Kubeflow
Data Science on Steroids

About us
saschagrunert



mail@
.de
About us
mbu93

The Evolution of Machine Learning
1950
Stochastic Neural Analog Reinforcement Calculator (SNARC) Maze Solver
2000





Data Science
Workflow
Data Source Handling
fixed set of technologies

Exploration
Regression?
Classification?
Supervised?
Unsupervised?
Baseline Modelling
Results
Trained Model
Results to share
Results
Deployment
Automation?
Infastructure?
Data Exploration
- select the data source
- collect statistical measures
- data cleaning
- typically done in notebook systems such as jupyter
import dill
with open("dataframe.dill", "rb") as fp:
dataframe = dill.load(fp)
dataframe.sample(frac=1).head(10)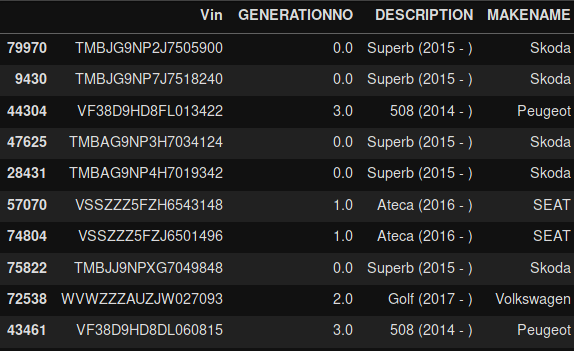
Data Munging
- creating datasets
- preparing data for cross validation
from fastai.vision import ImageDataBunch
dataset = ImageDataBunch.from_folder("dataset",
train="training",
valid="test")
datasetImageDataBunch;
Train: LabelList (3040 items)
x: ImageList
Image (3, 17, 33), Image (3, 17, 33),
Image (3, 17, 33), Image (3, 17, 33),
Image (3, 17, 33)
y: CategoryList
3,3,3,3,3
Path: dataset;
Valid: LabelList (760 items)
x: ImageList
Image (3, 17, 33), Image (3, 17, 33),
Image (3, 17, 33), Image (3, 17, 33),
Image (3, 17, 33)
y: CategoryList
3,3,3,3,3
Path: dataset;
Test: NoneBaseline modelling
- “Do we need supervised or unsupervised learning?”
- "Can we solve the problem with classification or regression?”
- most of the time experimental work
from fastai.vision import *
res18_learner = cnn_learner(dataset,
models.resnet18,
pretrained=False,
metrics=accuracy)
res18_learner.fit_one_cycle(5)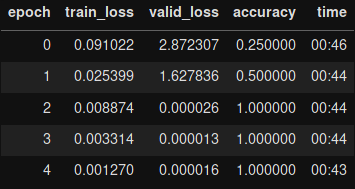
Evaluation and deployment
- check the models performance
- select and deploy a model
- create automated training process to update data and model
interp = res18_learner.interpret()
interp.plot_confusion_matrix(figsize=(8, 8))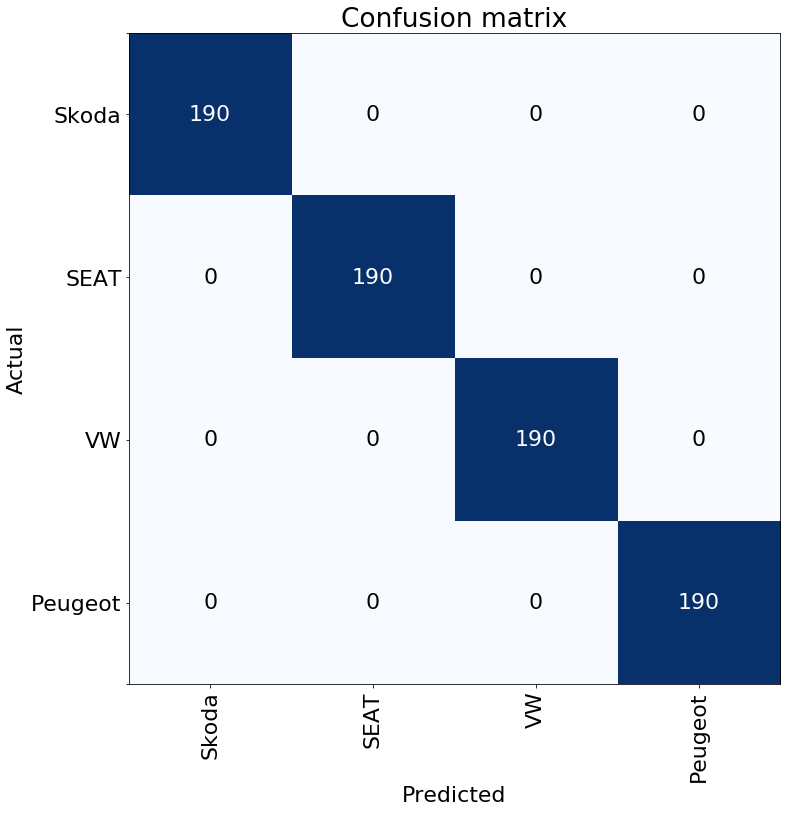
Yet Another Machine Learning Solution?
YES!
Cloud Native
Commercially available of-the-shelf (COTS) applications
The awareness of an application to run inside something like Kubernetes.
vs

“Can we improve
the classic data scientists workflow
by utilizing Kubernetes?”

announced 2017
abstracting machine learning best practices
Deployment and Test Setup



SUSE CaaS Platform
github.com/SUSE/skuba


> skuba cluster init --control-plane 172.172.172.7 caasp-cluster
> cd caasp-cluster> skuba node bootstrap --target 172.172.172.7 caasp-master> skuba node join --role worker --target 172.172.172.8 caasp-node-1
> skuba node join --role worker --target 172.172.172.24 caasp-node-2
> skuba node join --role worker --target 172.172.172.16 caasp-node-3> cp admin.conf ~/.kube/config
> kubectl get nodes -o wide
NAME STATUS ROLES AGE VERSION INTERNAL-IP EXTERNAL-IP OS-IMAGE KERNEL-VERSION CONTAINER-RUNTIME
caasp-master Ready master 2h v1.15.2 172.172.172.7 <none> SUSE Linux Enterprise Server 15 SP1 4.12.14-197.15-default cri-o://1.15.0
caasp-node-1 Ready <none> 2h v1.15.2 172.172.172.8 <none> SUSE Linux Enterprise Server 15 SP1 4.12.14-197.15-default cri-o://1.15.0
caasp-node-2 Ready <none> 2h v1.15.2 172.172.172.24 <none> SUSE Linux Enterprise Server 15 SP1 4.12.14-197.15-default cri-o://1.15.0
caasp-node-3 Ready <none> 2h v1.15.2 172.172.172.16 <none> SUSE Linux Enterprise Server 15 SP1 4.12.14-197.15-default cri-o://1.15.0Storage Provisioner
> helm install nfs-client-provisioner \
-n kube-system \
--set nfs.server=caasp-node-1 \
--set nfs.path=/mnt/nfs \
--set storageClass.name=nfs \
--set storageClass.defaultClass=true \
stable/nfs-client-provisioner
> kubectl -n kube-system get pods -l app=nfs-client-provisioner -o wide
NAME READY STATUS RESTARTS AGE IP NODE NOMINATED NODE READINESS GATES
nfs-client-provisioner-777997bc46-mls5w 1/1 Running 3 2h 10.244.0.91 caasp-node-1 <none> <none>Load Balancing
> kubectl apply -f https://raw.githubusercontent.com/google/metallb/v0.8.1/manifests/metallb.yaml---
apiVersion: v1
kind: ConfigMap
metadata:
namespace: metallb-system
name: config
data:
config: |
address-pools:
- name: default
protocol: layer2
addresses:
- 172.172.172.251-172.172.172.251> kubectl apply -f metallb-config.ymlDeploying Kubeflow
> kfctl init kfapp --config=https://raw.githubusercontent.com/kubeflow/kubeflow/1fa142f8a5c355b5395ec0e91280d32d76eccdce/bootstrap/config/kfctl_existing_arrikto.yaml> export KUBEFLOW_USER_EMAIL="sgrunert@suse.com"
> export KUBEFLOW_PASSWORD="my-strong-password"> cd kfapp
> kfctl generate all -V
> kfctl apply all -Vavailable command line tool called kfctl
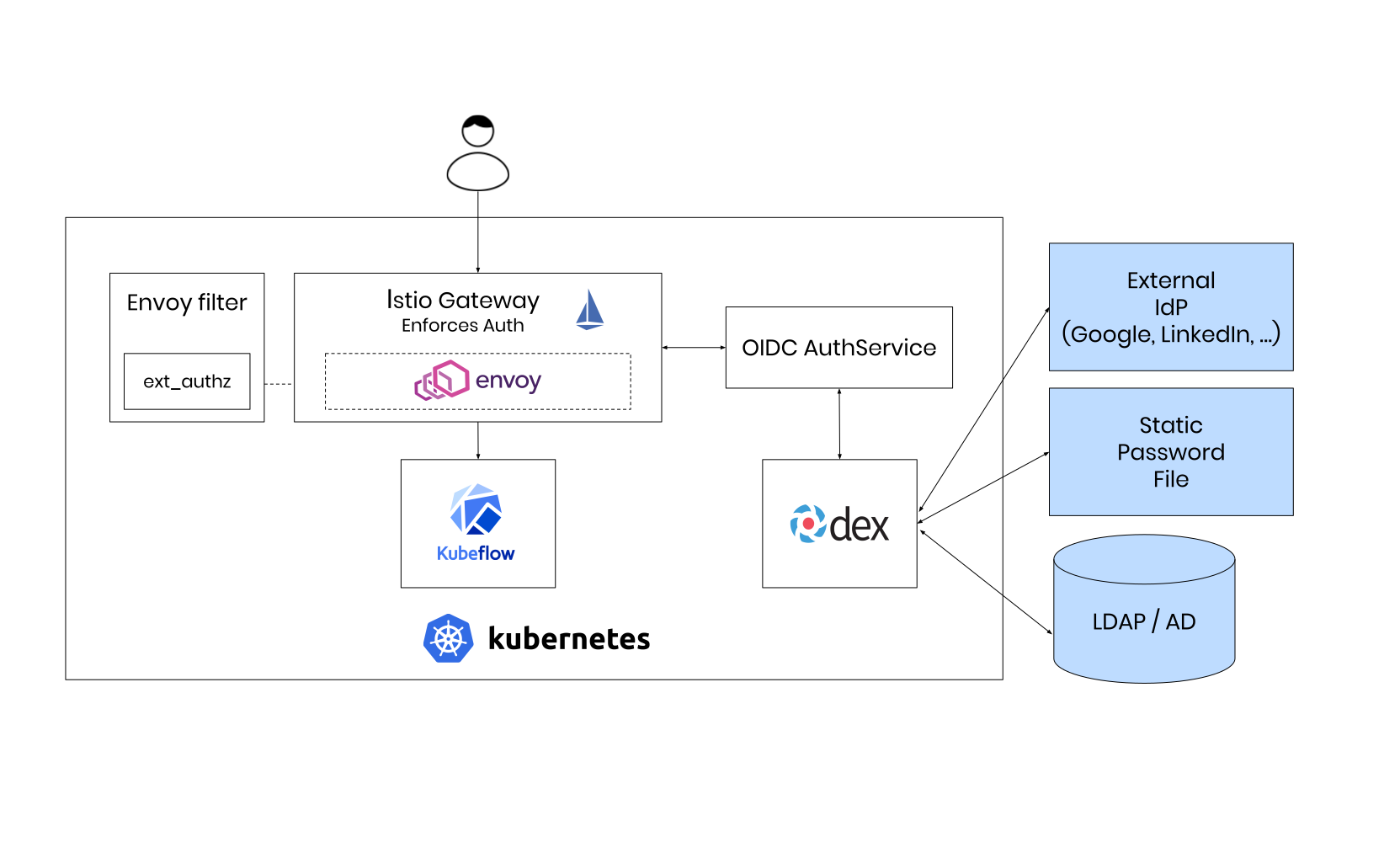
Improving the Data Scientists Workflow
Machine Learning Pipelines
- Kubeflow provides reusable end-to-end machine learning workflows via pipelines
- pipeline components are built using Kubeflows Python SDK
- Every pipeline step is executed directly in Kubernetes within its own pod
Basic component using ContainerOp
- simply executes a given command within a container
- input and output can be passed around
from kfp.dsl import ContainerOp
step1 = ContainerOp(name='step1',
image='alpine:latest',
command=['sh', '-c'],
arguments=['echo "Running step"'])Execution order
- Execution order can be made dependent
-
Arange components using op1.after(op2) syntax
step1 = ContainerOp(name='step1',
image='alpine:latest',
command=['sh', '-c'],
arguments=['echo "Running step"'])
step2 = ContainerOp(name='step2',
image='alpine:latest',
command=['sh', '-c'],
arguments=['echo "Running step"'])
step2.after(step1)Creating a pipeline
-
Pipelines are created using the @pipeline decorator and compiled afterwards via compile()
from kfp.compiler import Compiler
from kfp.dsl import ContainerOp, pipeline
@pipeline(name='My pipeline', description='')
def pipeline():
step1 = ContainerOp(name='step1',
image='alpine:latest',
command=['sh', '-c'],
arguments=['echo "Running step"'])
step2 = ContainerOp(name='step2',
image='alpine:latest',
command=['sh', '-c'],
arguments=['echo "Running step"'])
step2.after(step1)
if __name__ == '__main__':
Compiler().compile(pipeline)When should I use ContainerOp?
- Useful for deployment tasks
- Good option to query for new data
- Running of complex training scenarios (make use of training scripts)
Lightweight Python Components
- Directly run python functions in a container
- Limitations:
- need to be standalone (can't use code, imports or variables defined in another scope)
- Imported packages need to be available in the container image.
- Input parameters need to be set to str (default), int or float
Example: Sum of squares
- Code will be processed as a string and passed to the interpreter as below
from kfp.components import func_to_container_op
def sum_sq(a: int, b: int) -> int:
import numpy
dist = np.sqrt(a**2 + b**2)
return dist
add_op = func_to_container_op(sum_sq)(1, 2)python -u -c "def sum_sq(a: int, b:int):\n\
import numpy\n\
dist = np.sqrt(a**2 + b**2)\n\
return dist\n(...)"The need for storage
- Lightweight components only accept str, float or int inputs
- To pass objects around they must be stored/loaded
- For that purpose a volume must be attached
op = func_to_container_op(fit_squeezenet,
base_image="alpine:latest")("out/model", False,
"squeeze.model")
op.add_volume(
k8s.V1Volume(name='volume',
host_path=k8s.V1HostPathVolumeSource(
path='/data/out'))).add_volume_mount(
k8s.V1VolumeMount(name='volume', mount_path="out"))Up- and Downsides of Lightweight Components
- Very useful to run simple functions
- Can reduce the effort to create pipelines
- May not be used for complex functions
- Practical alternative: Google Cloud Platforms options
Getting the pipeline to run
- Pipelines are compiled using dsl-compile script
- They are then transformed to an Argo workflow and executed
> sudo pip install https://storage.googleapis.com/ml-pipeline/release/0.1.27/kfp.tar.gz
> dsl-compile --py pipeline.py --output pipeline.tar.gzGetting the pipeline to run
- Pipelines are compiled using dsl-compile script
- They are then transformed to an Argo workflow and executed
> kubectl get workflow
NAME AGE
my-pipeline-8lwcc 6m47sGetting the pipeline to run
- Pipelines are compiled using dsl-compile script
- They are then transformed to an Argo workflow and executed
> argo get my-pipeline-8lwcc
Name: my-pipeline-8lwcc
Namespace: kubeflow
ServiceAccount: pipeline-runner
Status: Succeeded
Created: Tue Aug 27 13:06:06 +0200 (4 minutes ago)
Started: Tue Aug 27 13:06:06 +0200 (4 minutes ago)
Finished: Tue Aug 27 13:06:22 +0200 (4 minutes ago)
Duration: 16 seconds
STEP PODNAME DURATION MESSAGE
✔ my-pipeline-8lwcc
├-✔ step1 my-pipeline-8lwcc-818794353 6s
└-✔ step2 my-pipeline-8lwcc-768461496 7sRunning pipelines from Notebooks
- Create and run pipelines by importing the kfp package
- Can be usefull to save notebook ressources
- Enable developers to combine prototyping with creating an automated training workflow
from kfp.compiler import Compiler
pipeline_func = training_pipeline
pipeline_filename = pipeline_func.__name__ + '.pipeline.yaml'
Compiler().compile(pipeline_func, pipeline_filename)Running pipelines from Notebooks
- Create and run pipelines by importing the kfp package
- Can be usefull to save notebook ressources
- Enable developers to combine prototyping with creating an automated training workflow
import kfp
client = kfp.Client()
try:
experiment = client.create_experiment("Prototyping")
except Exception:
experiment = client.get_experiment(experiment_name="Prototyping")Running pipelines from Notebooks
- Create and run pipelines by importing the kfp package
- Can be usefull to save notebook ressources
- Enable developers to combine prototyping with creating an automated training workflow
arguments = {'pretrained': 'False'}
run_name = pipeline_func.__name__ + ' test_run'
run_result = client.run_pipeline(experiment.id, run_name, pipeline_filename, arguments)Continuous Integration Lookout
Kubeflow provides a REST API
That’s it.
https://github.com/saschagrunert/
kubeflow-data-science-on-steroids
