
Crossing Over and Linkage Mapping
This is a live streamed presentation. You will automatically follow the presenter and see the slide they're currently on.
Loading

sbrady1
This is a live streamed presentation. You will automatically follow the presenter and see the slide they're currently on.



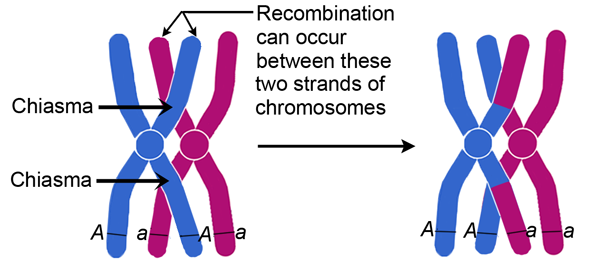
This close association between chromosomes can result in breakage and reformation known as crossing over.
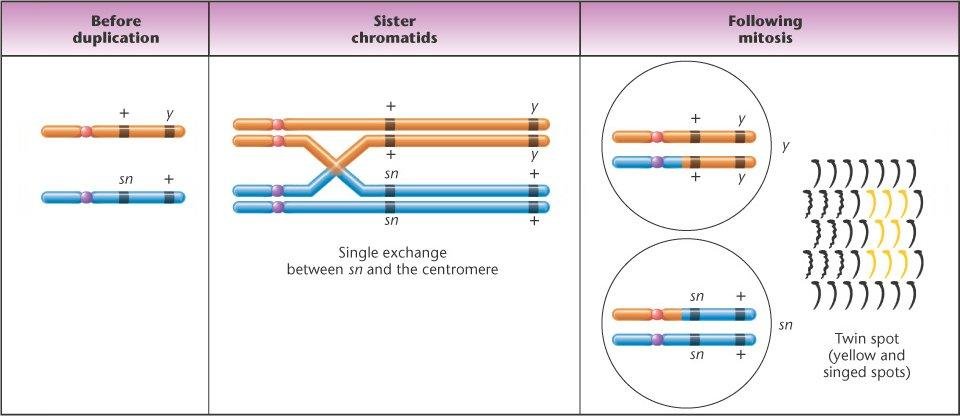







Map distance between 2 genes =
% SCO + % DCO *100/Total Offspring