Use BioFormats to read CZI image data into NumPy arrays or MATLAB matrices
(USE AT YOUT OWN RISK)
General Remarks
- All source code shown inside this presentation is usually just a snippet to illustrate the general idea
- It will will not work when just copied since important parts are missing
- Please use all tools at your own risk.
Prerequistes Python
- get the latest bioformats_package.jar
http://downloads.openmicroscopy.org/bio-formats/
- install python_bioformats and javabridge
https://pypi.python.org/pypi/python-bioformats
Prerequistes Python
- Get a python distro of your choice and the bfimage package:
https://www.continuum.io/why-anaconda
https://github.com/sebi06/BioFormatsRead
- The bfimage package contains the latest version of the bioformats_package.jar and additional scripts to read a Z-Stack or a TimeSeries from a multi-dimensional CZI data set.
- The package can read image data sets in general, not only CZI files thanks to BioFormats.
Use BioFormats from Python
- Register the latest bioformats_package.jar in order to use it
def set_bfpath(bfpackage_path=BFPATH):
# this function can be used to set the path to the package individually
global BFPATH
BFPATH = bfpackage_path
return BFPATH
def start_jvm(max_heap_size='4G'):
"""
Start the Java Virtual Machine, enabling BioFormats IO.
Optional: Specify the path to the bioformats_package.jar to your needs by calling.
set_bfpath before staring to read the image data
Parameters
----------
max_heap_size : string, optional
The maximum memory usage by the virtual machine. Valid strings
include '256M', '64k', and '2G'. Expect to need a lot.
"""
jars = jv.JARS + [BFPATH]
#jars = jv.JARS
jv.start_vm(class_path=jars, max_heap_size=max_heap_size)
VM_STARTED = TrueGet JavaMetaDataStore - 1
def get_java_metadata_store(imagefile):
if not VM_STARTED:
start_jvm()
if VM_KILLED:
jvm_error()
# get the actual image reader
rdr = bioformats.get_image_reader(None, path=imagefile)
# for "whatever" reason the number of total series can only be accessed here ...
try:
totalseries = np.int(rdr.rdr.getSeriesCount())
except:
totalseries = 1 # in case there is only ONE series
try:
for sc in range(0, totalseries):
rdr.rdr.setSeries(sc)
resolutioncount = rdr.rdr.getResolutionCount()
print('Resolution count for series #', sc, ' = ' + resolutioncount)
for res in range(0, resolutioncount):
rdr.rdr.setResolution(res)
print('Resolution #', res, ' dimensions = ', rdr.getSizeX(), ' x ', rdr.getSizeY())
except:
print('Multi-Resolution API not enabled yet.')
Get JavaMetaDataStore - 2
...
series_dimensions = []
# cycle through all the series and check the dimensions
for sc in range(0, totalseries):
rdr.rdr.setSeries(sc)
dimx = rdr.rdr.getSizeX()
dimy = rdr.rdr.getSizeY()
series_dimensions.append((dimx, dimy))
if len(series_dimensions) == 1:
multires = False
elif len(series_dimensions) > 1:
if series_dimensions[0] == series_dimensions[1]:
multires = False
if not series_dimensions[0] == series_dimensions[1]:
multires = True
# rdr.rdr is the actual BioFormats reader. rdr handles its lifetime
javametadata = jv.JWrapper(rdr.rdr.getMetadataStore())
imagecount = javametadata.getImageCount()
imageIDs = []
for id in range(0, imagecount):
imageIDs.append(id)
rdr.close()
return javametadata, totalseries, imageIDs, series_dimensions, multiresUse get_image6d
def get_image6d(imagefile, sizes):
"""
This function will read the image data and store them into a 6D numpy array.
The 6D array has the following dimension order: [Series, T, Z, C, X, Y].
"""
if not VM_STARTED:
start_jvm()
if VM_KILLED:
jvm_error()
rdr = bioformats.ImageReader(imagefile, perform_init=True)
img6d = np.zeros(sizes, dtype=BF2NP_DTYPE[rdr.rdr.getPixelType()])
readstate = 'OK'
readproblems = []
# main loop to read the images from the data file
for seriesID in range(0, sizes[0]):
for timepoint in range(0, sizes[1]):
for zplane in range(0, sizes[2]):
for channel in range(0, sizes[3]):
try:
img6d[seriesID, timepoint, zplane, channel, :, :] =\
rdr.read(series=seriesID, c=channel, z=zplane, t=timepoint, rescale=False)
except:
print('Problem reading data into Numpy for Series', seriesID, sys.exc_info()[1])
readstate = 'NOK'
readproblems = sys.exc_info()[1]
rdr.close()
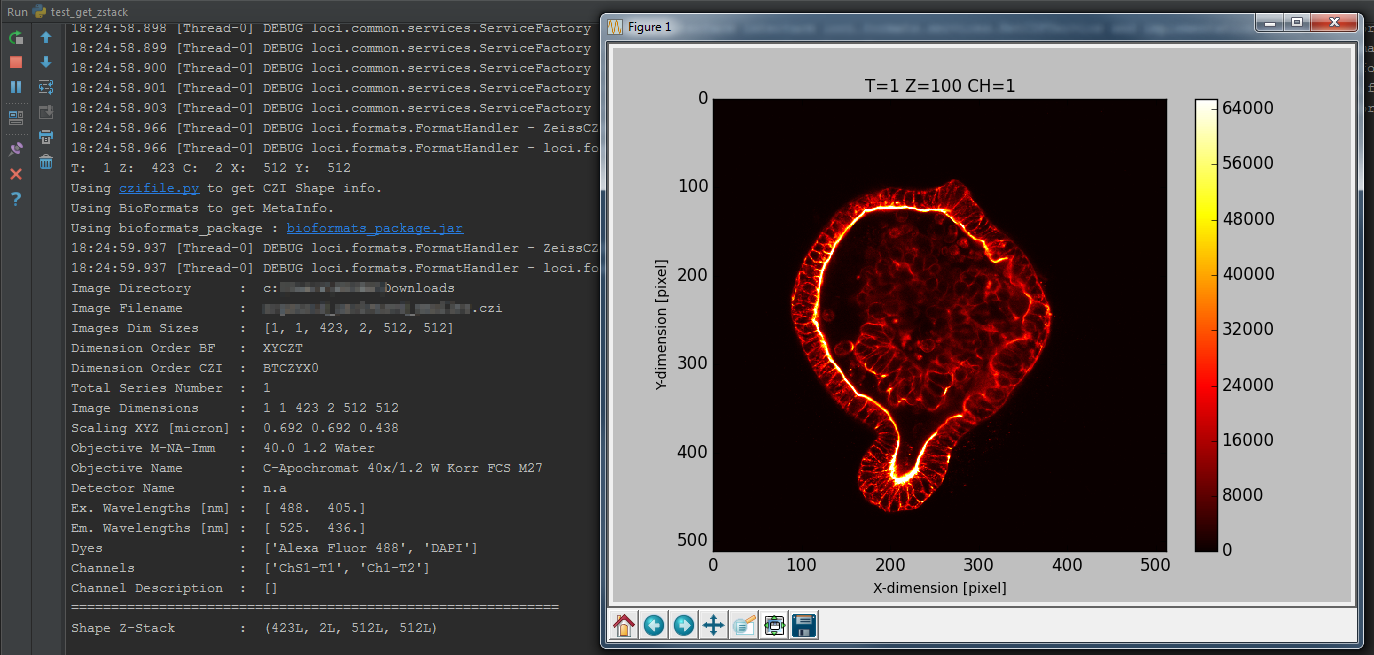
return img6d, readstateTest bfimage package
from __future__ import print_function
import numpy as np
import os
import bfimage as bf
import sys
# use for BioFormtas <= 5.1.10
urlnamespace = 'http://www.openmicroscopy.org/Schemas/OME/2015-01'
# use for BioFormtas > 5.2.0
#urlnamespace = 'http://www.openmicroscopy.org/Schemas/OME/2016-06'
# specify bioformats_package.jar to use if required
bfpackage = r'bioformats_package/5.1.10/bioformats_package.jar'
bf.set_bfpath(bfpackage)
# get image meta-information
MetaInfo = bf.bftools.get_relevant_metainfo_wrapper(filename, namespace=urlnamespace)
try:
img6d, readstate = bf.get_image6d(filename, MetaInfo['Sizes'])
arrayshape = np.shape(img6d)
except:
arrayshape = []
print('Could not read image data into NumPy array.')
# show relevant image Meta-Information
print('\n')
print('Testscript used : test_get_image6d.py')
print('OME NameSpace used : ', urlnamespace)
print('BF Version used : ', bfpackage)
print('Array Shape 6D : ', arrayshape)
print('Read Status : ', readstate)Test bfimage package
Extract MetaInfo from XML
import bfimage as bf
from lxml import etree as etl
def writeomexml(imagefile, method=1, writeczi_metadata=True):
# creates readable xml files from image data files. Default method should be = 1.
if method == 1:
# method 1
# Change File name and write XML file to same folder
xmlfile1 = imagefile[:-4] + '_MetaData1.xml'
try:
# get the actual OME-XML
omexml = get_OMEXML(imagefile)
# create root and tree from XML string and write "pretty" to disk
root = etl.fromstring(omexml)
tree = etl.ElementTree(root)
tree.write(xmlfile1, pretty_print=True, encoding='utf-8', method='xml')
print('Created OME-XML file for testdata: ', imagefile)
except:
print('Creating OME-XML failed for testdata: ', imagefile)
... (missing code part)Extract Metainformation from XML
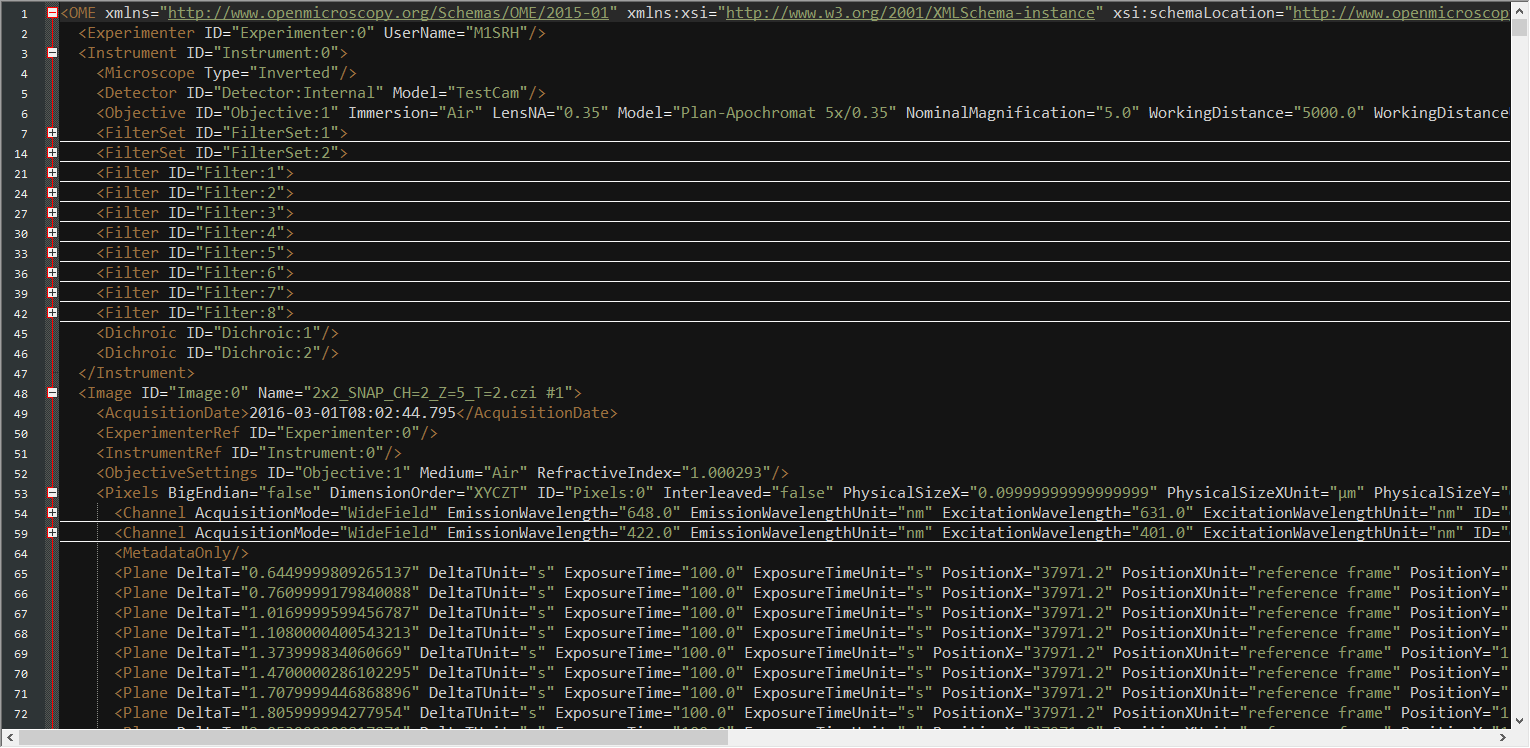
Create PlaneTables
def get_planetable(imagefile, writecsv=False, separator='\t'):
... (missing code part)
for imageIndex in range(0, max(MetaInfo['ImageIDs'])+1):
for planeIndex in range(0, MetaInfo['SizeZ'] * MetaInfo['SizeC'] * MetaInfo['SizeT']):
id.append(imageIndex)
plane.append(planeIndex)
theC.append(jmd.getPlaneTheC(imageIndex, planeIndex).getValue().intValue())
theZ.append(jmd.getPlaneTheZ(imageIndex, planeIndex).getValue().intValue())
theT.append(jmd.getPlaneTheT(imageIndex, planeIndex).getValue().intValue())
xpos.append(jmd.getPlanePositionX(imageIndex, planeIndex).value().doubleValue())
ypos.append(jmd.getPlanePositionY(imageIndex, planeIndex).value().doubleValue())
zpos.append(jmd.getPlanePositionZ(imageIndex, planeIndex).value().doubleValue())
dt.append(jmd.getPlaneDeltaT(imageIndex, planeIndex).value().doubleValue())
... (missing code part)
# create Pandas dataframe to hold the plane data
df = pd.DataFrame([np.asarray(id), np.asarray(plane), np.asarray(theT),
np.asarray(theZ), np.asarray(theC), xpos, ypos, zpos, dt])
df = df.transpose()
# give the planetable columns the correct names
df.columns = ['ImageID', 'Plane', 'TheT', 'TheZ', 'TheC', 'XPos', 'YPos', 'ZPos', 'DeltaT']
if writecsv:
csvfile = imagefile[:-4] + '_planetable.csv'
# use tab as separator and do not write the index to the CSV data table
df.to_csv(csvfile, sep=separator, index=False)
print('Writing CSV file: ', csvfile)
return df, csvfileAnalyze PlaneTables
Output:
Writing CSV file: testdata/Wellchamber_384_Comb.csv
IMAGEID Plane TheT TheZ TheC XPos YPos ZPos DeltaT
0 0 0 0 0 0 2250.9 6250.5 1253.2 0.000
1 1 0 0 0 0 6750.9 6250.5 1261.1 1.648
2 2 0 0 0 0 11250.9 6250.5 1256.9 3.403
3 3 0 0 0 0 15750.9 6250.5 1255.9 5.247
4 4 0 0 0 0 20250.9 6250.5 1262.1 7.049
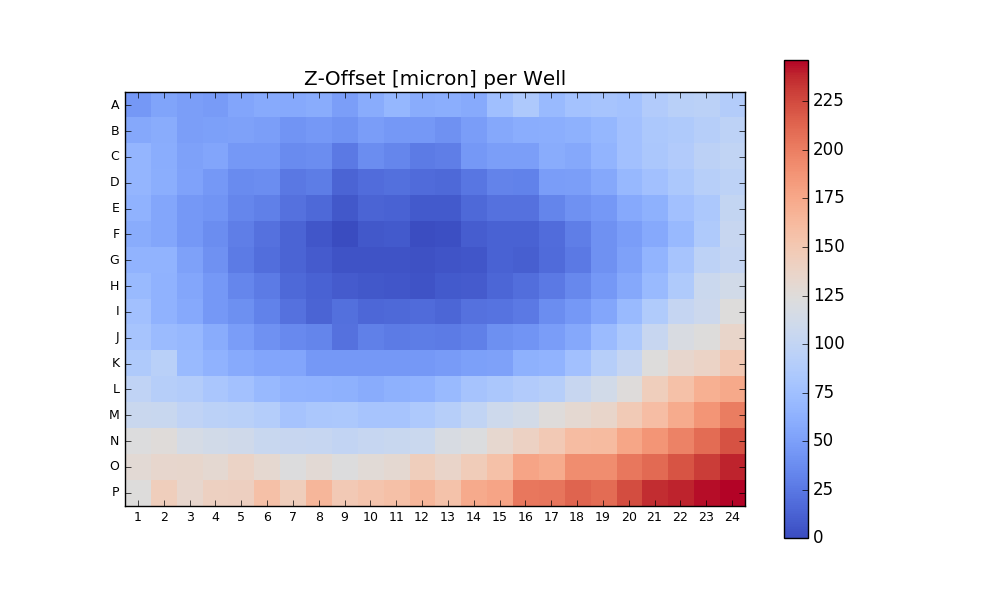
384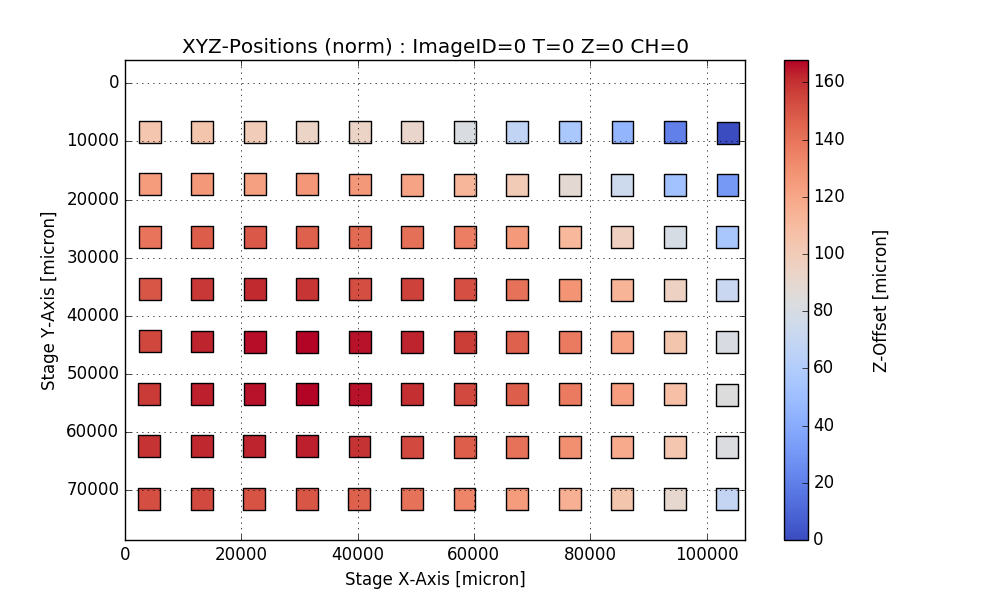
Prerequisites MATLAB
Obviously one needs a running MATLAB installation to perform the next steps.
- Get the MATLAB toolbox for BioFormats:
http://downloads.openmicroscopy.org/bio-formats/5.1.10/
- Follow the instruction given here:
http://www.openmicroscopy.org/site/support/bio-formats5.1/users/matlab/index.html
Prerequisites MATLAB
To make life a bit easier I created to simple m-files that can be used to read the CZI image data into MATLAB in an similar way explained for Python previously.
- Get the following m-files:
- GetOMEData.m
- ReadImage6D.m
Both files are simple wrapper to what the BioFormats MATLAB toolbox is already offering.
GetOMEData.m
function OMEData = GetOMEData(filename)
% Get OME Meta Information using BioFormats Library 5.1.8
% To access the file reader without loading all the data, use the low-level bfGetReader.m function:
reader = bfGetReader(filename);
% You can then access the OME metadata using the getMetadataStore() method:
omeMeta = reader.getMetadataStore();
% get ImageCount --> currently only reading one image is supported
imagecount = omeMeta.getImageCount();
imageID = imagecount - 1;
% get the actual metadata and store them in a structured array
[pathstr,name,ext] = fileparts(filename);
OMEData.FilePath = pathstr;
OMEData.Filename = strcat(name, ext);
% Get dimension order
OMEData.DimOrder = char(omeMeta.getPixelsDimensionOrder(imageID).getValue());
% Number of series inside the complete data set
OMEData.SeriesCount = reader.getSeriesCount();
% Dimension Sizes C - T - Z - X - Y
OMEData.SizeC = omeMeta.getPixelsSizeC(imageID).getValue();
OMEData.SizeT = omeMeta.getPixelsSizeT(imageID).getValue();
OMEData.SizeZ = omeMeta.getPixelsSizeZ(imageID).getValue();
OMEData.SizeX = omeMeta.getPixelsSizeX(imageID).getValue();
OMEData.SizeY = omeMeta.getPixelsSizeY(imageID).getValue();
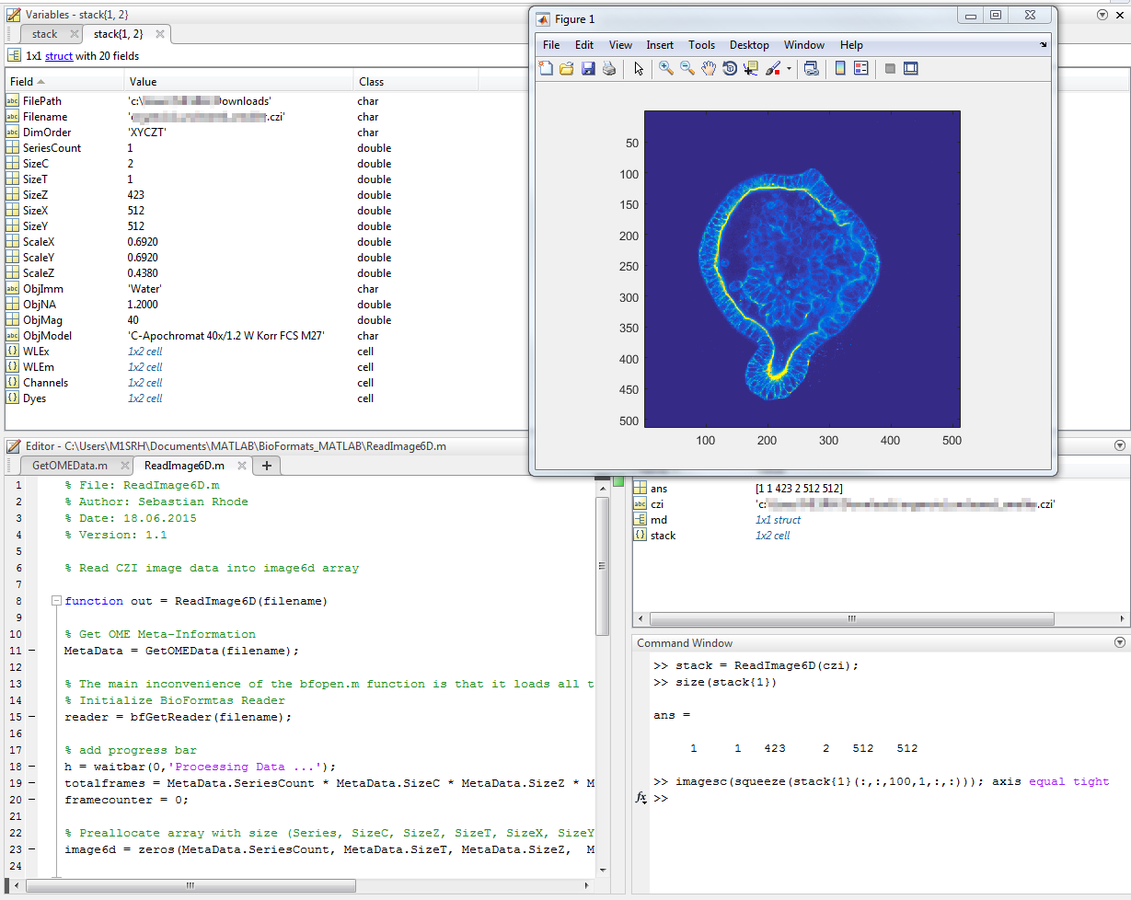
ReadImage6D.m
function out = ReadImage6D(filename)
% Get OME Meta-Information
MetaData = GetOMEData(filename);
% The main inconvenience of the bfopen.m function is that it loads all the content of an image regardless of its size.
% Initialize BioFormats reader.
reader = bfGetReader(filename);
% Preallocate array with size (Series, SizeC, SizeZ, SizeT, SizeX, SizeY)
image6d = zeros(MetaData.SeriesCount, MetaData.SizeT, MetaData.SizeZ, MetaData.SizeC, MetaData.SizeY, MetaData.SizeX);
for series = 1: MetaData.SeriesCount
% set reader to current series
reader.setSeries(series-1);
for timepoint = 1: MetaData.SizeT
for zplane = 1: MetaData.SizeZ
for channel = 1: MetaData.SizeC
% get linear index of the plane (1-based)
iplane = loci.formats.FormatTools.getIndex(reader, zplane - 1, channel - 1, timepoint -1) + 1;
% get frame for current series
image6d(series, timepoint, zplane, channel, :, :) = bfGetPlane(reader, iplane);
end
end
end
end
% close BioFormats Reader
reader.close();
% store image data and meta information in cell array
out = {};
out{1} = image6d;
out{2} = MetaData;Usage Example - MATLAB
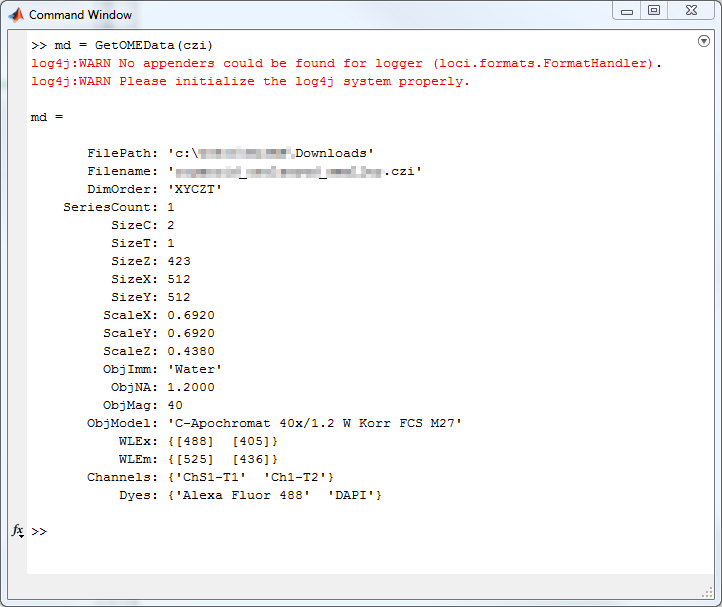
Usage Example - MATLAB