Stian Soiland-Reyes (helped by Robin Long & Michael Crusoe)
eScience lab, The University of Manchester
BioExcel Virtual Training
2020-04-29
This work is licensed under a
Creative Commons Attribution 4.0 International License.



This work has been done as part of the BioExcel CoE (www.bioexcel.eu), a project funded by the European Union contracts H2020-INFRAEDI-02-2018-823830, H2020-EINFRA-2015-1-675728

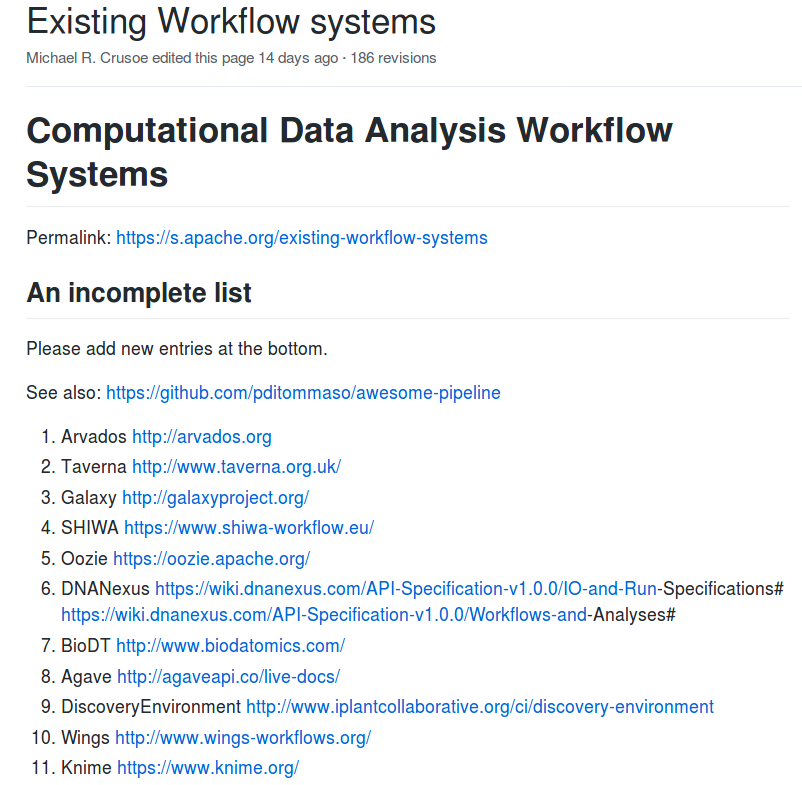
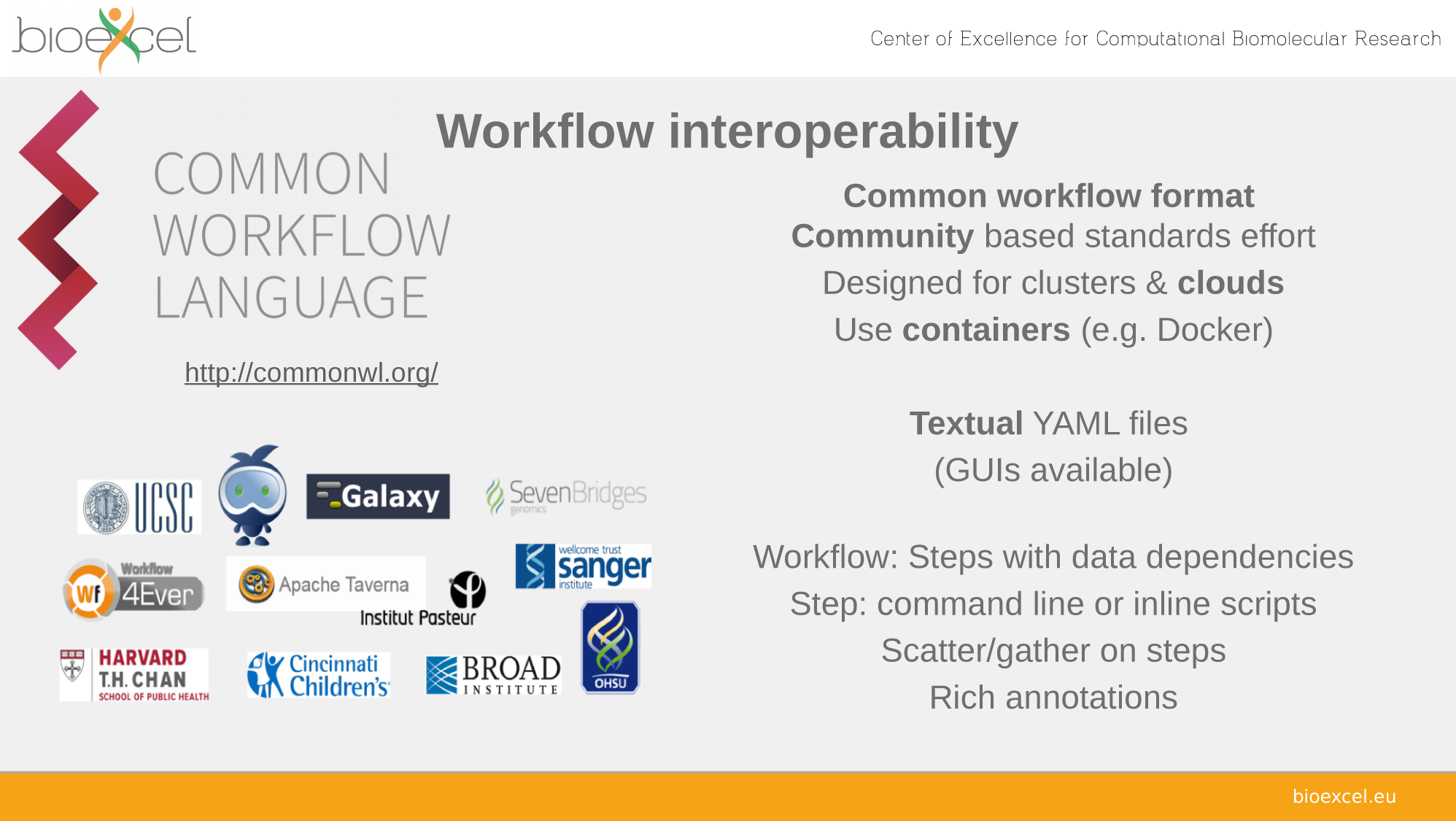
Reproducible analyses with
Common Workflow Language

BioExcel Cloud Portal

Findable
Accessible
Interoperable
Reusable

F1. (meta)data are assigned a globally unique and persistent identifier
F2. data are described with rich metadata (defined by R1 below)
F3. metadata clearly and explicitly include the identifier of the data it describes
F4. (meta)data are registered or indexed in a searchable resource
To be Findable:
A1. (meta)data are retrievable by their identifier using a
standardized communications protocol
A1.1 the protocol is open, free, and universally implementable
A1.2 the protocol allows for an authentication and authorization
procedure, where necessary
A2. metadata are accessible, even when the data are no longer available
To be Accessible:
I1. (meta)data use a formal, accessible, shared, and
broadly applicable language for knowledge representation.
I2. (meta)data use vocabularies that follow FAIR principles
I3. (meta)data include qualified references to other (meta)data
To be Interoperable:
R1. meta(data) are richly described with a plurality of
accurate and relevant attributes
R1.1. (meta)data are released with a clear and accessible data usage license
R1.2. (meta)data are associated with detailed provenance
R1.3. (meta)data meet domain-relevant community standards
To be Reusable:
Inner FAIR
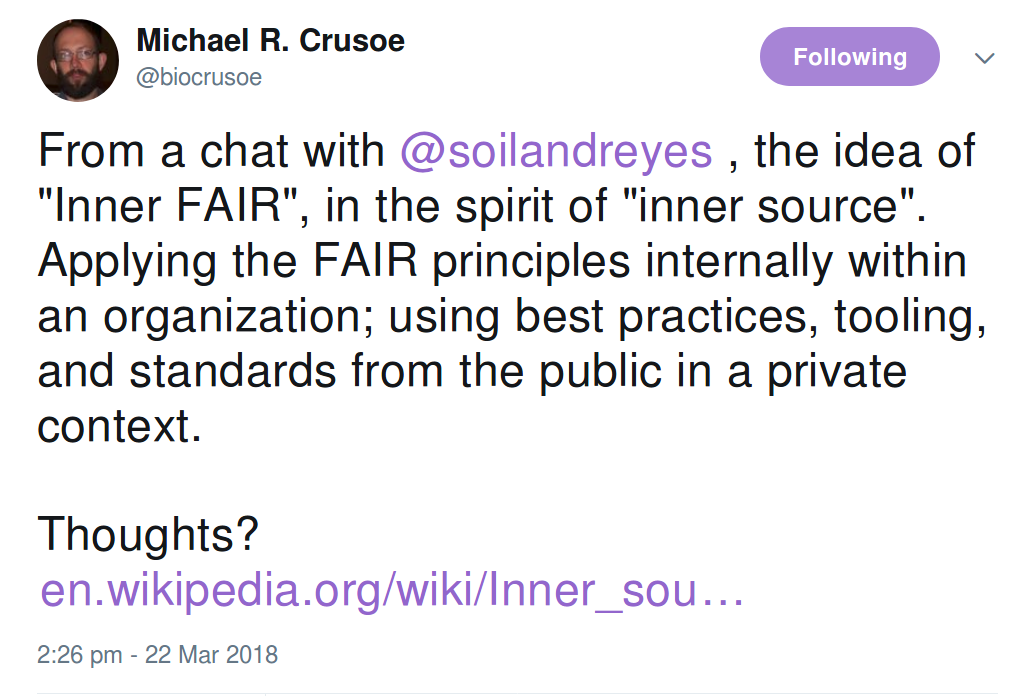
From a chat with @soilandreyes , the idea of "Inner FAIR", in the spirit of "inner source". Applying the FAIR principles internally within an organization; using best practices, tooling, and standards from the public in a private context.
— Michael R. Crusoe (@biocrusoe) March 22, 2018
Thoughts?https://t.co/ynkl47l07Y
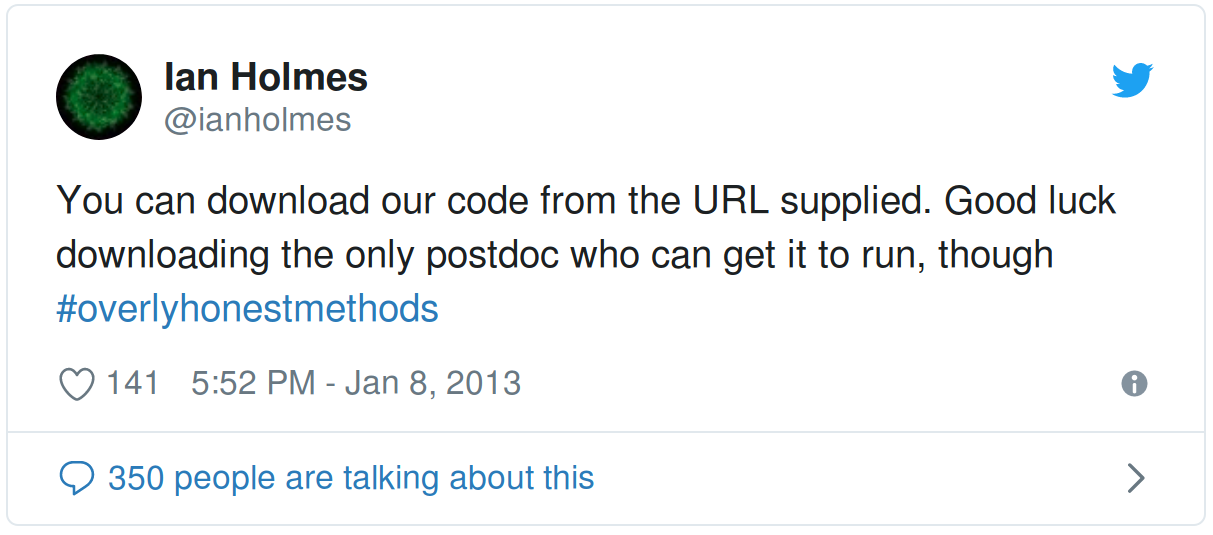
Workflows
You can download our code from the URL supplied. Good luck downloading the only postdoc who can get it to run, though #overlyhonestmethods
— Ian Holmes (@ianholmes) January 8, 2013
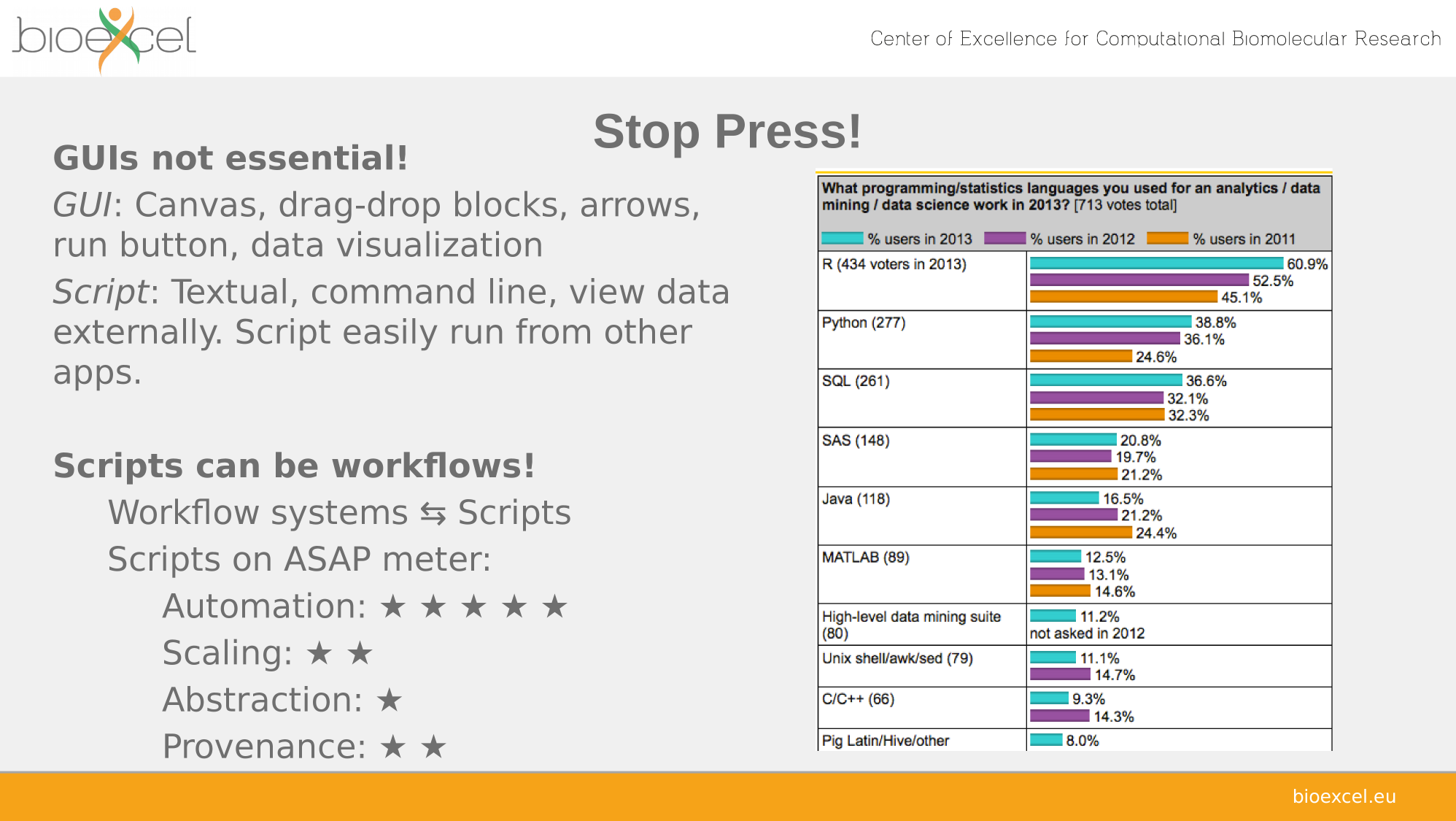
Automation
– Automate computational aspects
– Repetitive pipelines, sweep campaigns
Scaling—compute cycles
– Make use of computational infrastructure
– Handle large data
Abstraction—people cycles
– Shield complexity and incompatibilities
– Report, re-usue, evolve, share, compare
– Repeat—Tweak—Repeat
– First-class commodities
Provenance—reporting
– Capture, report and utilize log and data lineage
– Auto-documentation
– Tracable evolution, audit, transparency
– Reproducible science
Findable
Accessible
Interoperable
Reusable
(Reproducible)
Why use workflows?
Adapted from Bertram Ludäscher (2015)
https://www.slideshare.net/ludaesch/works-2015provenancemileage
https://doi.org/10.1007/s13222-012-0100-z
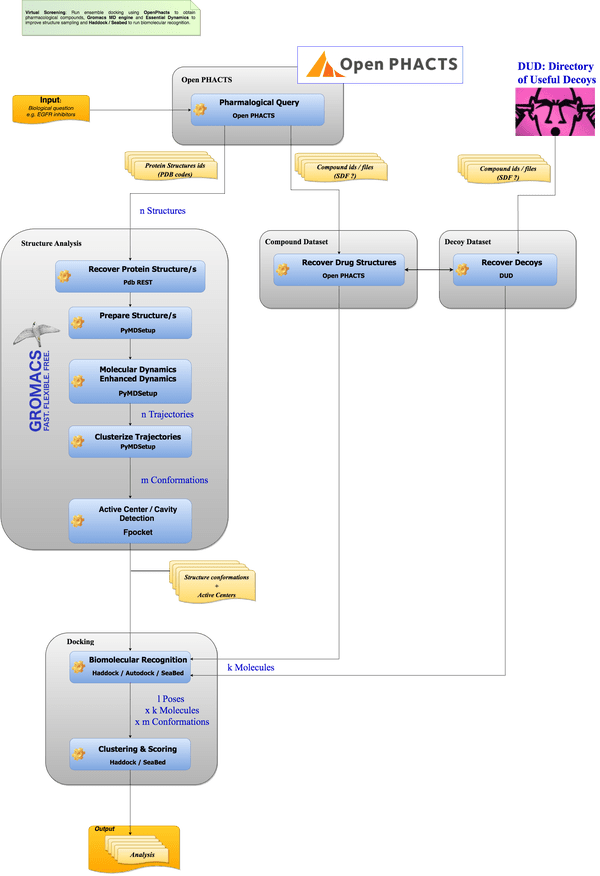

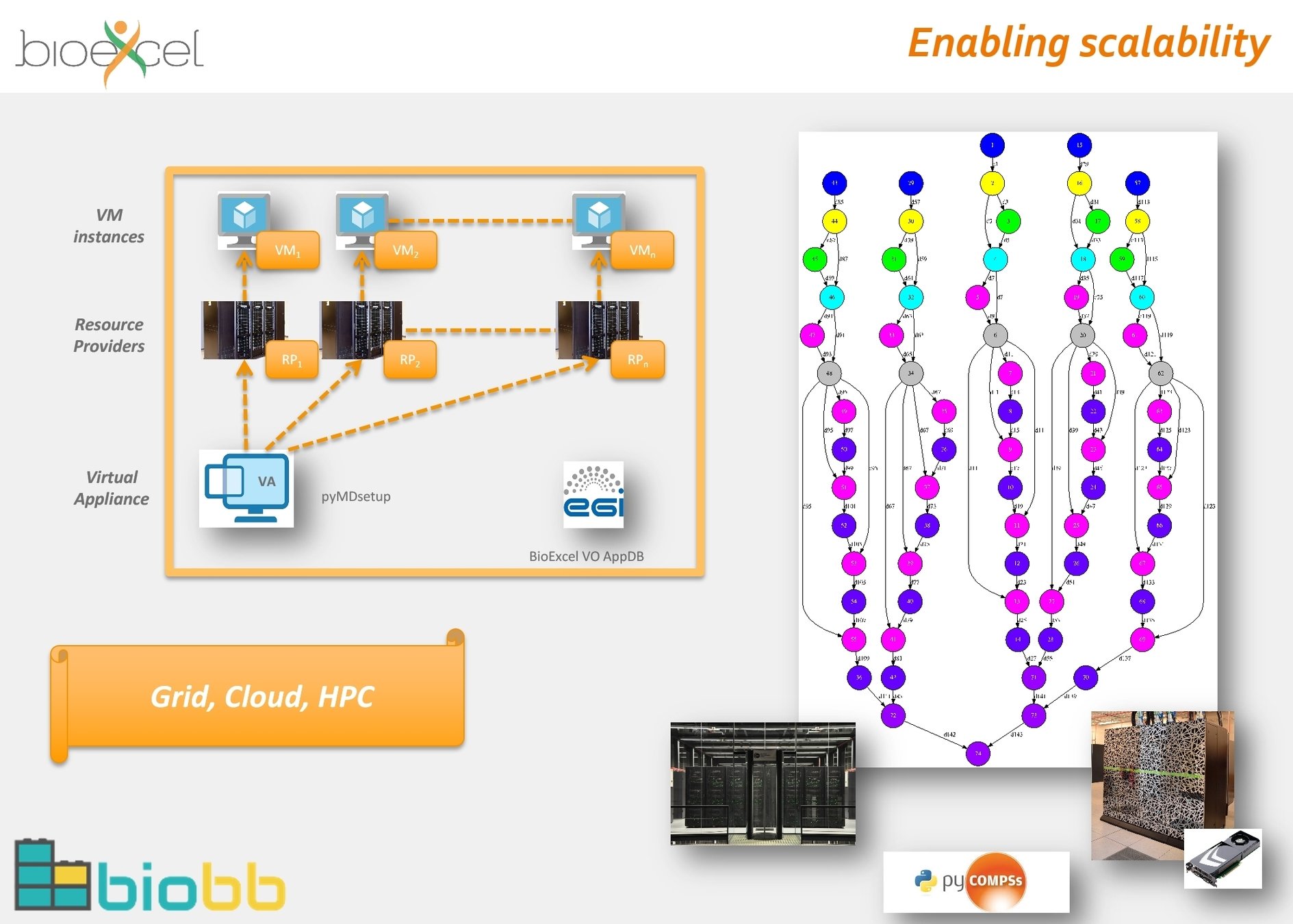
Adapted from Adam Hospital, BioExcel
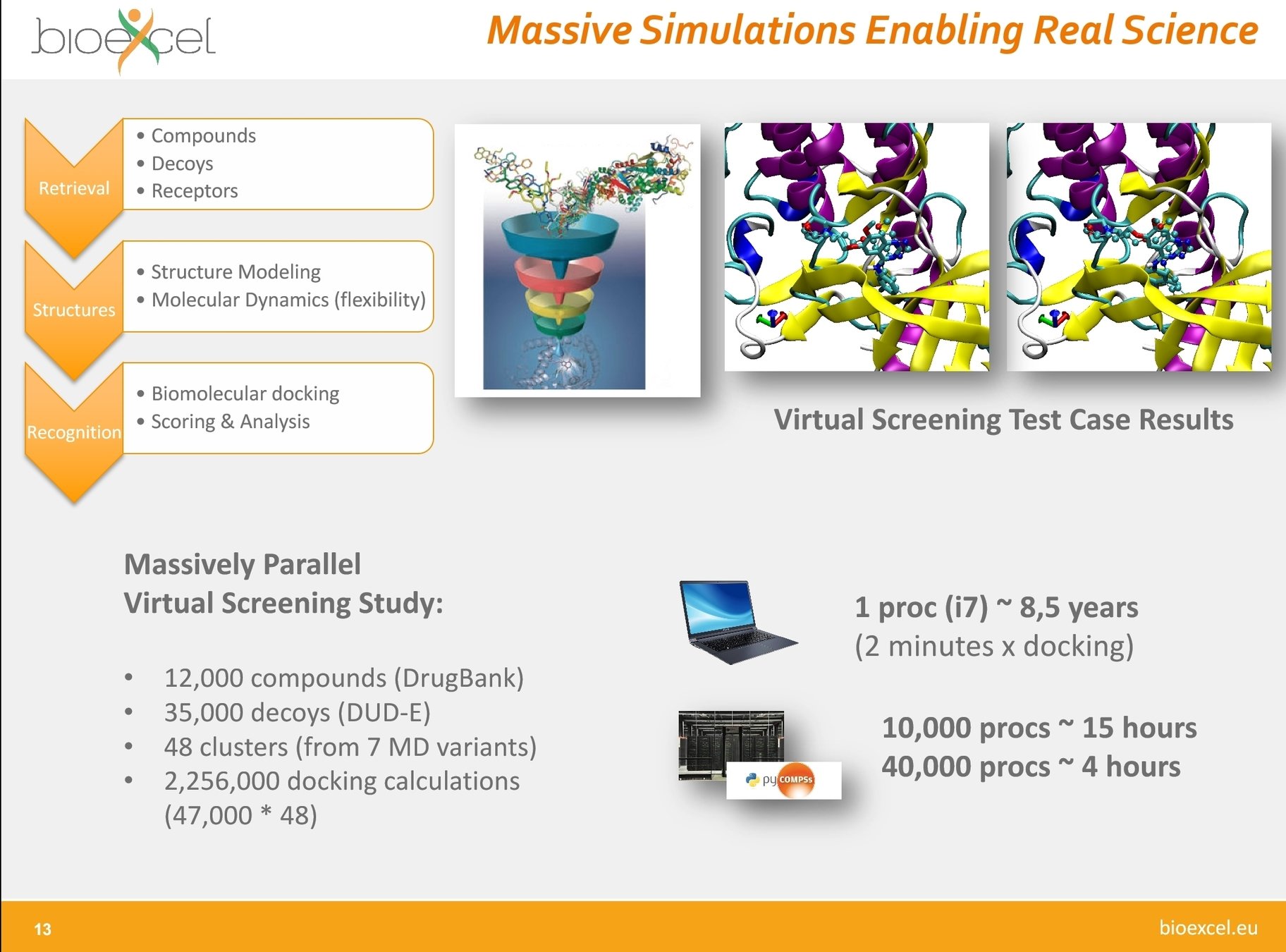
Adam Hospital, BioExcel
Adapted from Adam Hospital, BioExcel
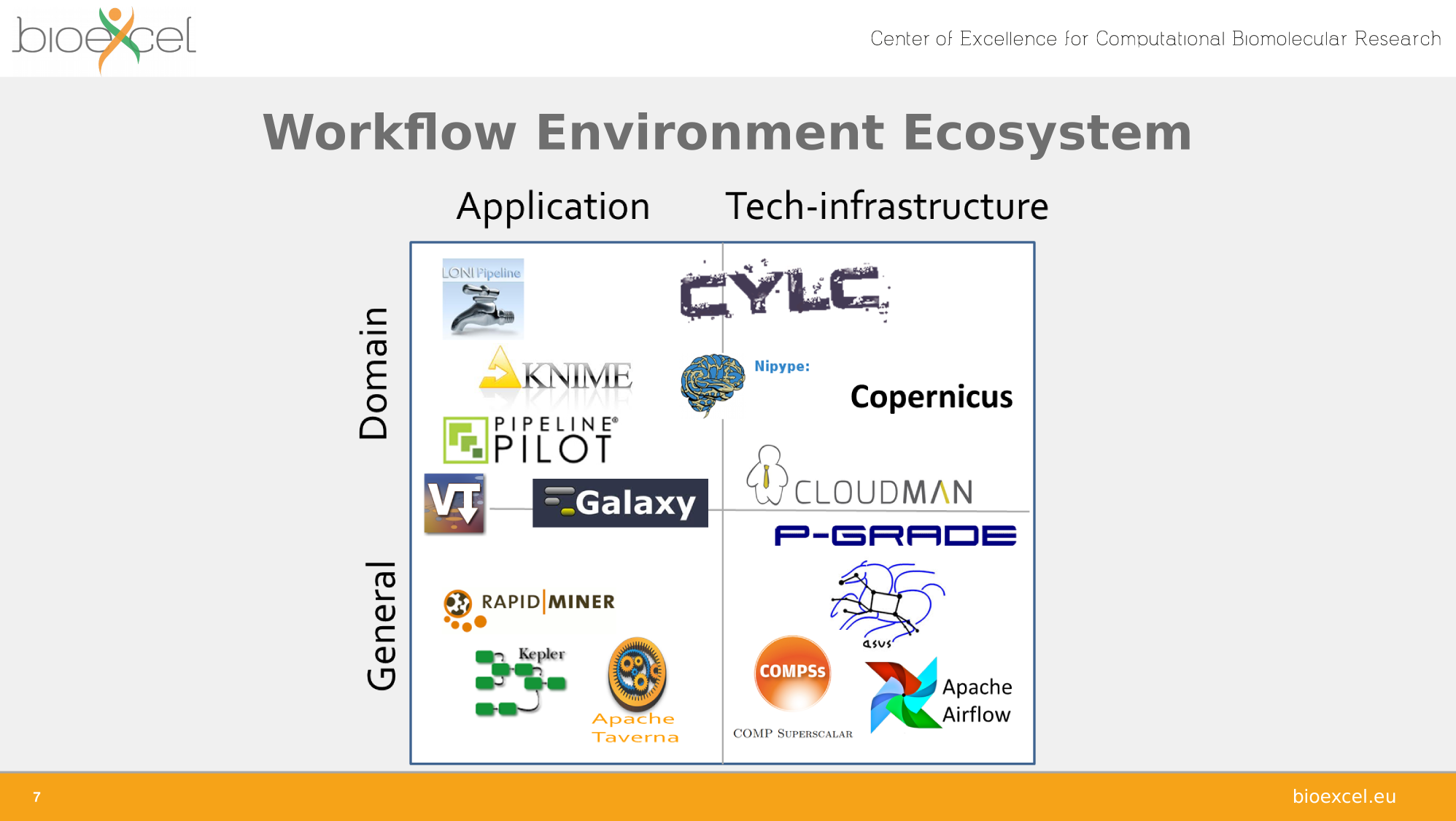

Adapted from Carole Goble





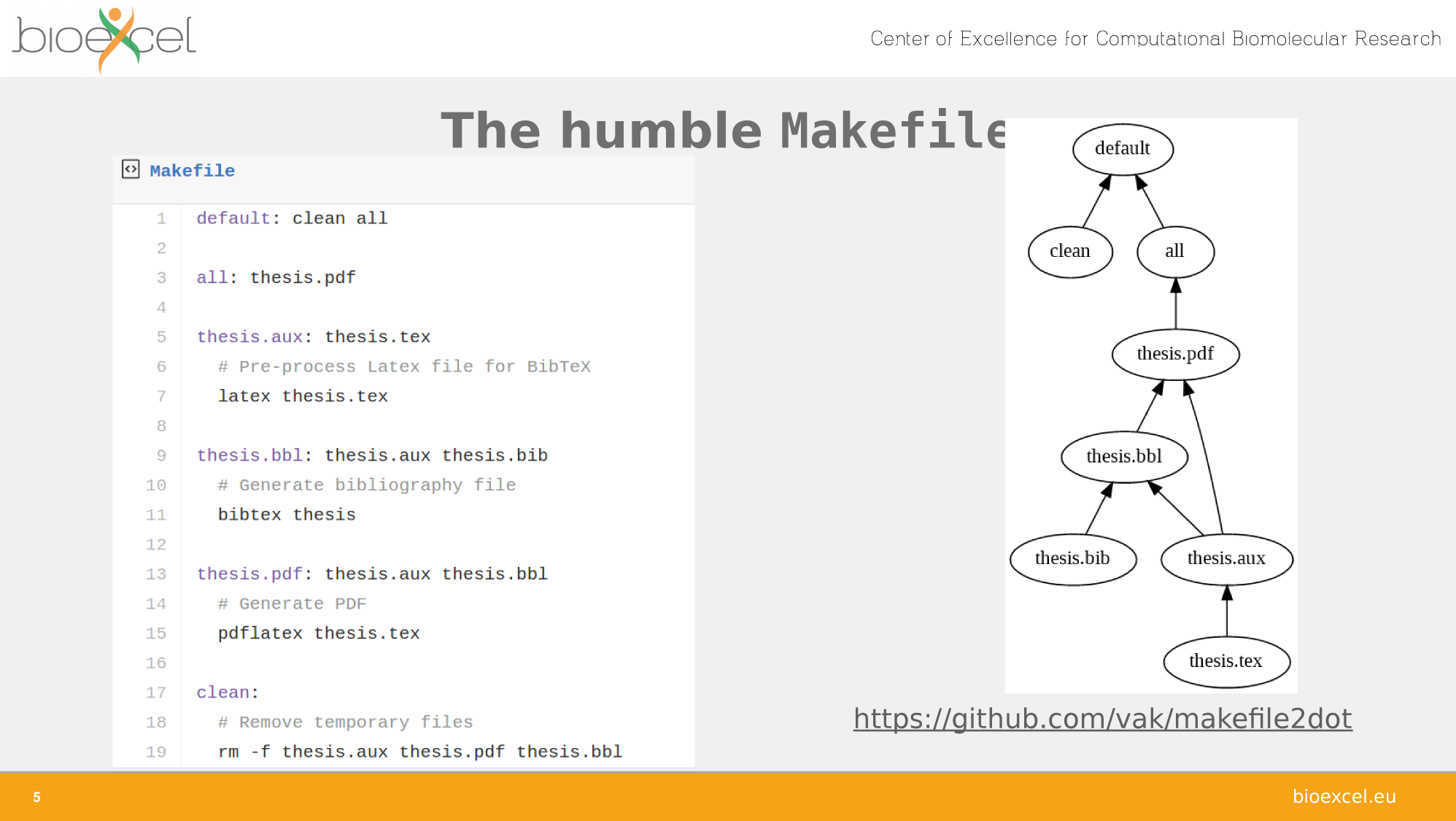
Adapted from Carole Goble


cwlVersion: v1.0
class: Workflow
inputs:
inp: File
ex: string
outputs:
classout:
type: File
outputSource: compile/classfile
steps:
untar:
run: tar-param.cwl
in:
tarfile: inp
extractfile: ex
out: [example_out]
compile:
run: arguments.cwl
in:
src: untar/example_out
out: [classfile]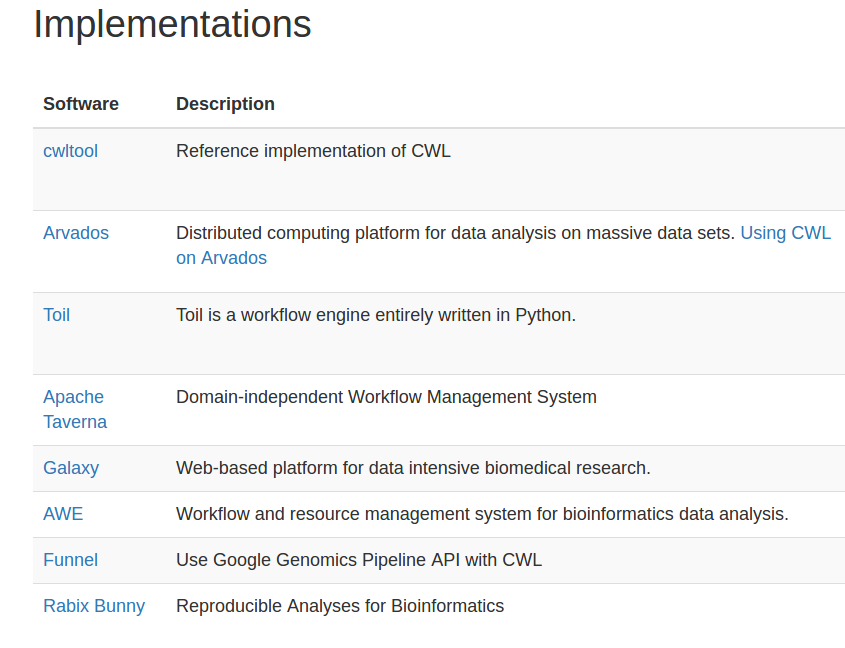

Nature 573, 149-150 (2019)
https://doi.org/10.1038/d41586-019-02619-z



Building CWL workflows
cwlVersion: v1.0
class: Workflow
inputs:
toConvert: File
outputs:
converted:
type: File
outputSource: convertMethylation/converted
combined:
type: File
outputSource: mergeSymmetric/combined
steps:
convertMethylation:
run: interconverter.cwl
in:
toConvert: toConvert
out: [converted]
mergeSymmetric:
run: symmetriccpgs.cwl
in:
toCombine: convertMethylation/converted
out: [combined]
cwlVersion: v1.0
class: CommandLineTool
inputs:
toConvert:
type: File
inputBinding:
prefix: -i
outputs:
converted:
type: File
outputBinding:
glob: "*.meth"
baseCommand: interconverter.sh
arguments: ["-d", $(runtime.outdir)]
hints:
- class: DockerRequirement
dockerPull: "quay.io/neksa/screw-tool"
cwlVersion: v1.0
class: CommandLineTool
inputs:
toCombine:
type: File
inputBinding:
prefix: -i
outputs:
combined:
type: File
outputBinding:
glob: "*.sym"
baseCommand: symmetriccpgs.sh
arguments: ["-d", $(runtime.outdir)]
hints:
- class: DockerRequirement
dockerPull: "quay.io/neksa/screw-tool"

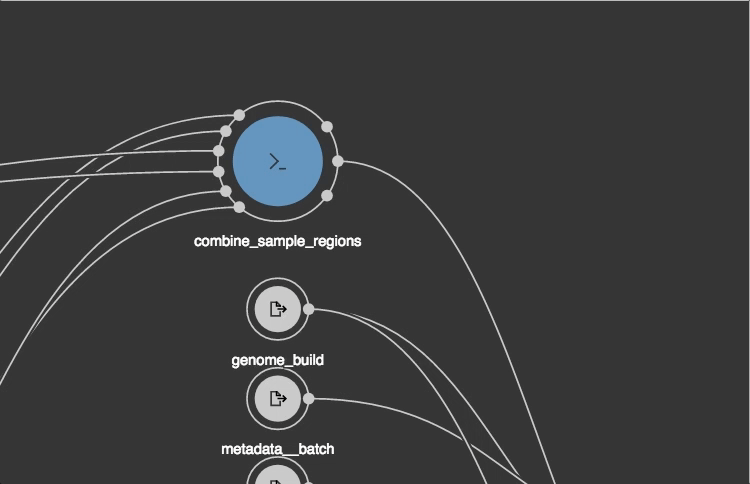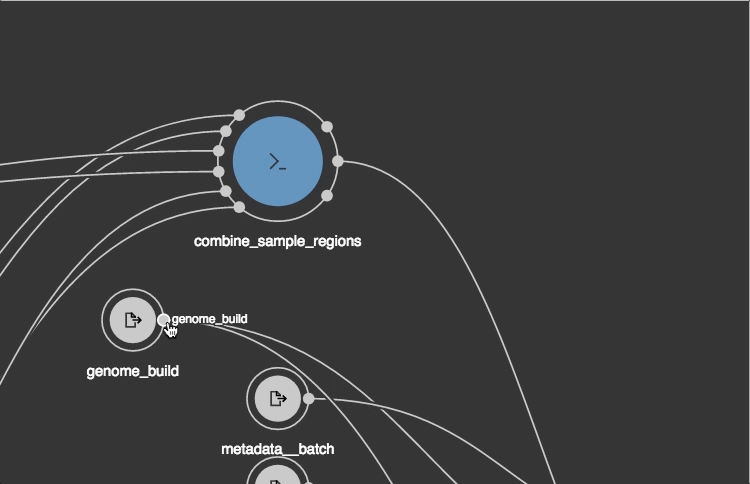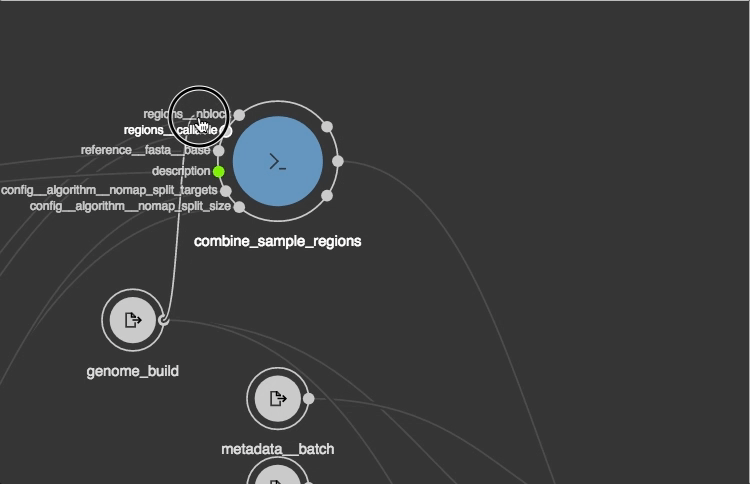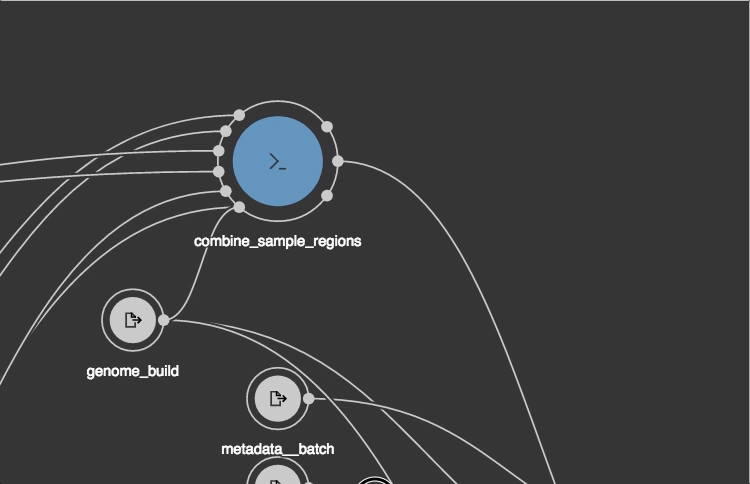

cwlVersion: v1.0
class: Workflow
label: EMG QC workflow, (paired end version). Benchmarking with MG-RAST expt.
requirements:
- class: SubworkflowFeatureRequirement
- class: SchemaDefRequirement
types:
- $import: ../tools/FragGeneScan-model.yaml
- $import: ../tools/trimmomatic-sliding_window.yaml
- $import: ../tools/trimmomatic-end_mode.yaml
- $import: ../tools/trimmomatic-phred.yaml
inputs:
reads:
type: File
format: edam:format_1930 # FASTQ
outputs:
processed_sequences:
type: File
outputSource: clean_fasta_headers/sequences_with_cleaned_headers
steps:
trim_quality_control:
doc: |
Low quality trimming (low quality ends and sequences with < quality scores
less than 15 over a 4 nucleotide wide window are removed)
run: ../tools/trimmomatic.cwl
in:
reads1: reads
phred: { default: '33' }
leading: { default: 3 }
trailing: { default: 3 }
end_mode: { default: SE }
minlen: { default: 100 }
slidingwindow:
default:
windowSize: 4
requiredQuality: 15
out: [reads1_trimmed]
convert_trimmed-reads_to_fasta:
run: ../tools/fastq_to_fasta.cwl
in:
fastq: trim_quality_control/reads1_trimmed
out: [ fasta ]
clean_fasta_headers:
run: ../tools/clean_fasta_headers.cwl
in:
sequences: convert_trimmed-reads_to_fasta/fasta
out: [ sequences_with_cleaned_headers ]
$namespaces:
edam: http://edamontology.org/
s: http://schema.org/
$schemas:
- http://edamontology.org/EDAM_1.16.owl
- https://schema.org/docs/schema_org_rdfa.html
s:license: "https://www.apache.org/licenses/LICENSE-2.0"
s:copyrightHolder: "EMBL - European Bioinformatics Institute"

Finding tools
Containers and packages

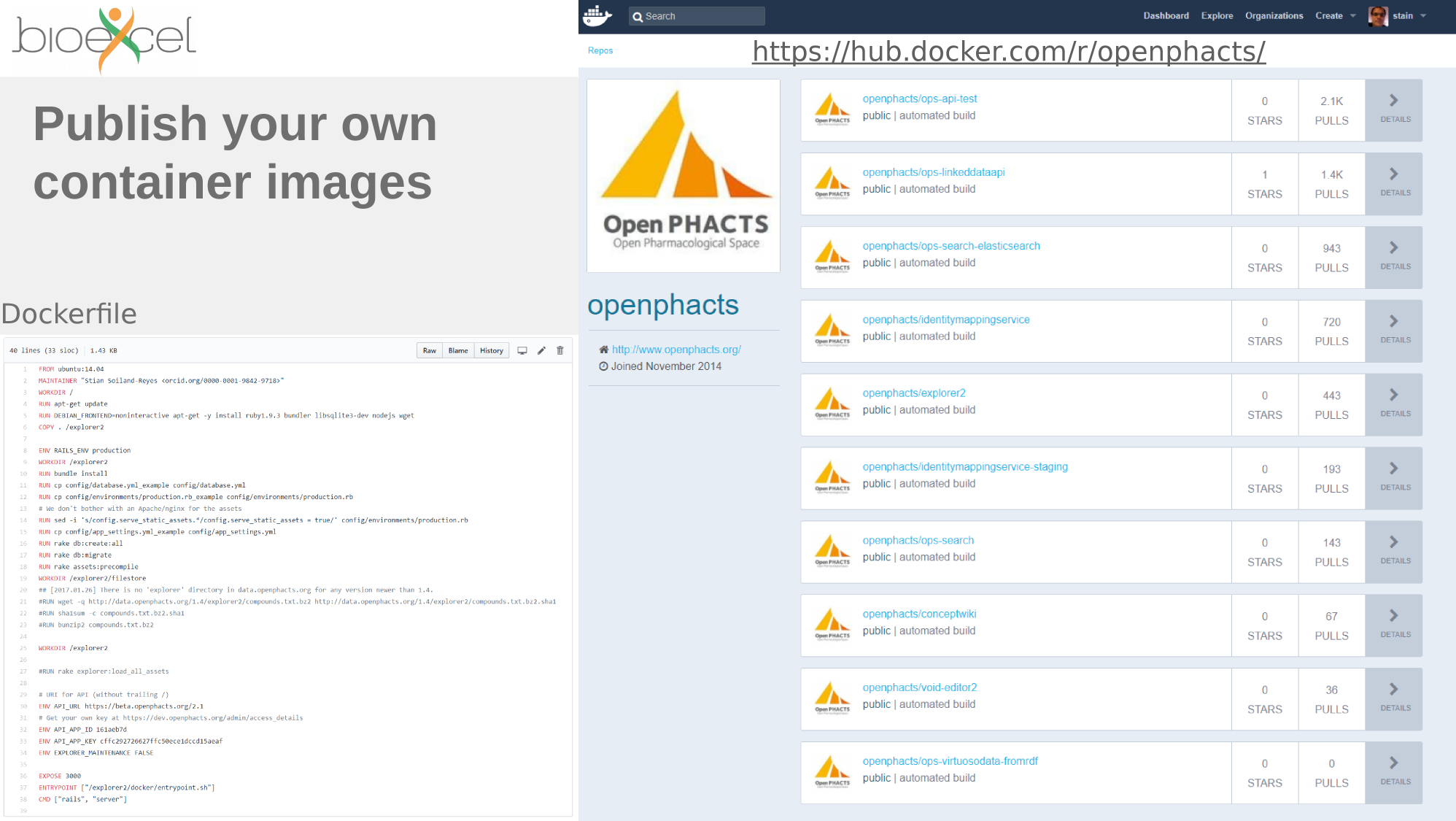


FROM ubuntu:14.04
MAINTAINER Stian Soiland-Reyes <soiland-reyes@cs.manchester.ac.uk>
# Install apache/PHP for REST API
RUN apt-get update && DEBIAN_FRONTEND=noninteractive apt-get install -y \
apache2 php5 memcached php5-memcached php5-memcache php5-curl \
supervisor \
unzip wget
RUN a2enmod rewrite
RUN rm -rf /var/www/html
#Install Linked Data API
RUN wget https://github.com/openphacts/OPS_LinkedDataApi/archive/1.5.0.zip -O /tmp/api.zip && \
cd /tmp && \
unzip api.zip && \
mv /tmp/OPS* /var/www/html
RUN sed -i '/<\/VirtualHost/ i\ <Directory /var/www/html/>\n AllowOverride All\n </Directory>' /etc/apache2/sites-enabled/000-default.conf
#RUN cat /etc/apache2/apache2.conf | tr "\n" "|||" | \
# sed 's,\(<Directory /var/www/html/>[^<]*\)AllowOverride None\([^<]*</Directory>\),\1AllowOverride All\2,' | \
# sed 's/|||/n/g' >/tmp/apache2 && \
# mv /tmp/apache2 /etc/apache2/apache2.conf
RUN mkdir /var/www/html/logs /var/www/html/cache && \
chmod 777 /var/www/html/logs /var/www/html/cache && \
chown -R www-data:www-data /var/www/html

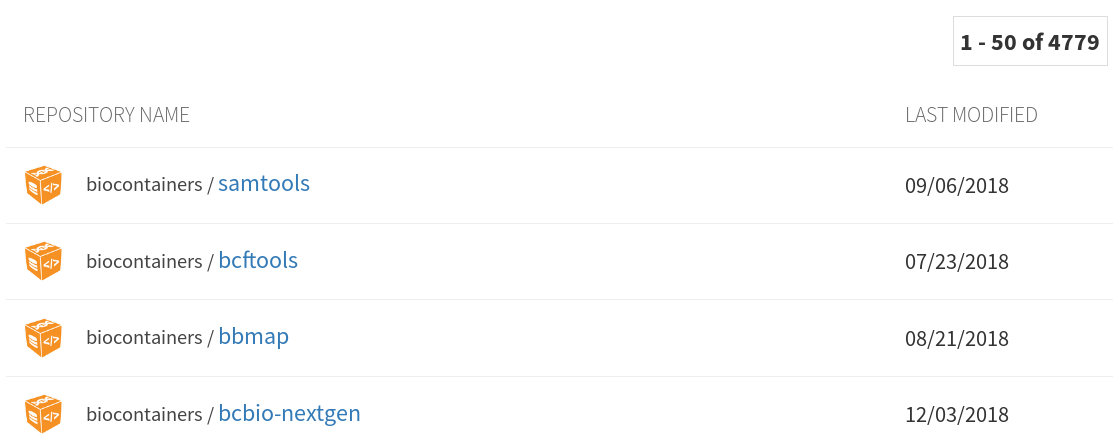
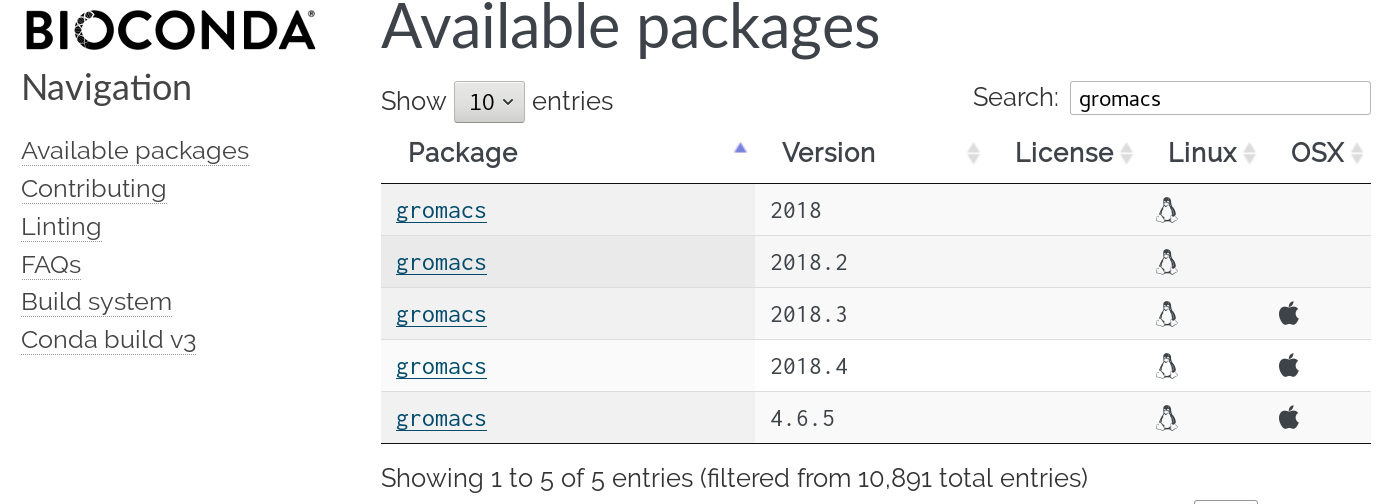
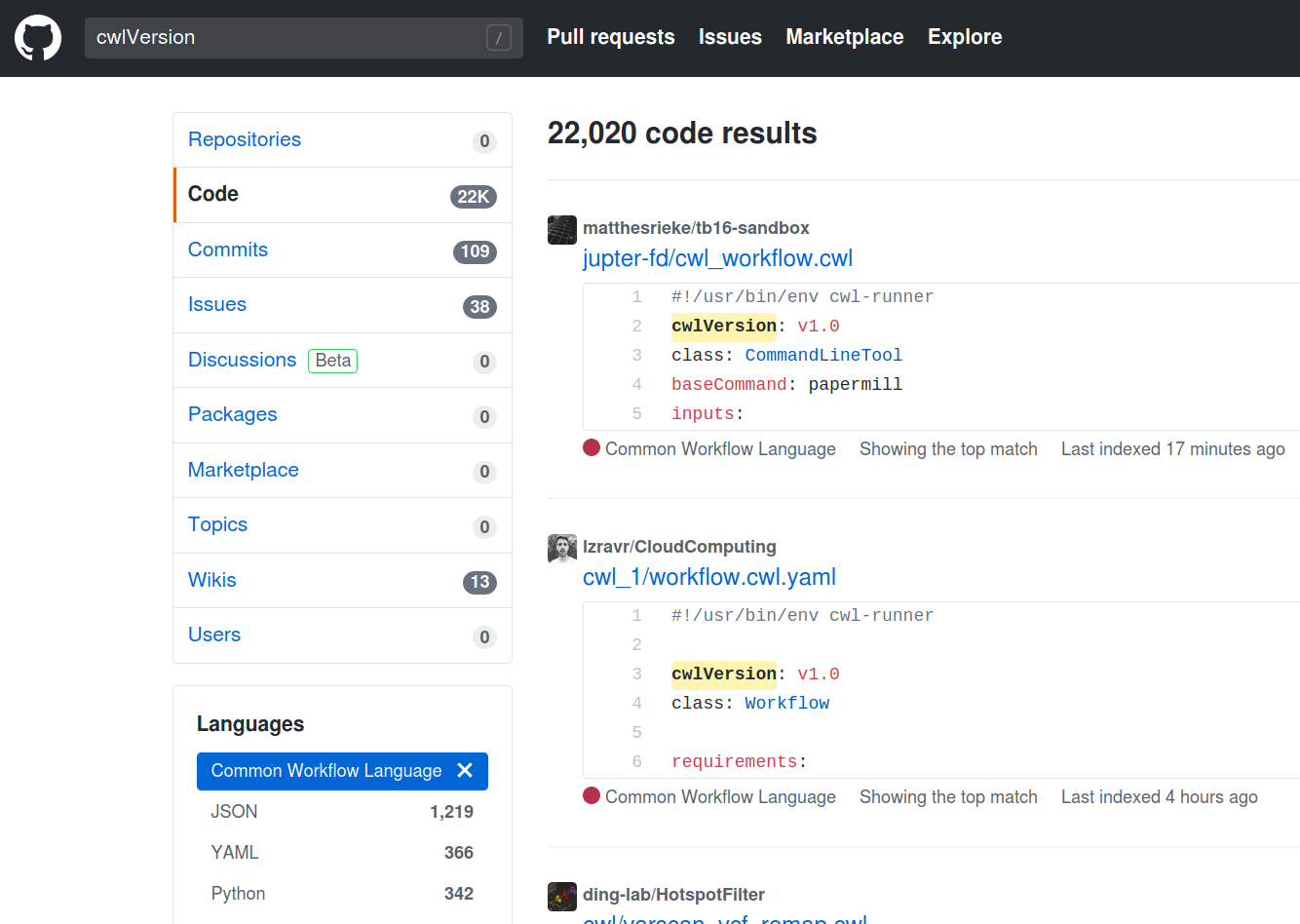
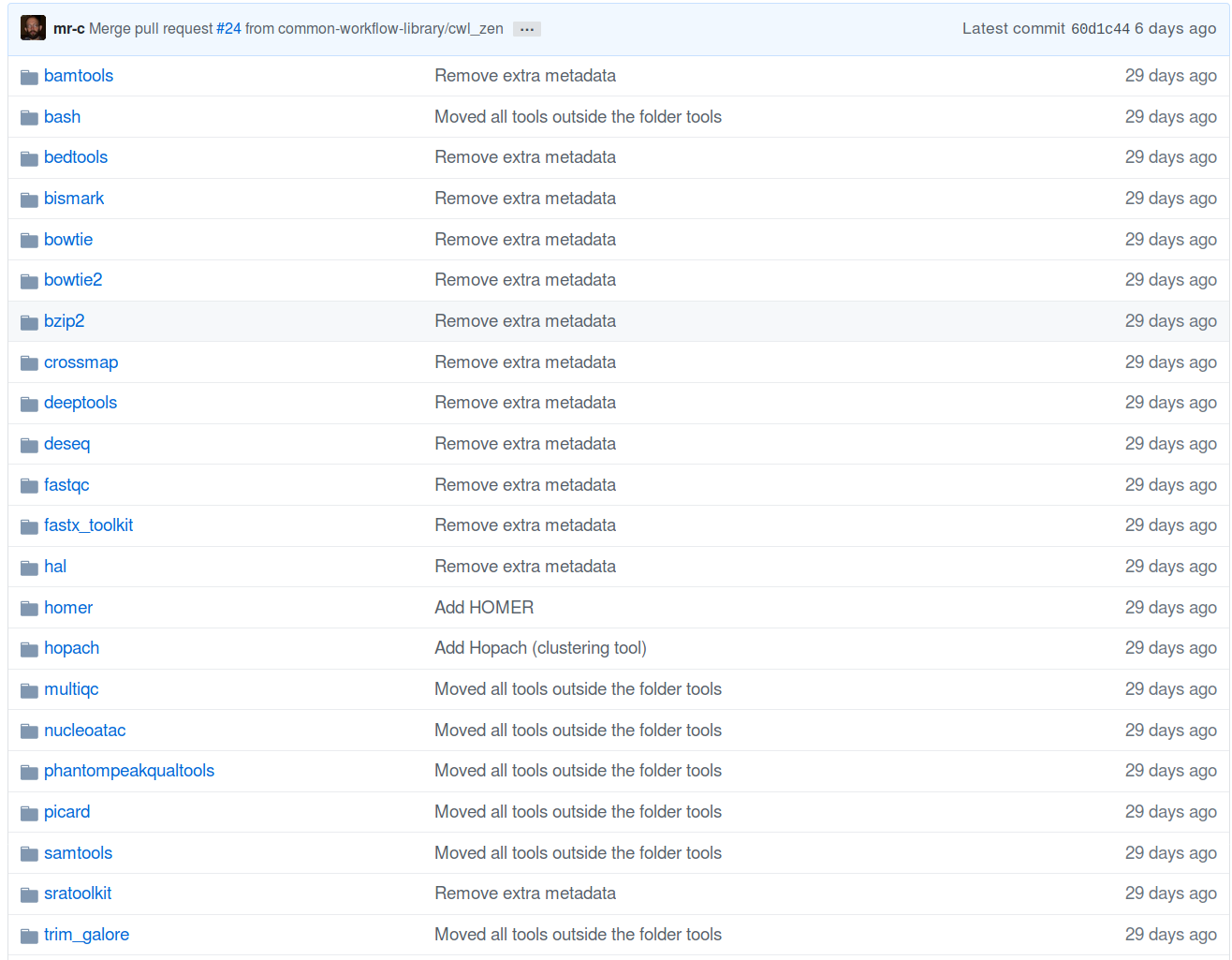
41 tools
89 operations
2020-04-28
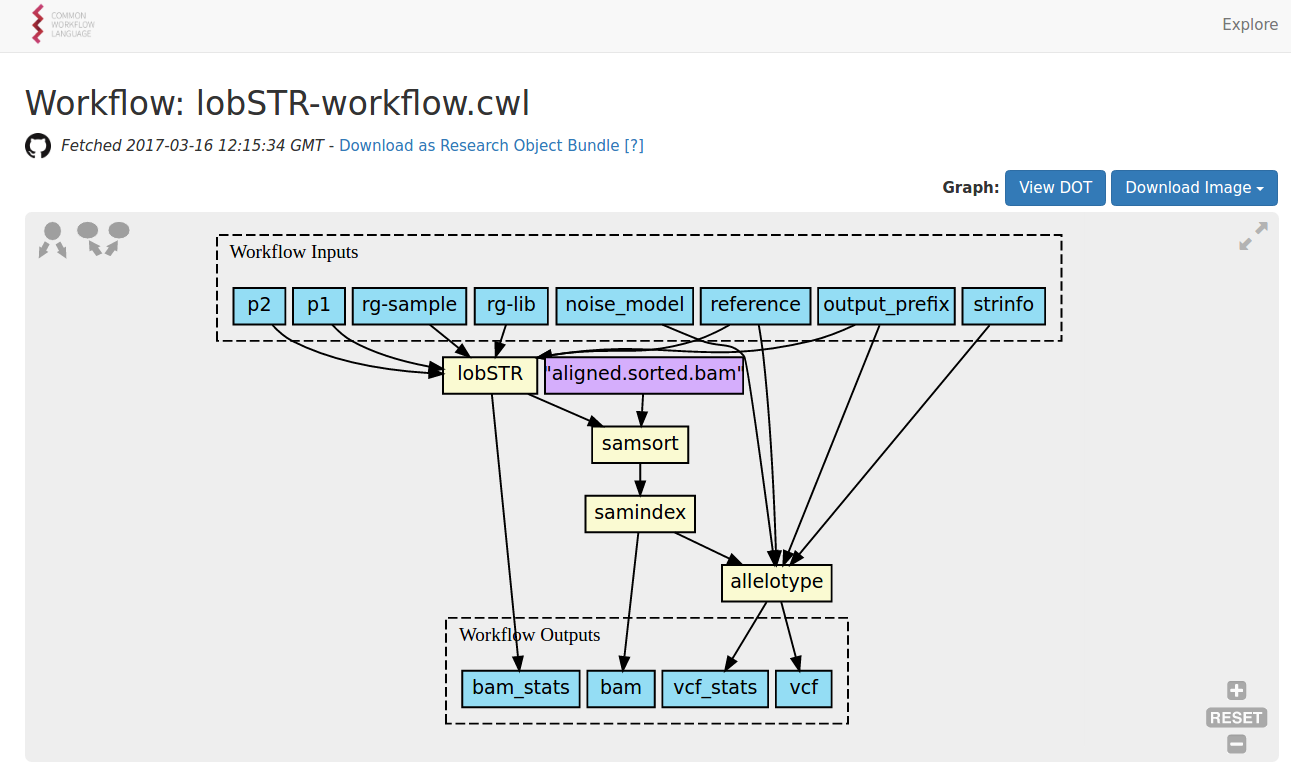
https://view.commonwl.org/workflows

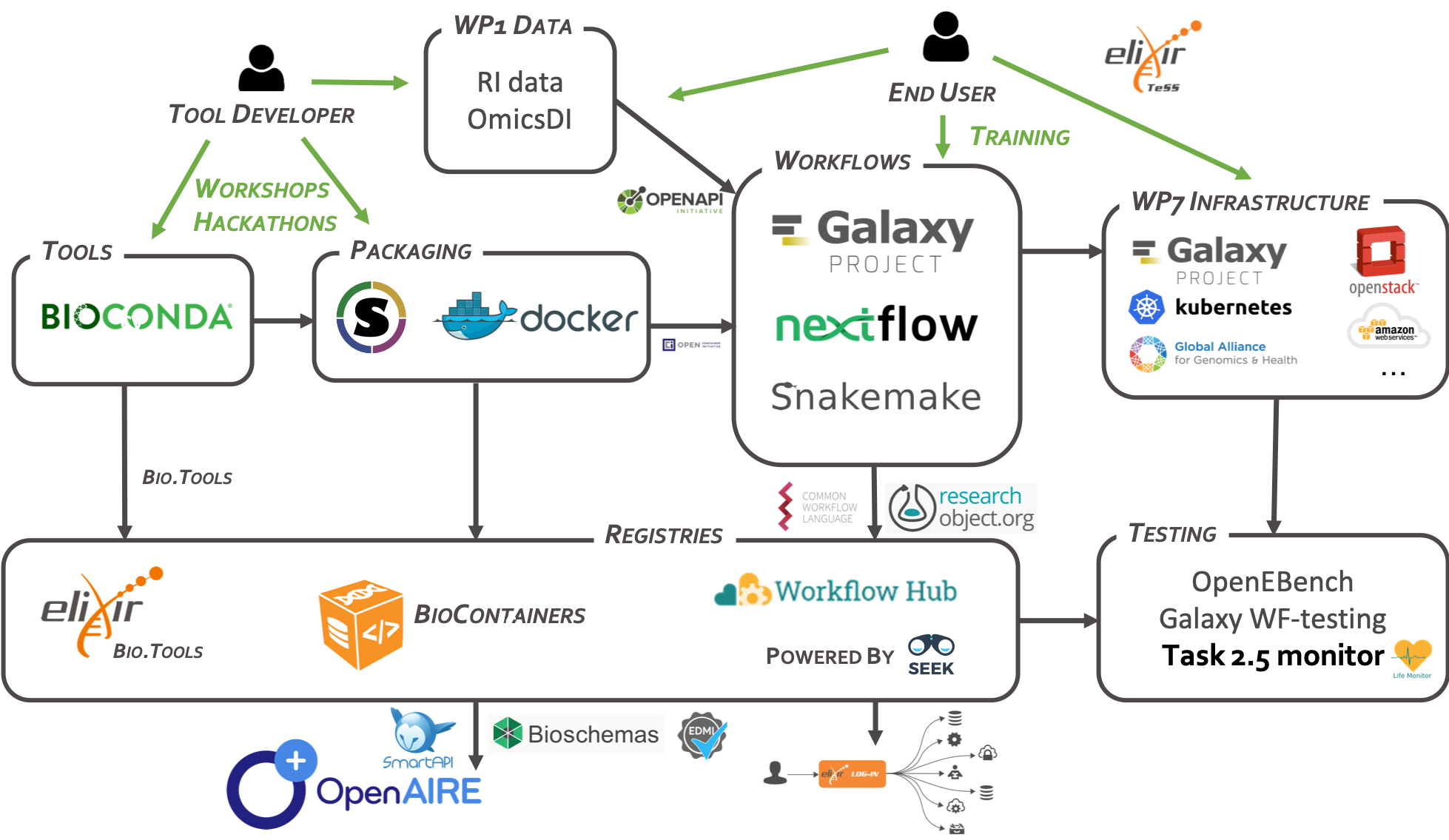




CWL Features
Interoperability
.. and portability



cwltool: local (Linux, OS X, Windows) — reference implementation
Arvados: AWS, GCP, Azure, Slurm, SaaS
Toil: local, AWS, Azure, GCP, SGE, LSF, Mesos, OpenStack, Slurm, PBS/Torque
CWL-Airflow: Linux, OS X
REANA: Kubernetes, CERN OpenStack
cromwell: local, HPC, Google, HtCondor
CWLEXEC: IBM Spectrum LSF
Which CWL engine runs where?
Galaxy: local (Linux), PBS/Torque, PBS Pro, LSF, Grid Engine, HTCondor, Slurm
Rabix Bunny: Local(Linux, OS X), GA4GH TES, SaaS
cwl-tes: Local, AliCloud, GCP, AWS, HTCondor, Grid Engine, PBS/Torque, Slurm
XENON: local, ssh, SLURM, Torque, Grid Engine
Consonance: AWS, OpenStack, Azure
Calrissian: Kubernetes
(..)
Portable command line tools
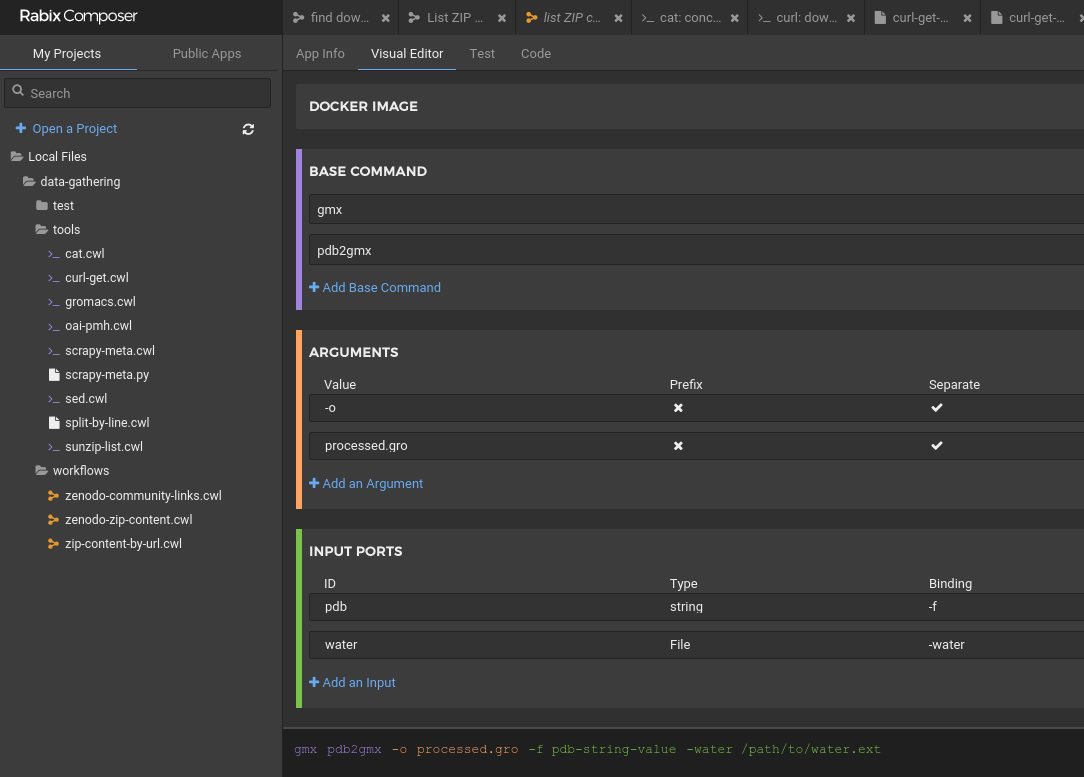

#!/usr/bin/env cwl-runner
cwlVersion: v1.0
class: CommandLineTool
baseCommand: ["gmx", "pdb2gmx"]
arguments: ["-o", "processed.gro"]
inputs:
pdb:
type: string
inputBinding:
prefix: -f
water:
type: File
inputBinding:
prefix: -water
default: spce
outputs:
processed_gro:
type: File
outputBinding:
glob: processed.groPortable command line tools


Where to find command line tools?

cwlVersion: v1.0
class: CommandLineTool
baseCommand: node
hints:
DockerRequirement:
dockerPull: gromacs/gromacs:2018.4
class: CommandLineTool
hints:
SoftwareRequirement:
packages:
samtools:
version: [ "0.1.19" ]
baseCommand: ["samtools", "index"]
#..Finding the tool
module load samtools/0.1.19apt-get install samtools=0.1.19*conda install samtools=0.1.19Let's add some identifiers!
hints:
SoftwareRequirement:
packages:
- package: gromacs
version:
- '2018.4'
specs:
- https://packages.debian.org/gromacs
- https://anaconda.org/bioconda/gromacs
- https://bio.tools/gromacs
- https://identifiers.org/rrid/RRID:SCR_014565
- https://hpc.example.edu/modules/gromacs/2018.4
DockerRequirement:
dockerPull: gromacs/gromacs:2018.4Going beyond CWL
#!/usr/bin/env cwl-runner
cwlVersion: v1.0
class: CommandLineTool
baseCommand: echo
requirements:
InlineJavascriptRequirement: {}
inputs: []
outputs:
example_out:
type: stdout
stdout: output.txt
arguments:
- prefix: -A
valueFrom: $(1+1)
- prefix: -B
valueFrom: $("/foo/bar/baz".split('/').slice(-1)[0])
- prefix: -C
valueFrom: |
${
var r = [];
for (var i = 10; i >= 1; i--) {
r.push(i);
}
return r;
}JavaScript expressions
#!/usr/bin/env cwl-runner
cwlVersion: v1.0
class: CommandLineTool
requirements:
DockerRequirement:
dockerPull: lukasheinrich/dummyanalysis
ShellCommandRequirement: {}
baseCommand: /bin/bash
arguments:
- valueFrom: |
source /usr/local/bin/thisroot.sh
cat $(runtime.outdir)/input_list | xargs hadd merged.root
prefix: -c
# (..)ShellCommandRequirement
#!/usr/bin/env cwl-runner
#
# Example score submission file
#
cwlVersion: v1.0
class: CommandLineTool
requirements:
- class: InlineJavascriptRequirement
- class: InitialWorkDirRequirement
listing:
- entryname: score.py
entry: |
#!/usr/bin/env python
import synapseclient
import argparse
import os
import json
parser = argparse.ArgumentParser()
parser.add_argument("-f", "--submissionfile", required=True, help="Submission File")
# ..
args = parser.parse_args()
score = 3
prediction_file_status = "SCORED"
result = {'score':score, 'prediction_file_status':prediction_file_status}
with open(args.results, 'w') as o:
o.write(json.dumps(result))
baseCommand: python
arguments:
- valueFrom: score.py
- valueFrom: $(inputs.inputfile.path)
prefix: -f
inputs:
- id: inputfile
type: File
outputs:
- id: results
type: File
outputBinding:
glob: results.jsonExample: Embedding Python script
CWL patterns
Some patterns commonly encountered when writing CWL workflows
Encouraging best practice
Not just files
Secondary files (e.g. *.bam.bai)
Inline files/strings
Directories
Arrays of files
Enums
JSON records (string, int, array, dict)
CWL implementations can decide how to handle data management, e.g. data store or file copying
Dataflow features
Optional inputs (tool reuse)
Sub-workflows (reuse workflow as a tool)
Task Parallelization (step execute when data is ready)
Scattering (multiple tasks from single array)
Merge (multiple inputs to single array)
Provenance
Who ran it?
When did it run?
Where did it run?
What workflow ran?
Which tool versions?
What data was created?
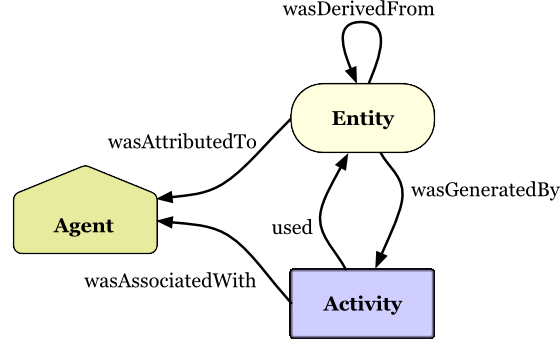
Copyright © 2013 W3C® (MIT, ERCIM, Keio, Beihang), All Rights Reserved. W3C liability, trademark and document use rules apply.
PROV Model Primer
W3C Working Group Note 30 April 2013

Khan et al,
Sharing interoperable workflow provenance: A review of best practices and their practical application in CWLProv
GigaScience (in print)
https://doi.org/10.5281/zenodo.3336124


(venv3) stain@biggie:~/src/cwlprov-py/test/nested-cwlprov-0.3.0$ cwlprov run
2018-08-08 22:44:06.573330 Flow 39408a40-c1c8-4852-9747-87249425be1e [ Run of workflow/packed.cwl#main
2018-08-08 22:44:06.691722 Step 4f082fb6-3e4d-4a21-82e3-c685ce3deb58 Run of workflow/packed.cwl#main/create-tar (0:00:00.010133)
2018-08-08 22:44:06.702976 Step 0cceeaf6-4109-4f08-940b-f06ac959944a * Run of workflow/packed.cwl#main/compile (unknown duration)
2018-08-08 22:44:12.680097 Flow 39408a40-c1c8-4852-9747-87249425be1e ] Run of workflow/packed.cwl#main (0:00:06.106767)
Legend:
[ Workflow start
* Nested provenance, use UUID to explore: cwlprov run 0cceeaf6-4109-4f08-940b-f06ac959944a
] Workflow end
(venv3) stain@biggie:~/src/cwlprov-py/test/nested-cwlprov-0.3.0$ cwlprov run 0cceeaf6-4109-4f08-940b-f06ac959944a
2018-08-08 22:44:06.607210 Flow 0cceeaf6-4109-4f08-940b-f06ac959944a [ Run of workflow/packed.cwl#main
2018-08-08 22:44:06.707070 Step 83752ab4-8227-4d4a-8baa-78376df34aed Run of workflow/packed.cwl#main/untar (0:00:00.008149)
2018-08-08 22:44:06.718554 Step f56d8478-a190-4251-84d9-7f69fe0f6f8b Run of workflow/packed.cwl#main/argument (0:00:00.532052)
2018-08-08 22:44:07.251588 Flow 0cceeaf6-4109-4f08-940b-f06ac959944a ] Run of workflow/packed.cwl#main (0:00:00.644378)
Legend:
[ Workflow start
] Workflow endstain@biggie:~/src/cwlprov-py/test/nested-cwlprov-0.3.0$ cwlprov outputs 4f082fb6-3e4d-4a21-82e3-c685ce3deb58 --format=files
Output tar:
data/c0/c0fd5812fe6d8d91fef7f4f1ba3a462500fce0c5
stain@biggie:~/src/cwlprov-py/test/nested-cwlprov-0.3.0$ tar tfv `cwlprov -q outputs 4f082fb6-3e4d-4a21-82e3-c685ce3deb58 --format=files`
-rw-r--r-- stain/stain 115 2018-08-08 23:44 Hello.java
Inspecting step runs
$ cwlprov --help
usage: cwlprov [-h] [--version] [--directory DIRECTORY] [--relative]
[--absolute] [--output OUTPUT] [--verbose] [--quiet] [--hints]
[--no-hints]
{validate,info,who,prov,inputs,outputs,run,runs,rerun,derived,runtimes}
...
cwlprov explores Research Objects containing provenance of Common Workflow
Language executions. <https://w3id.org/cwl/prov/>
commands:
{validate,info,who,prov,inputs,outputs,run,runs,rerun,derived,runtimes}
validate Validate the CWLProv Research Object
info show research object Metadata
who show Who ran the workflow
prov export workflow execution Provenance in PROV format
inputs list workflow/step Input files/values
outputs list workflow/step Output files/values
run show workflow Execution log
runs List all workflow executions in RO
rerun Rerun a workflow or step
derived list what was Derived from a data item, based on
activity usage/generation
runtimes calculate average step execution Runtimes
Work in progress
CWL 1.2 draft
Conditional branching (~switch statement)
towards CWL 2.x
Looping
usability
CWL integrations: Galaxy, KNIME
Ansible scripts for installation (ansible-cwl)
Text editor language support (benten)
flexibility
alternative script-like grammar (cwl-ex)
findability
Federated workflow repository (workflowhub.eu)
Tool collection (common-workflow-library)
training
Introductory user guide
(cwl-novice-tutorial)
Ask the CWL community
Join the CWL community
Talk to the CWL community
Weekly CWL Community telcons
Announced at
https://groups.google.com/forum/#!forum/common-workflow-language
"Meet" us at BOSC
Next in Virtual Training session:
https://slides.com/soilandreyes/2020-04-29-cwl-tutorial/
..or if learning at your own pace:
Break
Tip: Start your VMs early to avoid wait