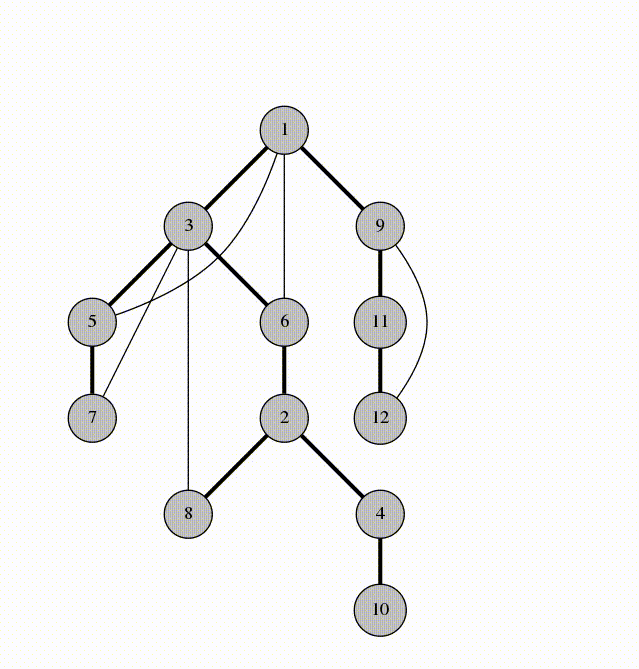
We will visit by depth, the idea is similar to using a stack
animation from codeforces

Depth First Search
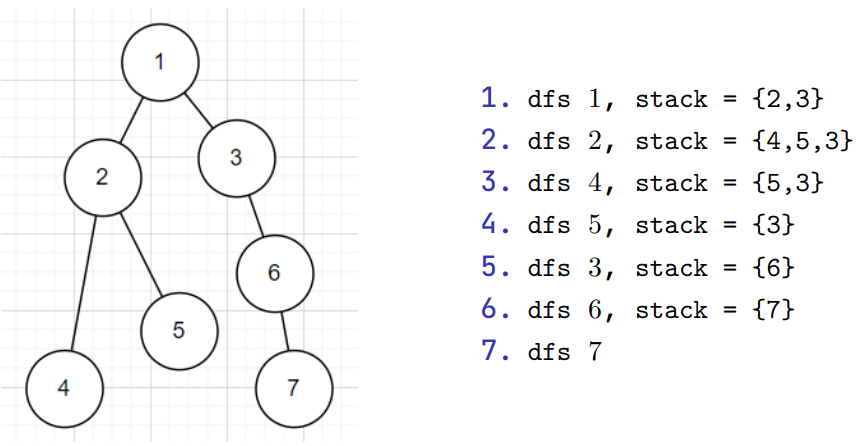

Depth First Search
void dfs(int u){
visited[u] = true;
for (int v = 1; v <= n; v++){
if (!adj[u][v] || visited[v]) continue;
dfs(v, u);
}
}DFS using an adjacency matrix
Time Complexity: \(O(|V|^2)\)

Depth First Search
void dfs(int u){
visited[u] = true;
for (int v : adj[u]){
if (visited[v]) continue;
dfs(v, u);
}
}DFS using an adjacency list
Time Complexity: \(O(|V| + |E|)\)

Breath First Search
For breath first search, we search the vertices closest to the starting vertex
We usually use a queue to do this
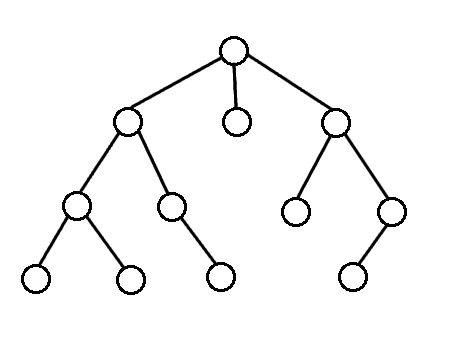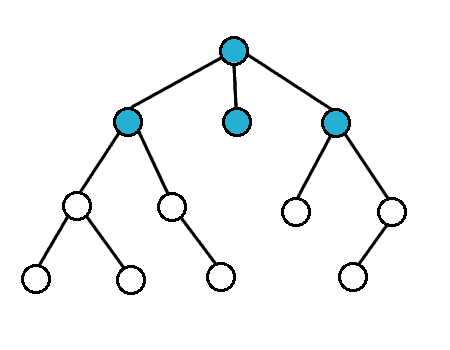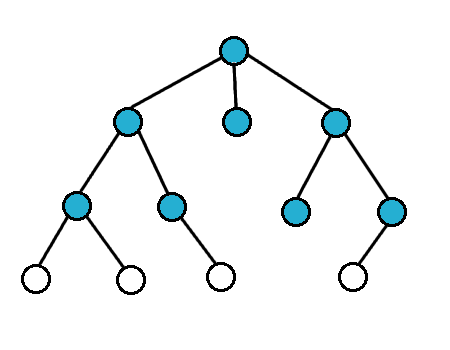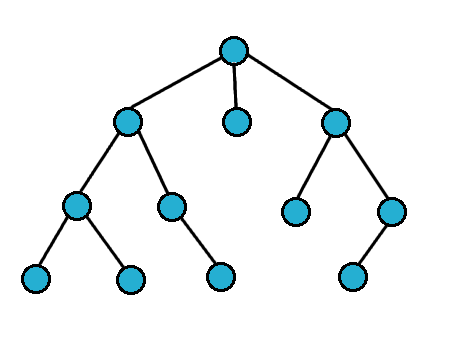

Breath First Search
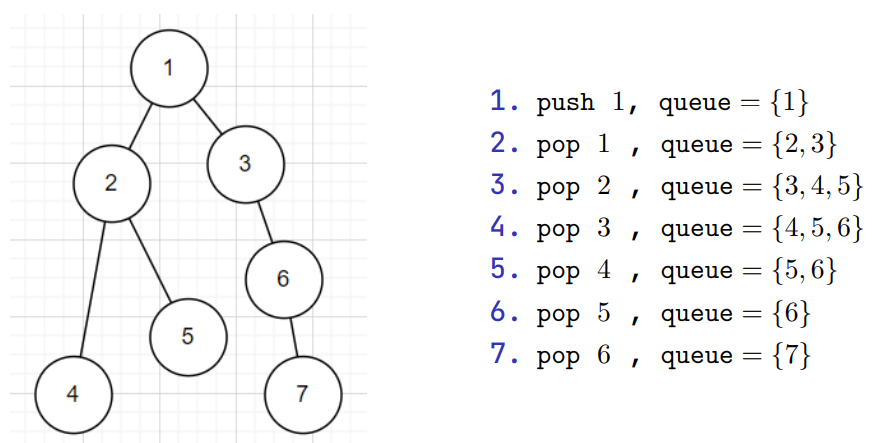

Breath First Search
queue<int> q;
q.push(1);
while(!q.empty()){
int u = q.front(); q.pop();
for(int v = 1; v <= n; v++){
if(!adj[u][v] || visited[v]) continue;
q.push(v);
}
}BFS with adjacency matrix
Time Complexity: \(O(|V|^2)\)

Breath First Search
queue<int> q;
q.push(1);
while(!q.empty()){
int u = q.front(); q.pop();
for(int v : adj[u]){
if(visited[v]) continue;
q.push(v);
}
}BFS with adjacency list
Time Complexity: \(O(|V| + |E|)\)

Some Practice Problems


Bipartite Coloring
A bipartite graph is a graph where you can color the vertices into two colors where no adjacent vertices has same color
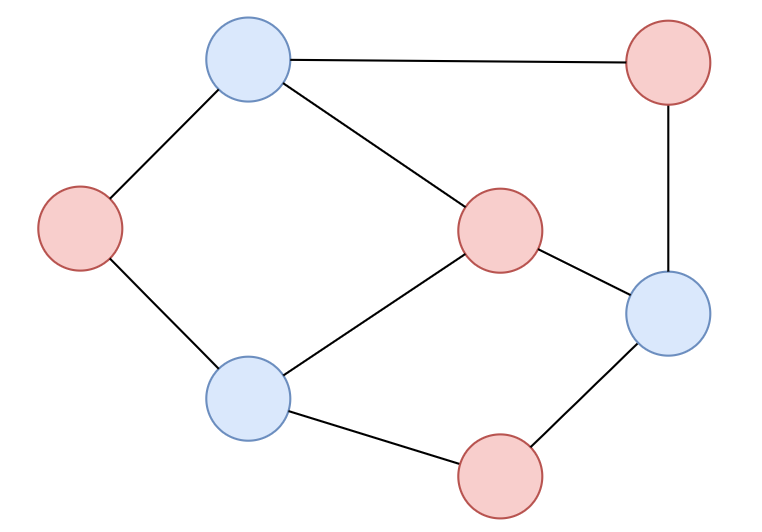

Bipartite Coloring
Fix a vertex, and try coloring the vertices
If there exists some conflicts, then the graph is not bipartite

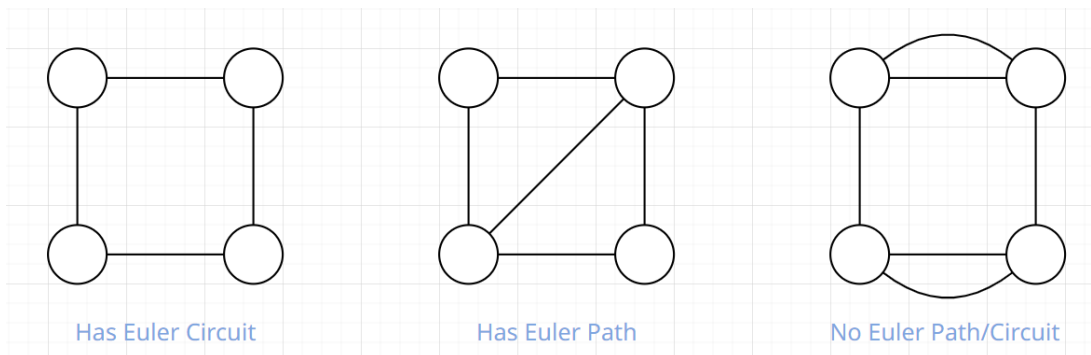
Eulerian & Hamiltonian Circuit

Eulerian Graph
Can you draw a graph in one pen stroke without going through same edges
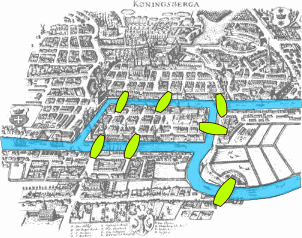

Eulerian Graph
- Eulerian Path: A path that visits all edge but does no have repeating edges
- Eulerian Circuit: Eulerian Path but starts and ends at same vertex
- Chinese Postman Problem: Find the shortest eulerian path (weighted graph)

Eulerian Graph
You can prove a conclusion
- If a connected graph has exactly \(0\) odd degree vertices \(\implies\) Has Eulerian Circuit
- If a connected graph has exactly \(2\) odd degree vertices \(\implies\) Has Eulerian Path
- If a connected graph has \(> 2\) odd degree vertices \(\implies\) not Eulerian

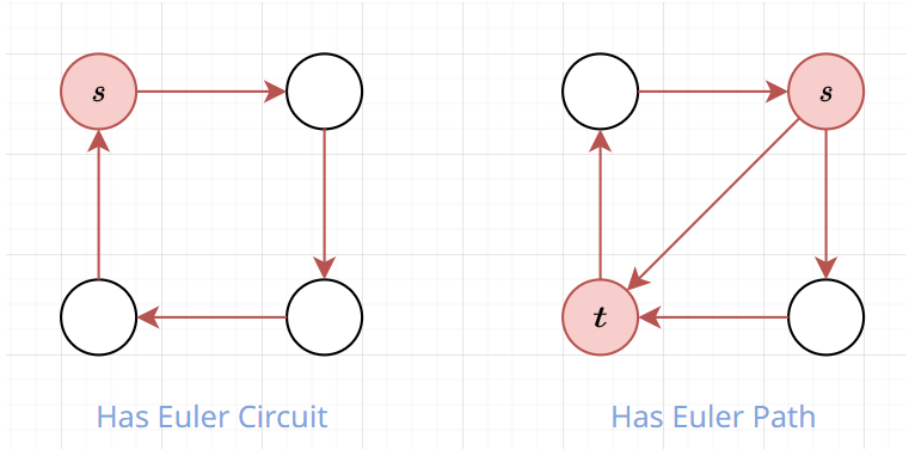
How to Find Eulerian Path?
- If there are exactly \(0\) odd degree vertices \(\implies\) start from any vertex, you will always find an Eulerian circuit
- If there are exactly \(2\) odd degree vertices \(\implies\) start from any odd degree vertex, you will always find an Eulerian path

Hamiltonian Graph
- Hamiltonian Path: A path that visits all vertices but does no have repeating vertex
- Hamiltonian Circuit: Hamiltonian Path but starts and ends at same vertex
- Travelling Saleman Problem: Find the shortest Hamiltonian Path (weighted graph)
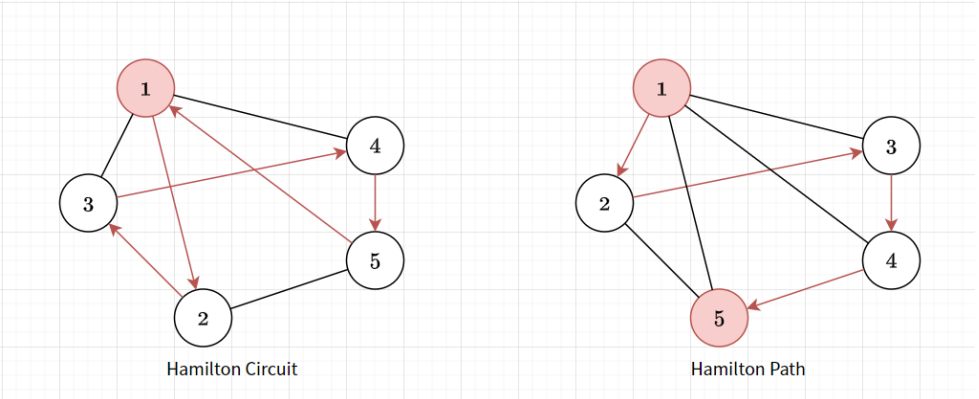

Hamiltonian Graph
There is no easy way to find Hamiltonian Path/Circuit
Usually we will have to use Bitmask DP to solve problems related to this

DAG & Topological Sort

Topological Order of DAG
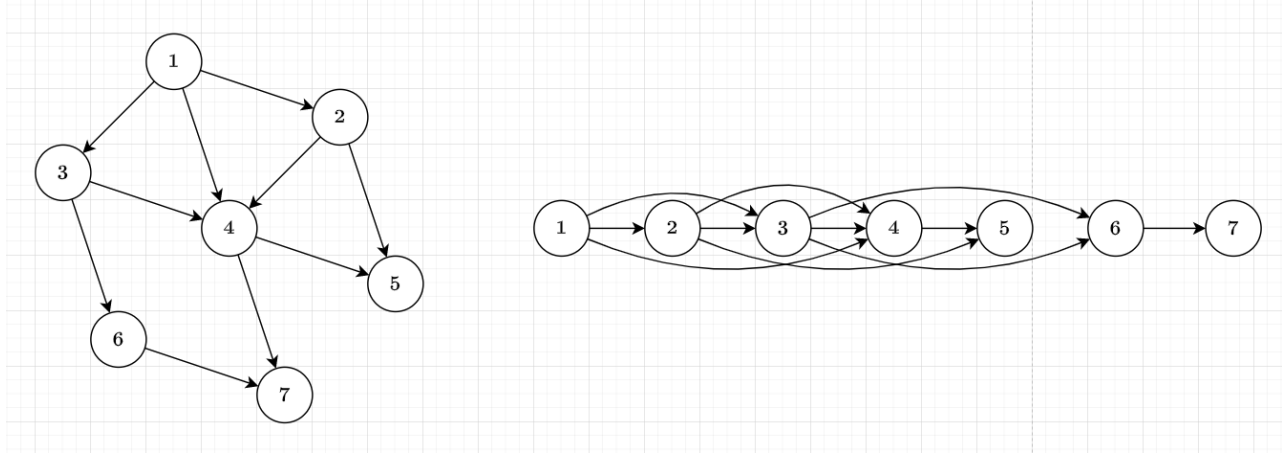

Topological Order of DAG
Think about taking courses in university
Some classes have prerequisites, then we can draw it into a graph
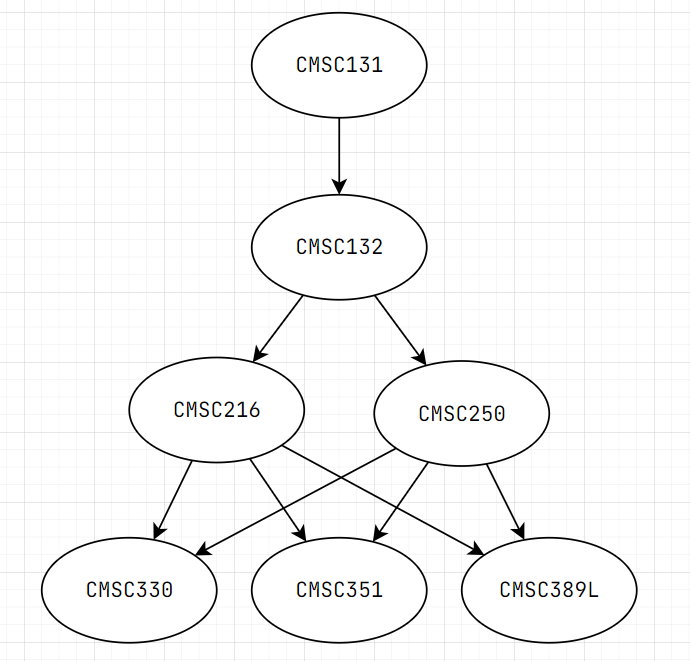

Topological Order of DAG
Therefore, we have the idea of sorting the vertices by their order
The edges will only go from the vertices in front of the order to back

Topological Order of DAG
The algorithm
- Start from the vertices with
indeg[u] = 0 - We will do BFS, push all the vertices with indeg 0 into the queue
- Remove the vertex, and decrement the
indeg[v] - If the indegree of \(v\) becomes \(0\), then push it into a queue
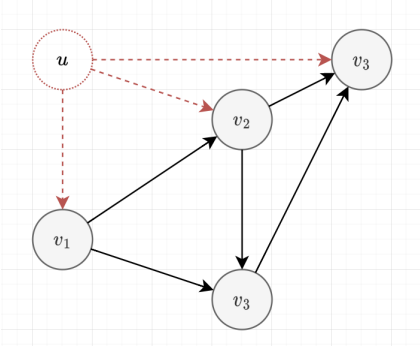

Topological Order of DAG
queue<int> q;
vector<int> topo;
for(int i = 1; i <= n; i++){
if(deg[i] == 0)
q.push(i);
}
while(!q.empty()){
int u = q.front(); q.pop();
topo.push_back(u);
for(int v : adj[u]){
deg[v]--;
if(deg[v] == 0)
q.push(v);
}
}
//if topo.size() == n, then the graph is a DAG

DP On DAGs

Disjoint Set Union

We want a data structure that can
- Connect two vertices \(u, v\)
- Answer if \(u,v\) are connected with each other
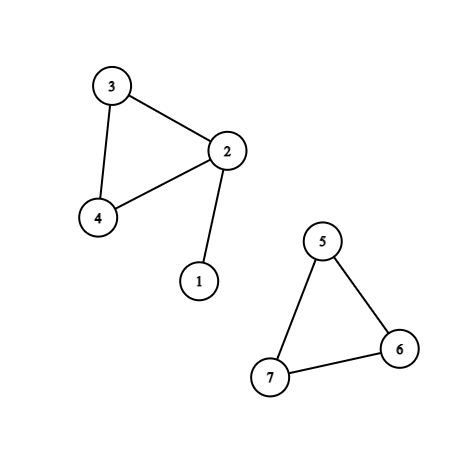

Disjoint Set Union
We introduce the data structure Disjoint Set Union (DSU)
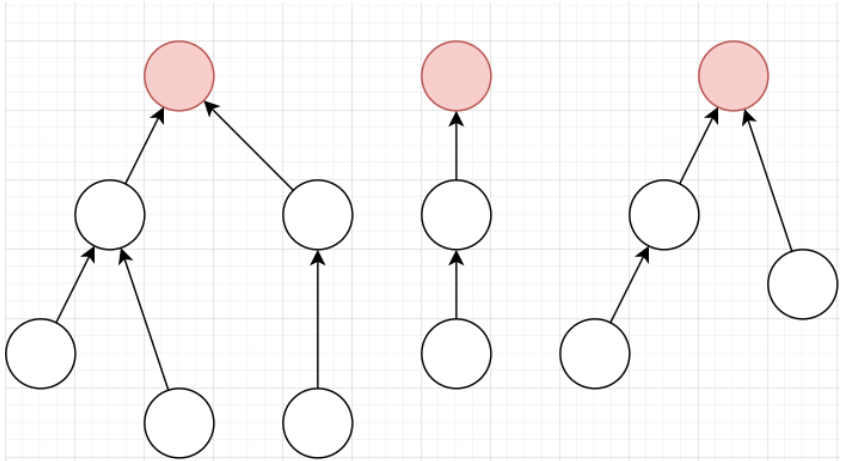

Disjoint Set Union
For an undirected graph, we connect each vertices in the same connected components with a directed arrow


Disjoint Set Union
When we want to know whether two vertices are connected
We check if their roots are the same


Disjoint Set Union
When we want to know whether two vertices are connected
We check if their roots are the same


Disjoint Set Union
We will declare an array dsu, where dsu[u]is the vertex \(u\) points to


Disjoint Set Union
Initialize the data structure
for(int i = 1; i <= n; i++)
dsu[i] = i;
//or you can do
iota(dsu, dsu+MAXN, 0);Time Complexity: \(O(|V|)\)

Disjoint Set Union
To find the root of each vertex
void find(int u) {
if(dsu[u] == u) return u;
return find(dsu[u]);
}Time Complexity: \(O(|V|)\)

Disjoint Set Union
To connect two vertices, we connect the roots of both vertex
void unite(int u, int v){
u = find(v), v = find(v);
if(dsu[u] == dsu[v]) continue;
dsu[u] = v;
}Time Complexity: \(O(1)\)

Disjoint Set Union
However, notice that finding whether \(u,v\) are connected is same as just doing DFS/BFS from one vertex
How can we speed this up?

Disjoint Set Union
The problem for finding is that, every time we will have to go through all vertices on the chain
What if we shorten the chain?

Path Compression
Each time we query a vertex, we connect all the vertices on the path to the root
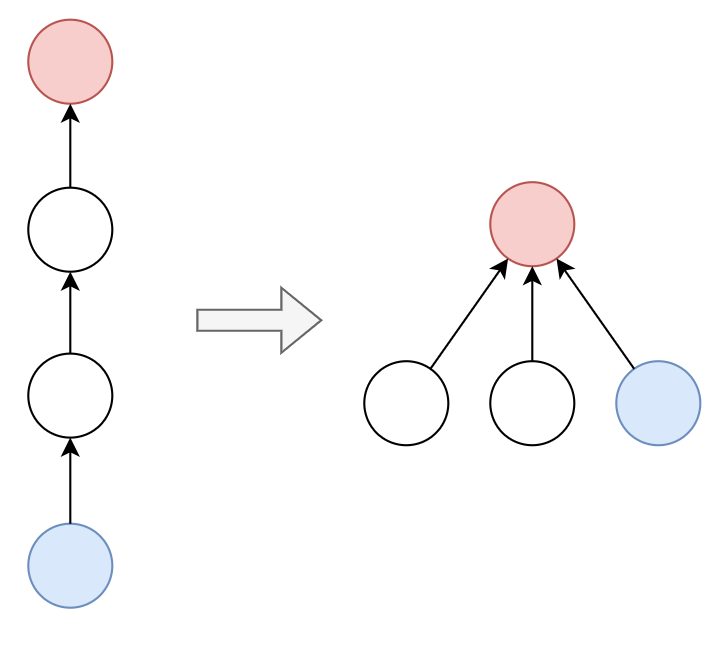

Path Compression
void find(int u) {
if(dsu[u] == u) return u;
return dsu[u] = find(dsu[u]);
}Time Complexity: \(O(\log n)\) amortized

Union By Size
There is another way to speed up DSU operations
We maintain the size of each component in sz[rt]

Union By Size
There is another way to speed up DSU operations
We maintain the size of each component in sz[rt]
void unite(int u, int v){
u = find(u), v = find(v);
if(u == v) return;
dsu[u] = v;
sz[v] += sz[u];
}
Union By Size
We connect the smaller to the larger one

Union By Size
Doing this technique, you can prove the complexity will become \(O(\log n)\) amortized

Union By Size
void unite(int u, int v){
u = find(u), v = find(v);
if(u == v) return;
if(sz[u] > sz[v]) swap(u, v);
dsu[u] = v;
sz[v] += sz[u];
}Time Complexity: \(O(\log n)\) amortized

Using Both Optimizations
Both optimizations can be used independently
If you use both optimizations, the time complexity of each operations becomes
\(O(\alpha(n))\) amoritzed
The \(\alpha\) here is the inverse Ackermann function
You can consider it almost as a constant

Problems
