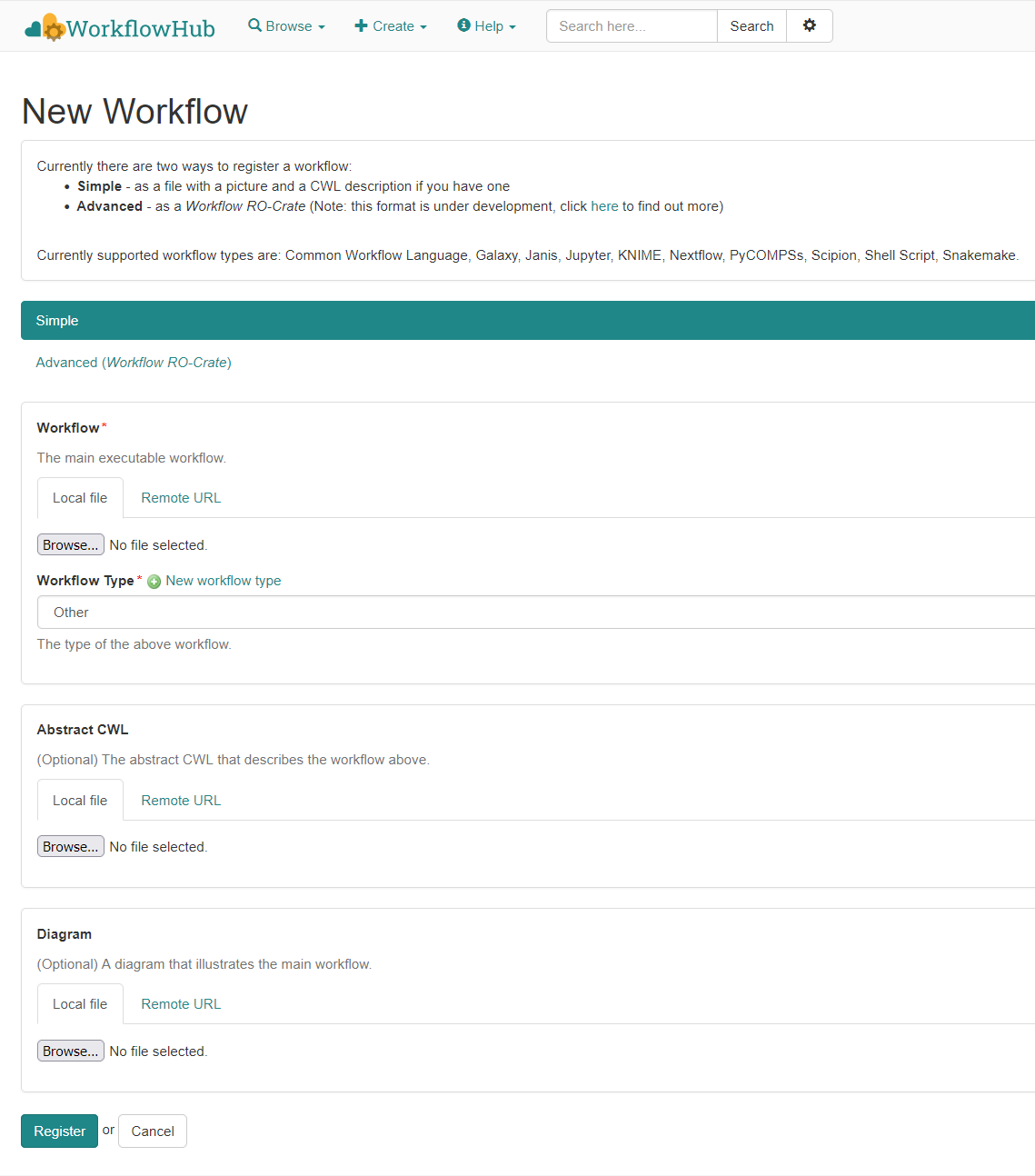
RO-Crate
A quick introduction
Stian Soiland-Reyes
eScience lab, The University of Manchester
INDElab, University of Amsterdam
FAIR-IMPACT Support Offer #2
Workshop #1
2023-09-25
This work is licensed under a
Creative Commons Attribution 4.0 International License.







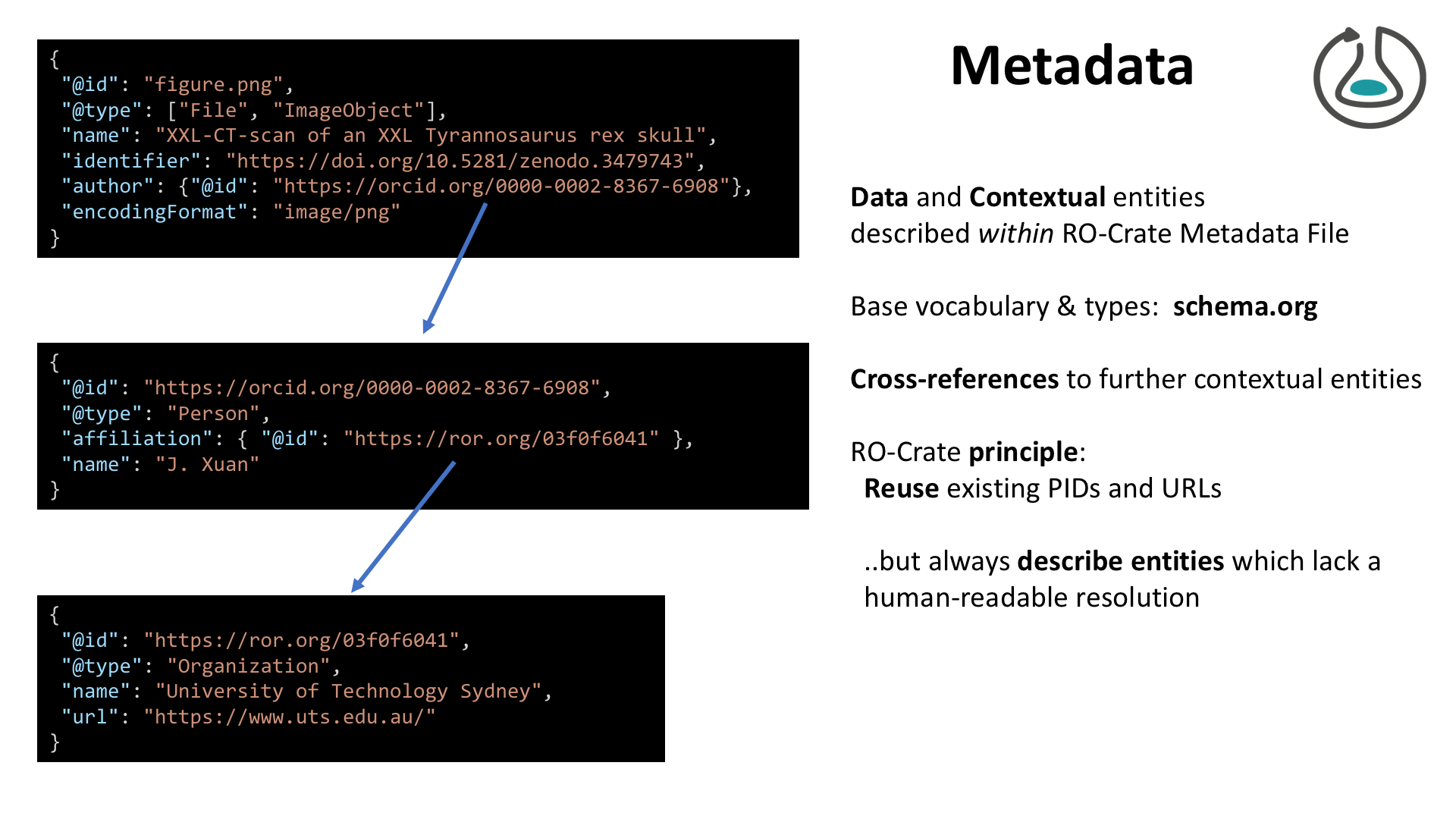
{
"@context": "https://w3id.org/ro/crate/1.1/context",
"@graph": [
{
"@id": "ro-crate-metadata.json",
"@type": "CreativeWork",
"about": {
"@id": "./"
},
"conformsTo": [
{
"@id": "https://w3id.org/ro/crate/1.1"
},
{
"@id": "https://about.workflowhub.eu/Workflow-RO-Crate/"
}
]
},
{
"@id": "ro-crate-preview.html",
"@type": "CreativeWork",
"about": {
"@id": "./"
}
},
{
"@id": "./",
"@type": "Dataset",
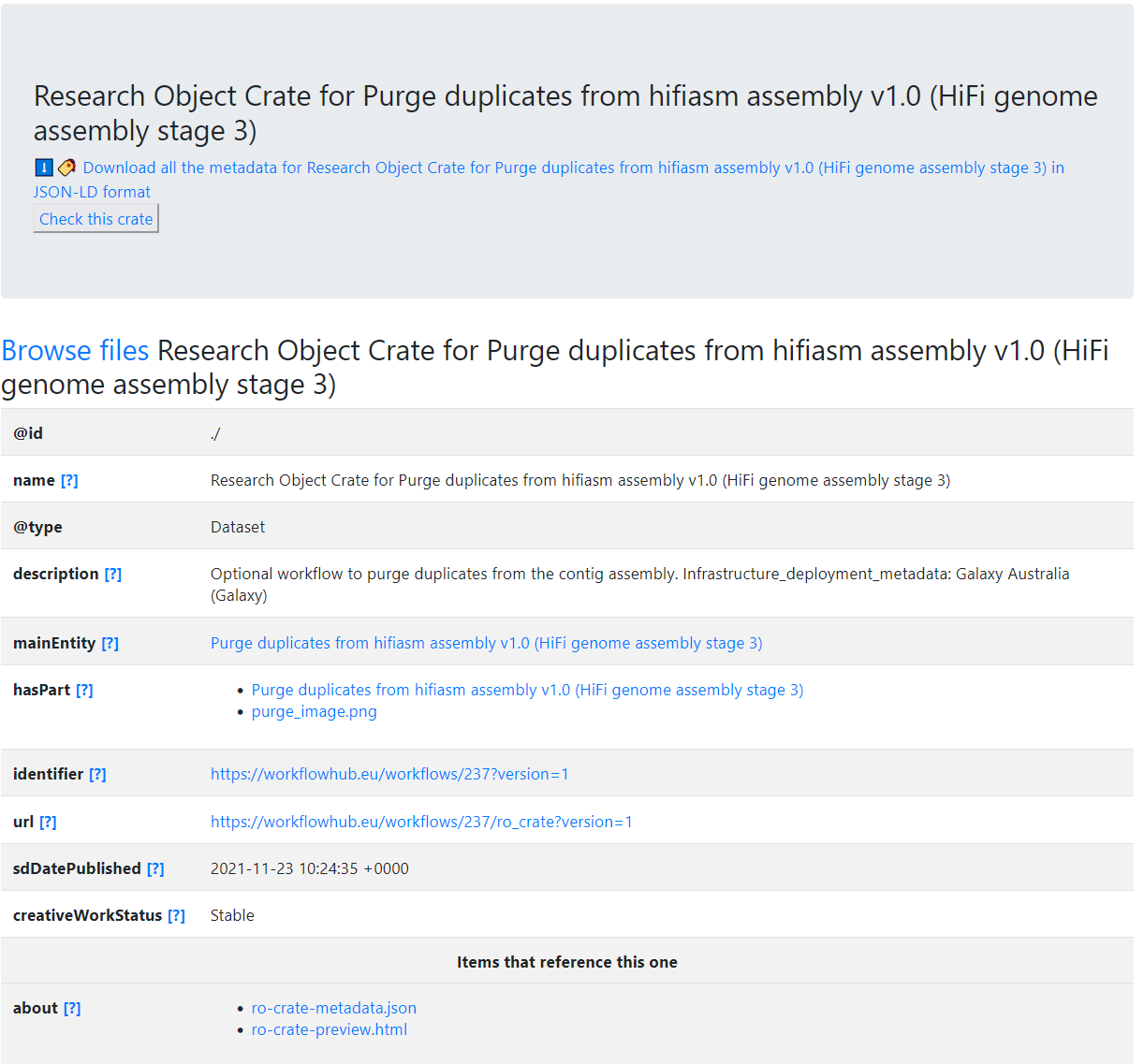
"mainEntity": {
"@id": "Galaxy-Workflow-Purge_duplicates_from_hifiasm_assembly_v1.0.ga"
},
"hasPart": [
{
"@id": "Galaxy-Workflow-Purge_duplicates_from_hifiasm_assembly_v1.0.ga"
},
{
"@id": "purge_image.png"
}
],
"identifier": "https://workflowhub.eu/workflows/237?version=1",
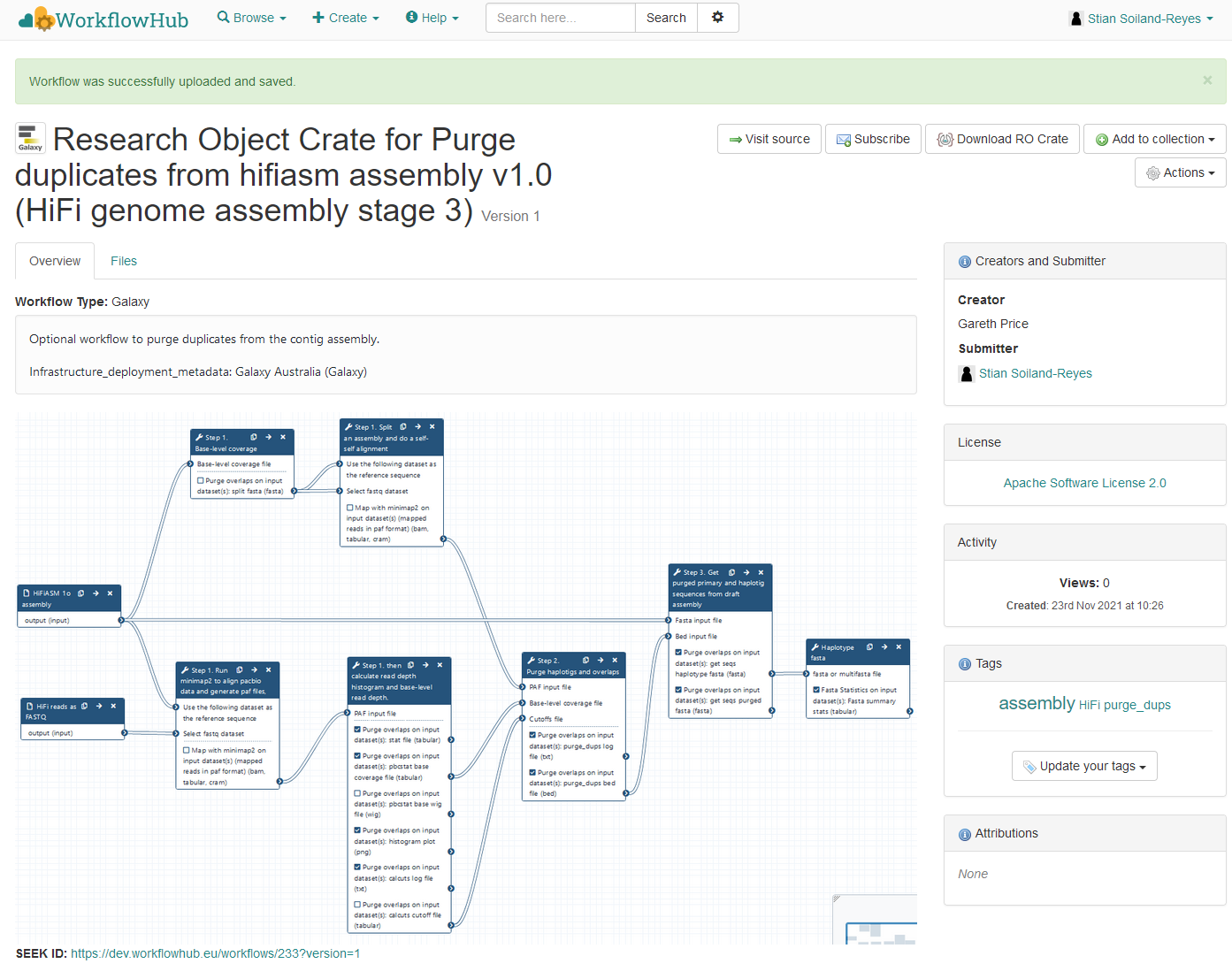
"url": "https://workflowhub.eu/workflows/237/ro_crate?version=1",
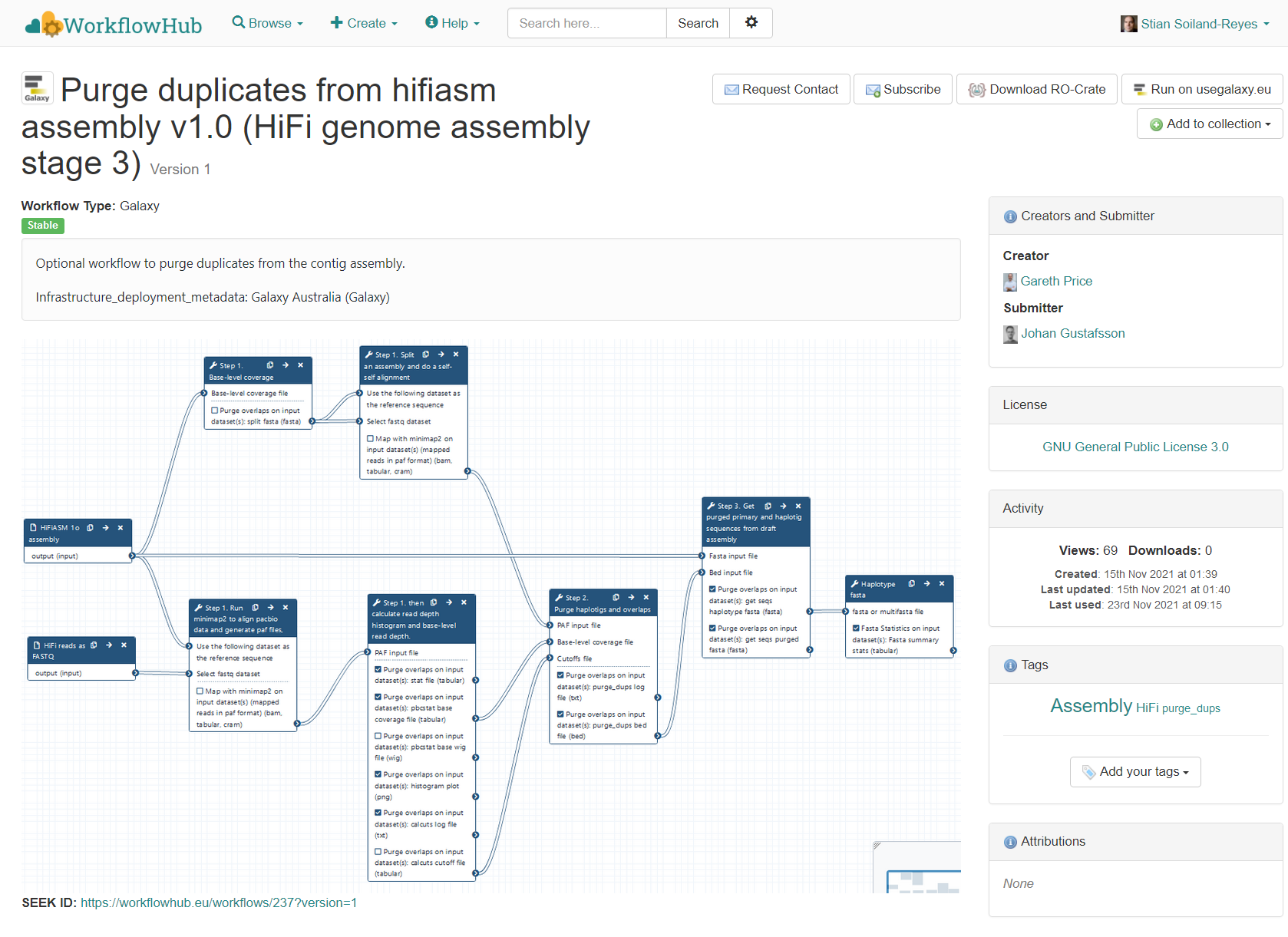
"name": "Research Object Crate for Purge duplicates from hifiasm assembly v1.0 (HiFi genome assembly stage 3)",
"description": "Optional workflow to purge duplicates from the contig assembly.\r\n\r\nInfrastructure_deployment_metadata: Galaxy Australia (Galaxy)",
"sdDatePublished": "2021-11-23 10:44:32 +0000",
"creativeWorkStatus": "Stable"
},
{
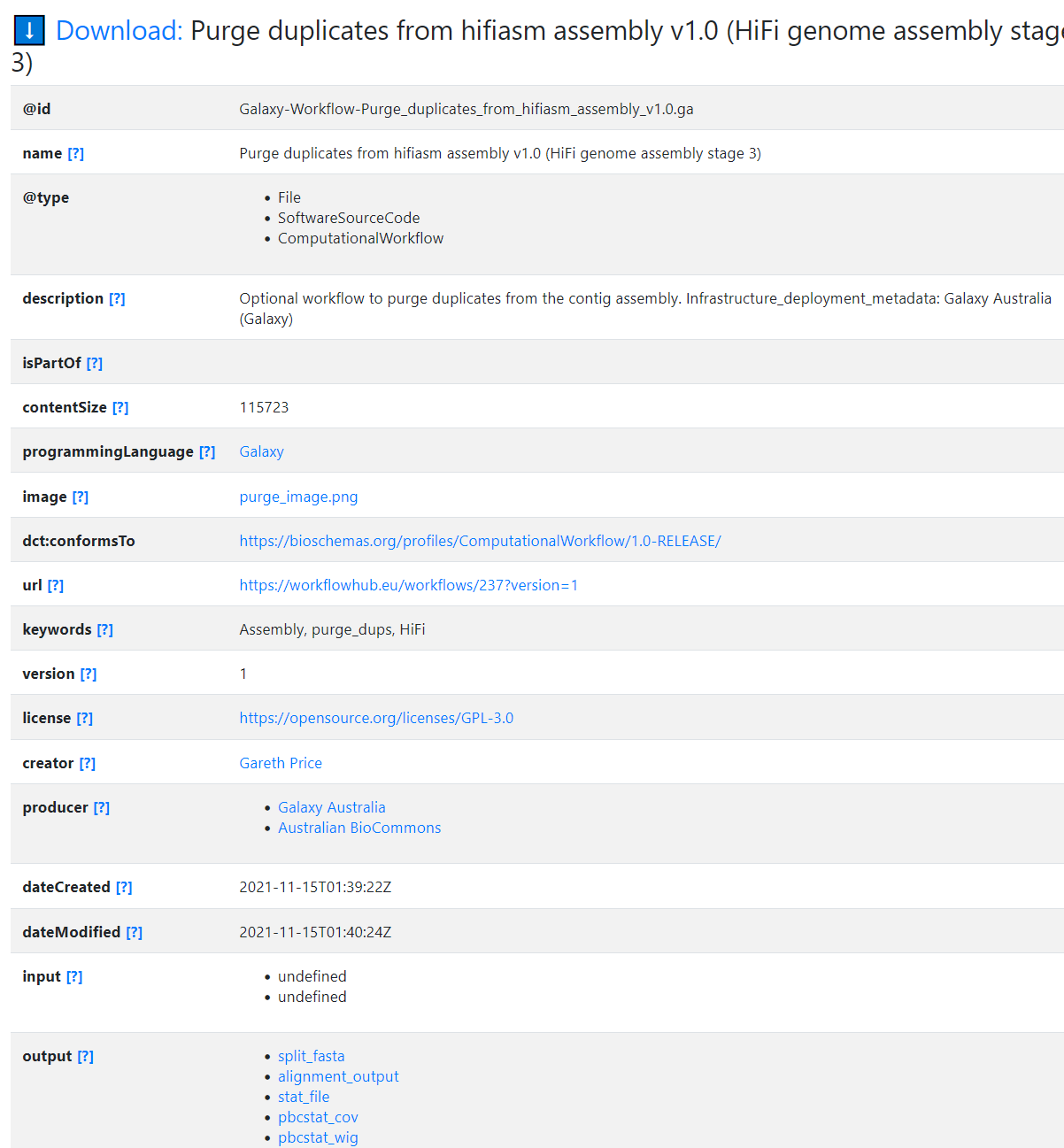
"@id": "Galaxy-Workflow-Purge_duplicates_from_hifiasm_assembly_v1.0.ga",
"@type": [
"File",
"SoftwareSourceCode",
"ComputationalWorkflow"
],
"programmingLanguage": {
"@id": "#galaxy"
},
"image": {
"@id": "purge_image.png"
},
"contentSize": 115723,
"dct:conformsTo": "https://bioschemas.org/profiles/ComputationalWorkflow/1.0-RELEASE/",
"description": "Optional workflow to purge duplicates from the contig assembly.\r\n\r\nInfrastructure_deployment_metadata: Galaxy Australia (Galaxy)",
"name": "Purge duplicates from hifiasm assembly v1.0 (HiFi genome assembly stage 3)",
"url": "https://workflowhub.eu/workflows/237?version=1",
"keywords": "Assembly, purge_dups, HiFi",
"version": 1,
"license": "https://opensource.org/licenses/GPL-3.0",
"creator": {
"@id": "https://workflowhub.eu/people/139"
},
"producer": [
{
"@id": "https://workflowhub.eu/projects/54"
},
{
"@id": "https://workflowhub.eu/projects/30"
}
],
"dateCreated": "2021-11-15T01:39:22Z",
"dateModified": "2021-11-15T01:40:24Z",
"isPartOf": [
],
"input": [
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFiASM 1o assembly"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFi reads as FASTQ"
}
],
"output": [
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-split_fasta"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-alignment_output"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stat_file"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_cov"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_wig"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-hist"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_log"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_tab"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_log"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_bed"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_hap"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_purged"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stats"
}
],
"sdPublisher": {
"@id": "http://about.workflowhub.eu"
}
},
{
"@id": "purge_image.png",
"@type": [
"File",
"ImageObject",
"WorkflowSketch"
],
"contentSize": 118673
},
{
"@id": "https://about.workflowhub.eu/Workflow-RO-Crate/",
"@type": "CreativeWork",
"name": "Workflow RO-Crate Profile",
"version": "0.2.0"
},
{
"@id": "https://workflowhub.eu/people/139",
"@type": "Person",
"name": "Gareth Price"
},
{
"@id": "https://workflowhub.eu/projects/54",
"@type": [
"Project",
"Organization"
],
"name": "Galaxy Australia"
},
{
"@id": "https://workflowhub.eu/projects/30",
"@type": [
"Project",
"Organization"
],
"name": "Australian BioCommons"
},
{
"@id": "#galaxy",
"@type": "ComputerLanguage",
"name": "Galaxy",
"identifier": {
"@id": "https://galaxyproject.org/"
},
"url": {
"@id": "https://galaxyproject.org/"
}
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFiASM%201o%20assembly",
"@type": "FormalParameter",
"name": "HiFiASM 1o assembly",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFi%20reads%20as%20FASTQ",
"@type": "FormalParameter",
"name": "HiFi reads as FASTQ",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-split_fasta",
"@type": "FormalParameter",
"name": "split_fasta",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-alignment_output",
"@type": "FormalParameter",
"name": "alignment_output",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stat_file",
"@type": "FormalParameter",
"name": "stat_file",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_cov",
"@type": "FormalParameter",
"name": "pbcstat_cov",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_wig",
"@type": "FormalParameter",
"name": "pbcstat_wig",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-hist",
"@type": "FormalParameter",
"name": "hist",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_log",
"@type": "FormalParameter",
"name": "calcuts_log",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_tab",
"@type": "FormalParameter",
"name": "calcuts_tab",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_log",
"@type": "FormalParameter",
"name": "purge_dups_log",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_bed",
"@type": "FormalParameter",
"name": "purge_dups_bed",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_hap",
"@type": "FormalParameter",
"name": "get_seqs_hap",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_purged",
"@type": "FormalParameter",
"name": "get_seqs_purged",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stats",
"@type": "FormalParameter",
"name": "stats",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "http://about.workflowhub.eu",
"@type": "Organization",
"name": "WorkflowHub",
"url": "http://about.workflowhub.eu"
}
]
}
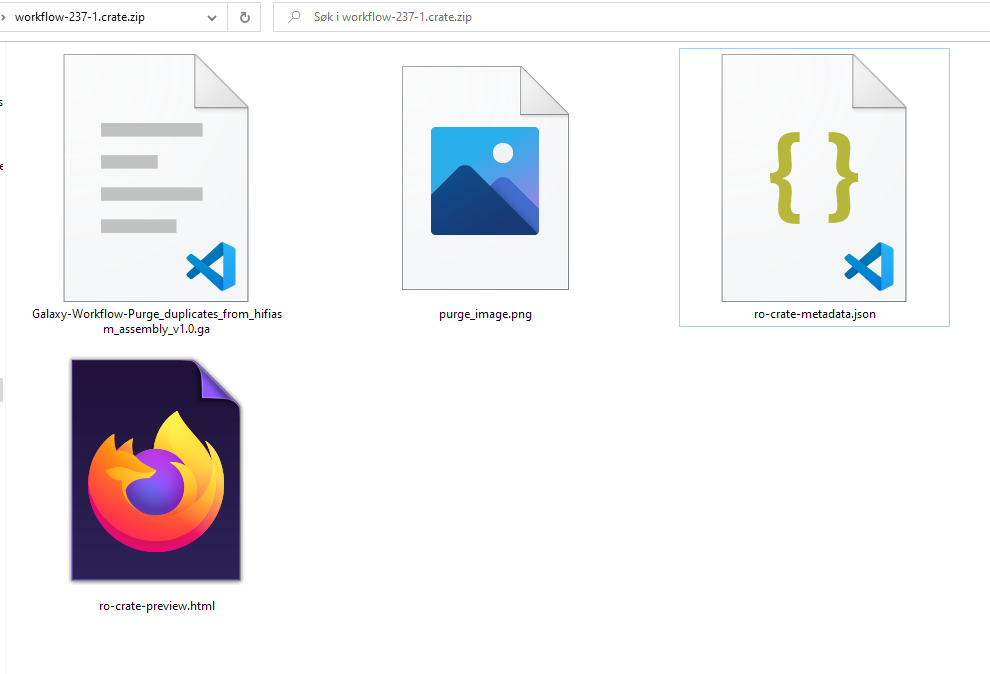
ro-crate-metadata.json


Text
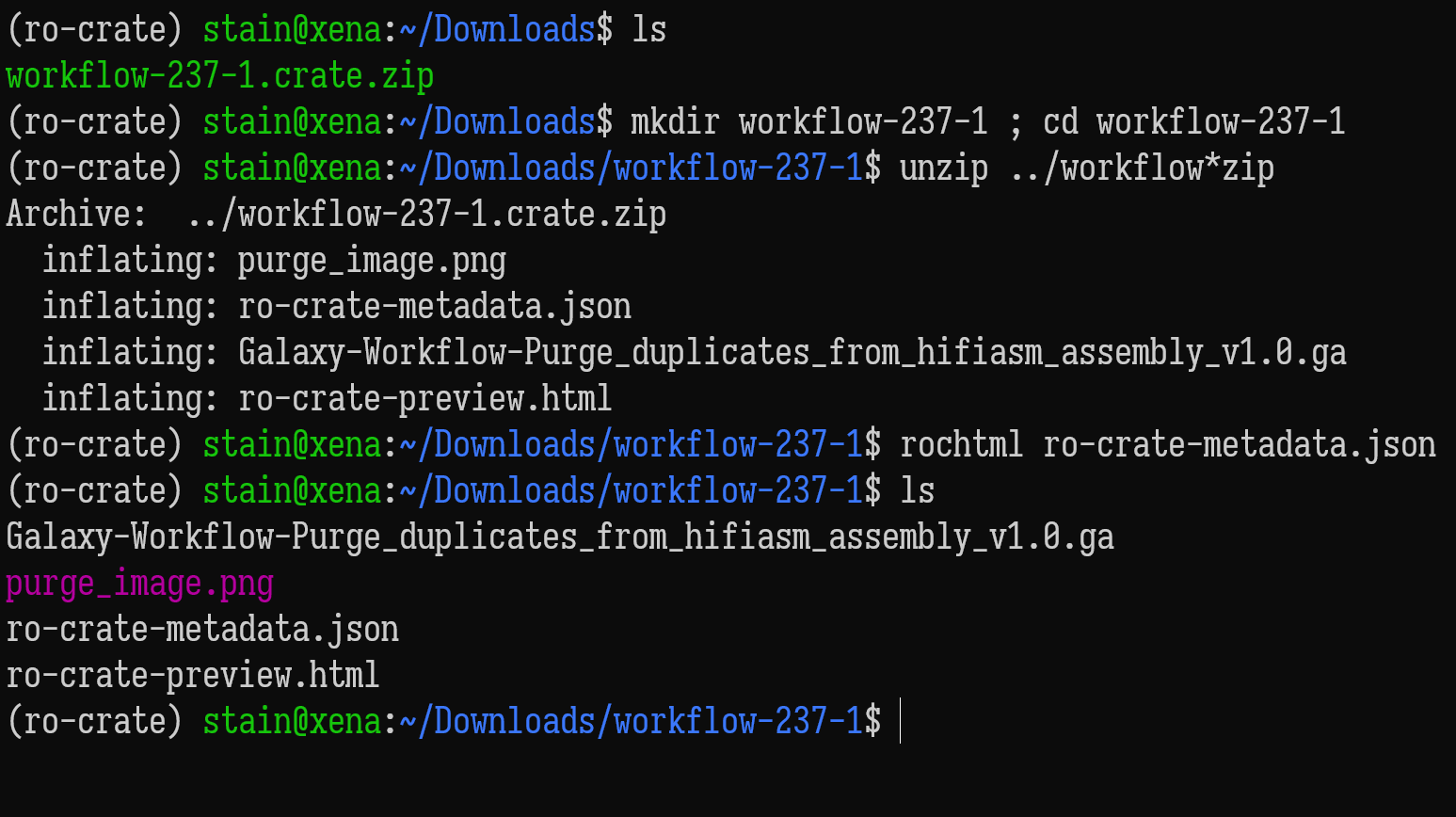
Generate a HTML preview from JSON-LD






Techie deep-dive!
Warning: JSON ahead














RO-Crate tutorials

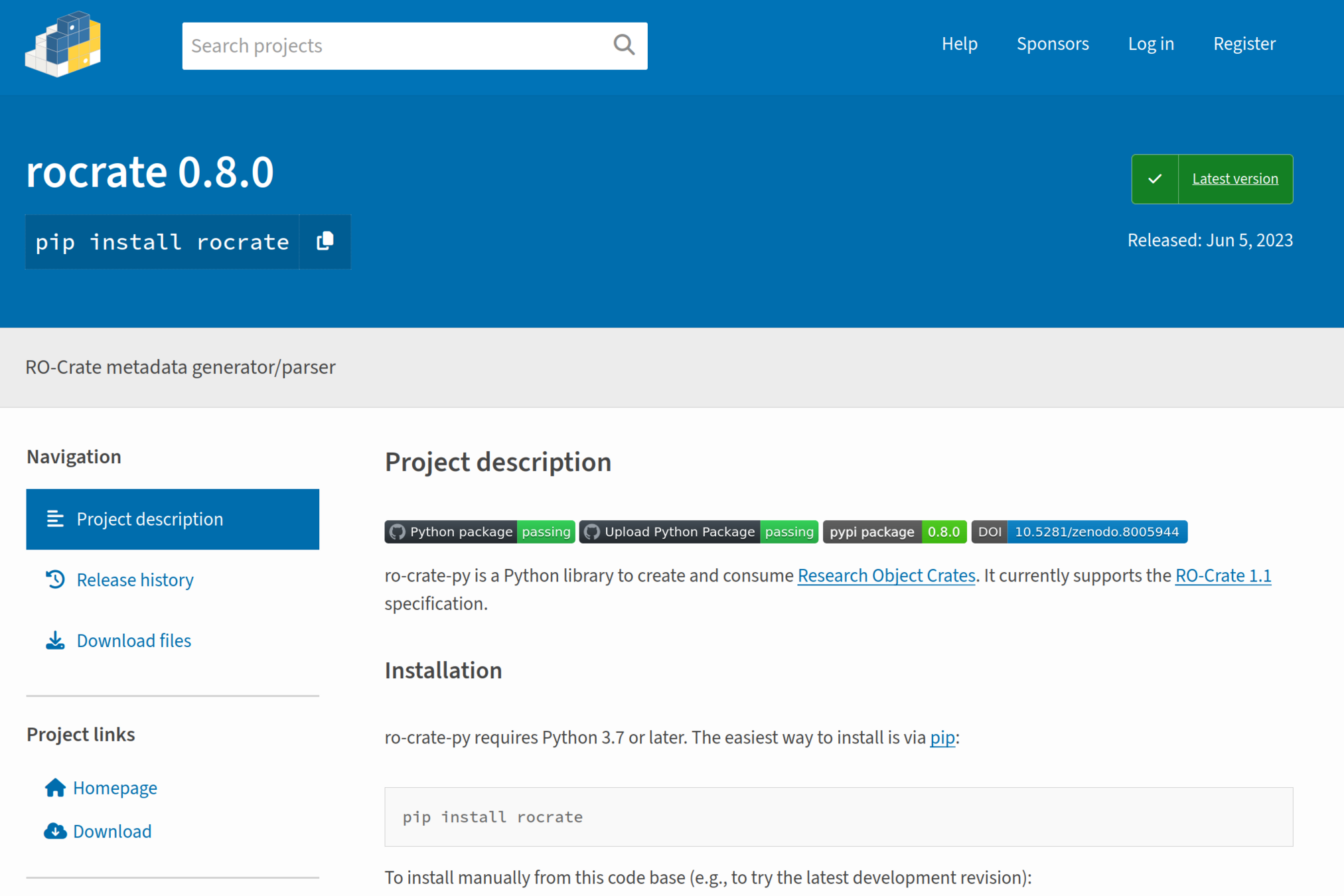
Making your own RO-Crate with Crate-O
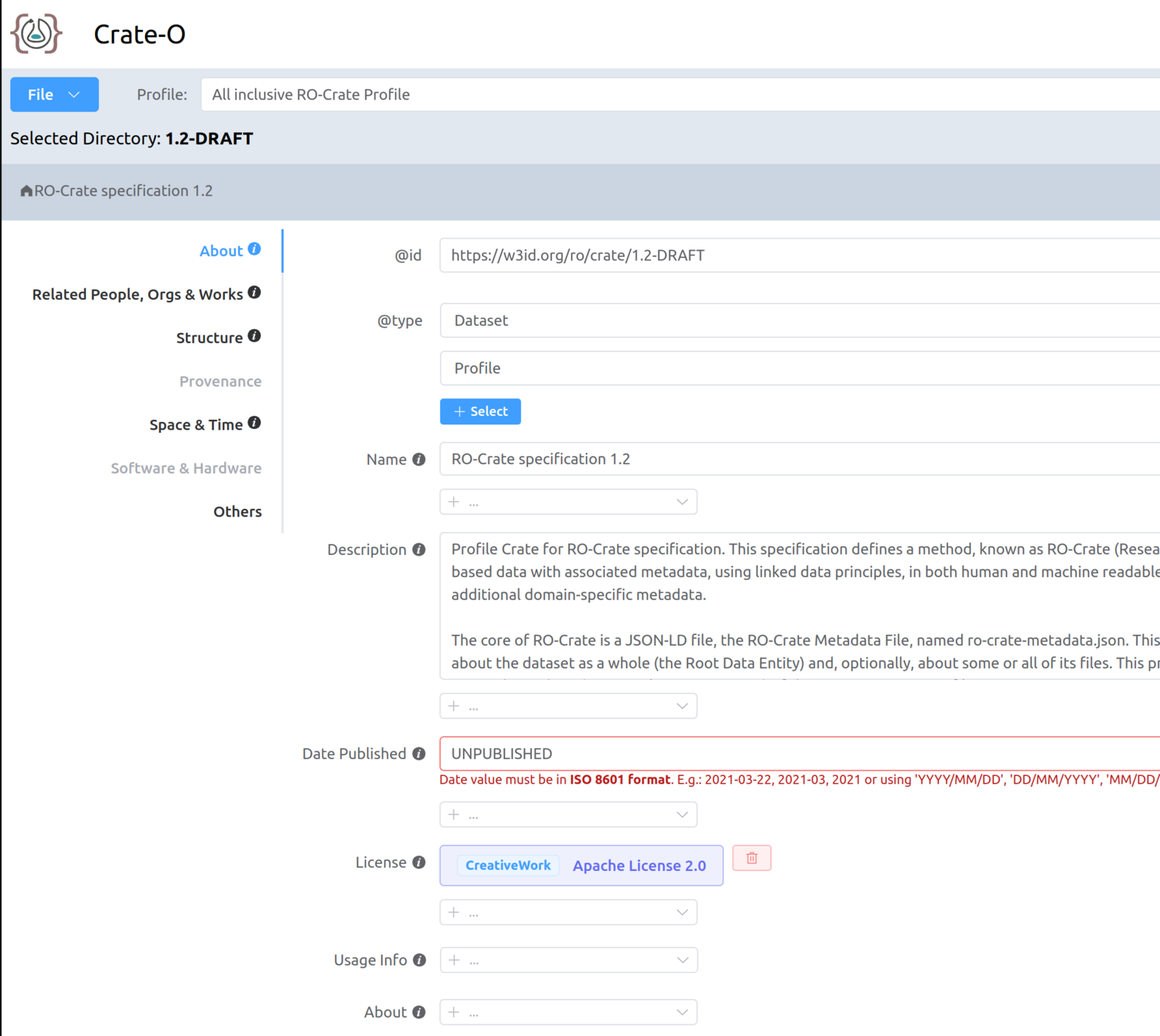
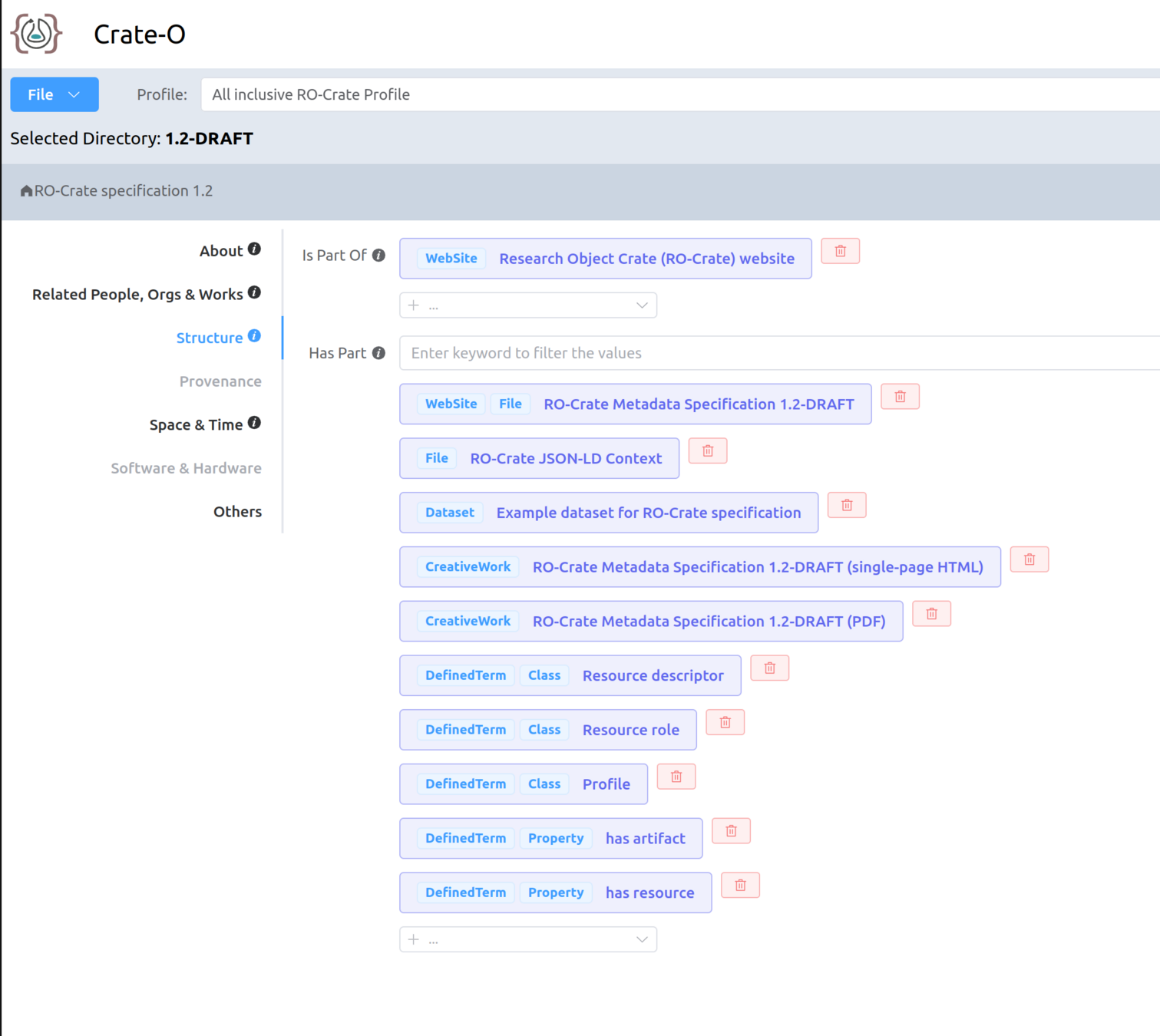
How should we resolve an
RO-Crates from a DOI?


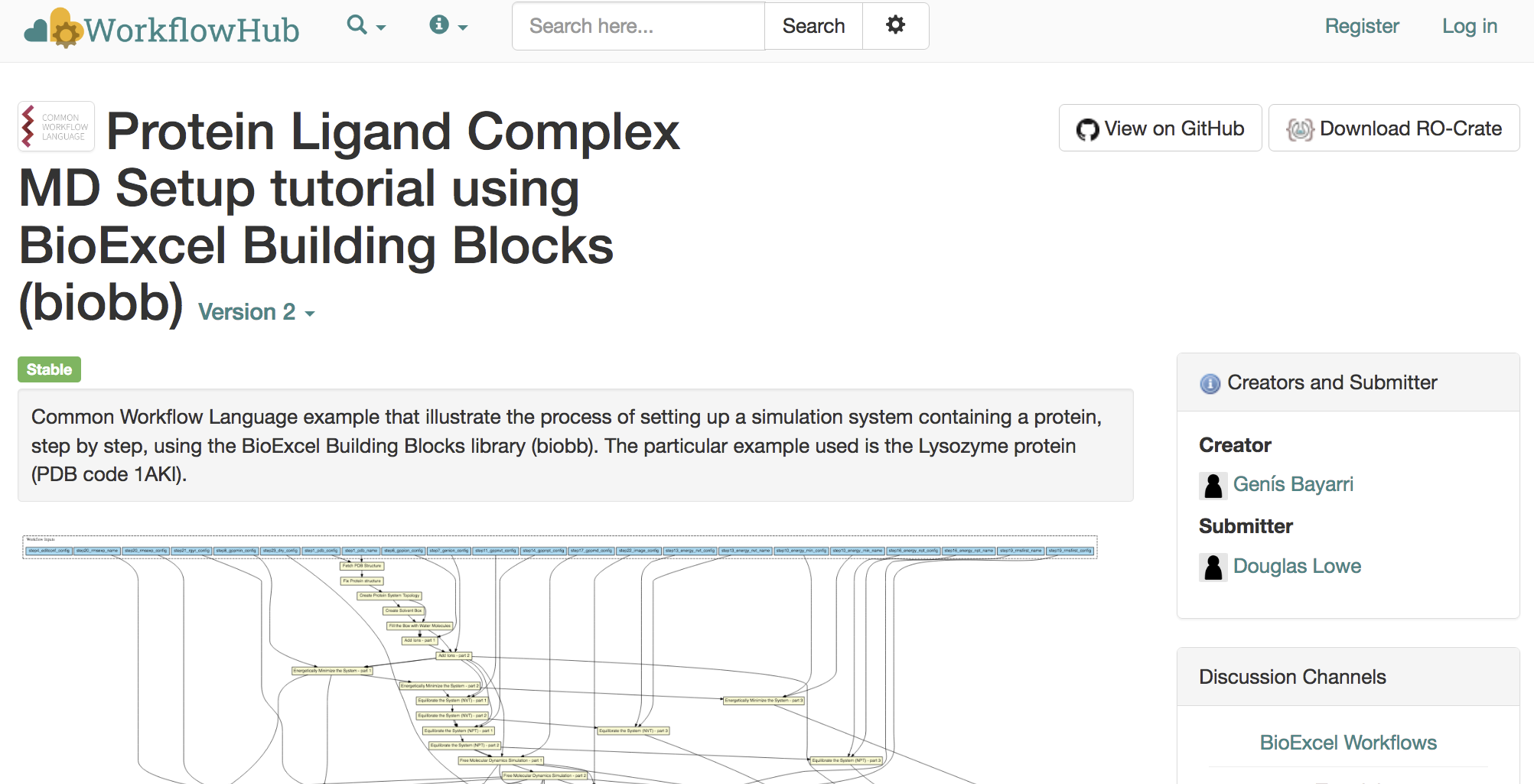

302 Found Location: https://workflowhub.eu/workflows/29?version=2
Accept: text/html
Resolving an RO-Crate with content-negotiation


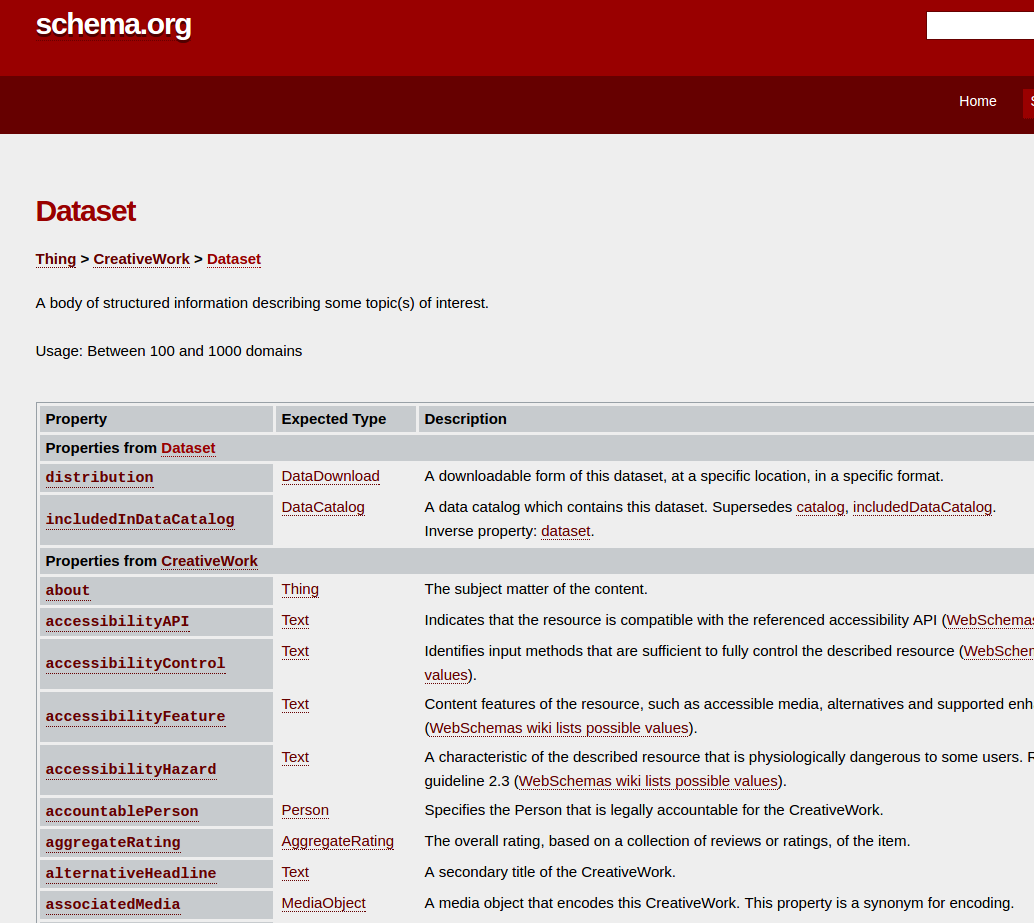
ComputationalWorkflow




302 Found Location: https://workflowhub.eu/workflows/29?version=2
Accept: text/html

Accept: application/zip



Resolving an RO-Crate with content-negotiation






302 Found Location: https://workflowhub.eu/workflows/29?version=2
Accept: text/html
Accept: application/ld+json; profile=https://w3id.org/ro/crate



Accept: application/zip





Resolving an RO-Crate with content-negotiation




302 Found Location: https://workflowhub.eu/workflows/29?version=2
Accept: application/ld+json; profile=https://w3id.org/ro/crate




Downside: Indirection to find core metadata and content

author
@type

ComputationalWorkflow

license
hasPart
isBasedOn
Parse JSON, find the right node


HEAD https://doi.org/10.48546/workflowhub.workflow.29.2 302 Found Location: https://workflowhub.eu/workflows/29?version=2
HEAD https://workflowhub.eu/workflows/29?version=2 200 OK Link: <https://doi.org/10.48546/workflowhub.workflow.29.2>;rel=cite-as Link: <https://workflowhub.eu/workflows/29/ro_crate?version=2>;rel=item type="application/zip" ; profile="https://w3id.org/ro/crate" Link: <https://orcid.org/0000-0003-0513-0288>;rel=author Link: <https://workflowhub.eu/workflows/29?version=2> ; rel="describedby" ; type="application/ld+json" Link: <https://workflowhub.eu/workflows/29?version=2> ; rel="describedby" ; type="application/vnd.datacite.datacite+xml" …



Resolving an RO-Crate with FAIR Signposting
rel=item
rel=describedby;
type="application/ld+json"
rel=cite-as

rel=item;
type="application/zip"

Attached vs Detached RO-Crate
Attached RO-Crate are used when there is some kind of folder-structure, typically archived as a ZIP file,
or on a traditional website.
Absolute URLs may still be listed as parts, but typically the parts are "files" and "directories" using relative paths.
{ "@context": "https://w3id.org/ro/crate/1.2-DRAFT/context",
"@graph": [
{
"@type": "CreativeWork",
"@id": "ro-crate-metadata.json",
"conformsTo": {"@id": "https://w3id.org/ro/crate/1.2-DRAFT"},
"about": {"@id": "./"}
},
{
"@id": "./",
"@type": [
"Dataset"
],
"hasPart": [
{
"@id": "cp7glop.ai"
},
{
"@id": "lots_of_little_files/"
}
]
},
{
"@id": "cp7glop.ai",
"@type": "File",
"name": "Diagram showing trend to increase",
"contentSize": "383766",
"description": "Illustrator file for Glop Pot",
"encodingFormat": "application/pdf"
},
{
"@id": "lots_of_little_files/",
"@type": "Dataset",
"name": "Too many files",
"description": "This directory contains many small files, that we're not going to describe in detail."
}
]
}Detached RO-Crate has a standalone JSON-LD metadata file, e.g. returned by an API.
All the resources have absolute URIs
This style may also be suitable for a repository exposing the metadata from an RO-Crate ZIP file without exposing its individual files.
{ "@context": "https://w3id.org/ro/crate/1.2-DRAFT/context",
"@graph": [
{
"@type": "CreativeWork",
"@id": "ro-crate-metadata.json",
"conformsTo": {"@id": "https://w3id.org/ro/crate/1.2-DRAFT"},
"about": {"@id": "https://example.com/item/15?crate"}
},
{
"@id": "https://example.com/item/15?crate",
"@type": [
"Dataset"
],
"hasPart": [
{
"@id": "https://example.com/data/12312/cp7glop.ai"
}
]
},
{
"@id": "http://example.com/data/12312/cp7glop.ai",
"@type": "File",
"name": "Diagram showing trend to increase",
"contentSize": "383766",
"description": "Illustrator file for Glop Pot",
"encodingFormat": "application/pdf"
}
]
}