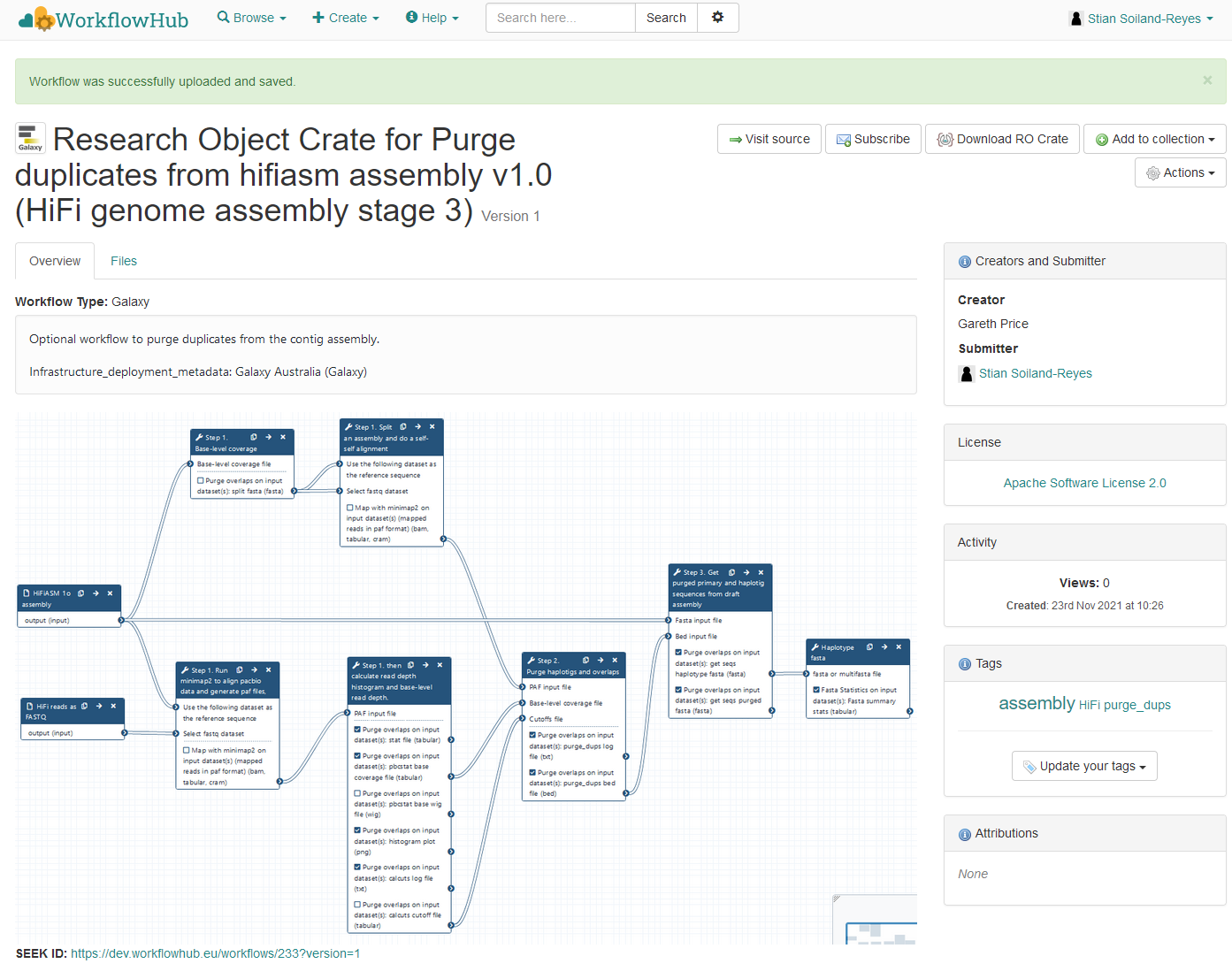
RO-Crate
Use cases in biodiversity
Stian Soiland-Reyes
eScience lab, The University of Manchester
INDElab, University of Amsterdam
ERGA ITIC meeting
2023-11-23
This work is licensed under a
Creative Commons Attribution 4.0 International License.








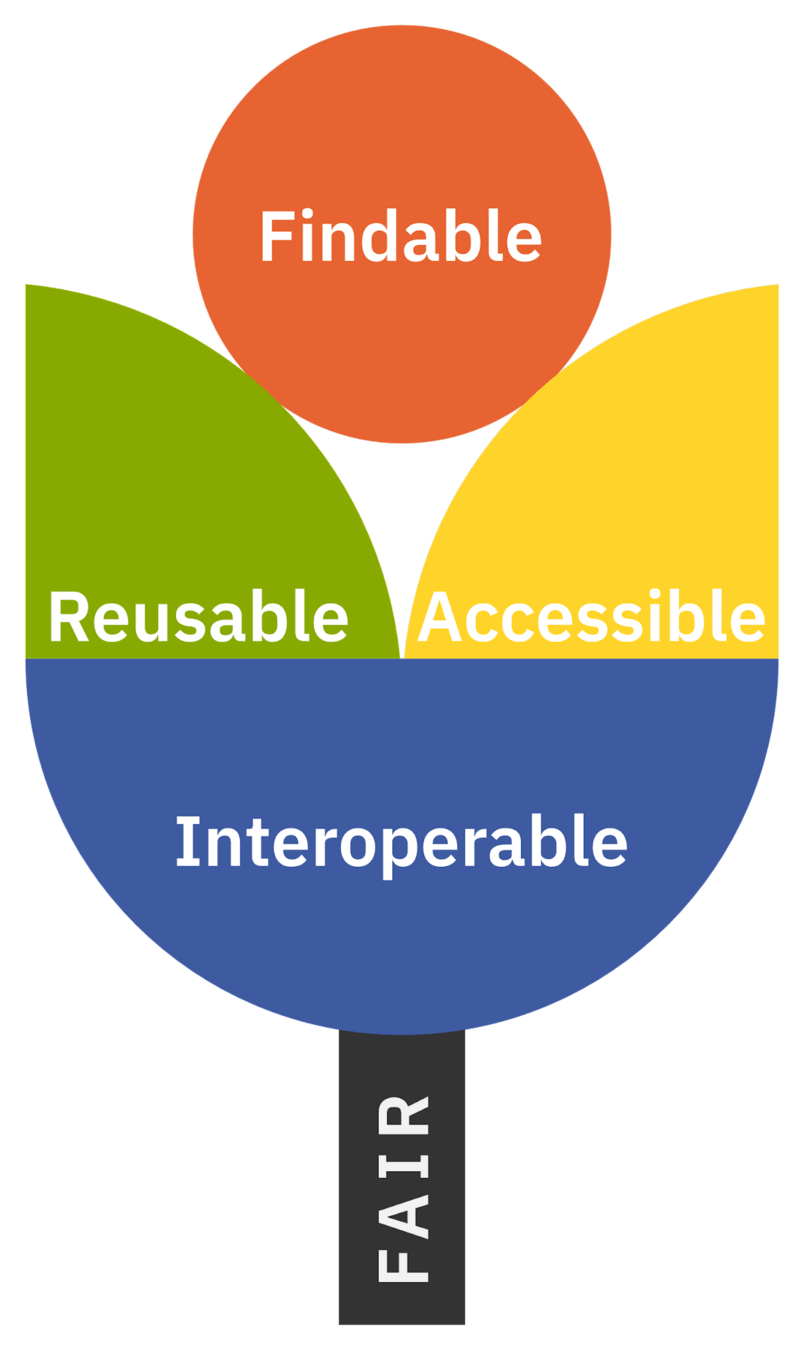
Aims of FAIR Research Objects
Describe and package data collections, datasets, software etc. with their metadata
Platform-independent object exchange between repositories and services
Support reproducibility and analysis: link data with codes and workflows
Transfer of sensitive/large distributed datasets with persistent identifiers
Aggregate citations and persistent identifiers
Propagate provenance and existing metadata
Publish and archive mixed objects and references
Reuse existing standards, but hide their complexity

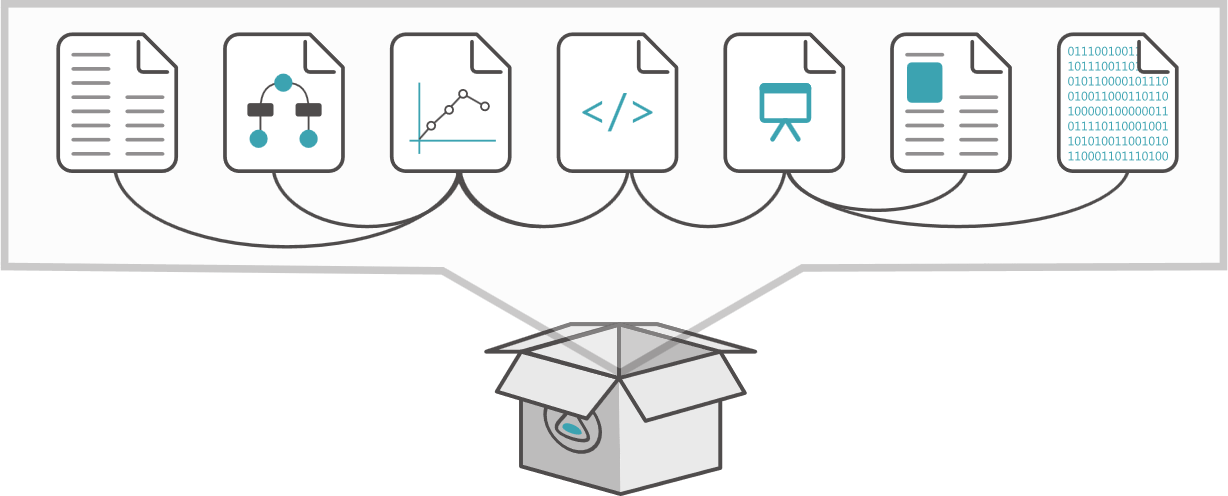


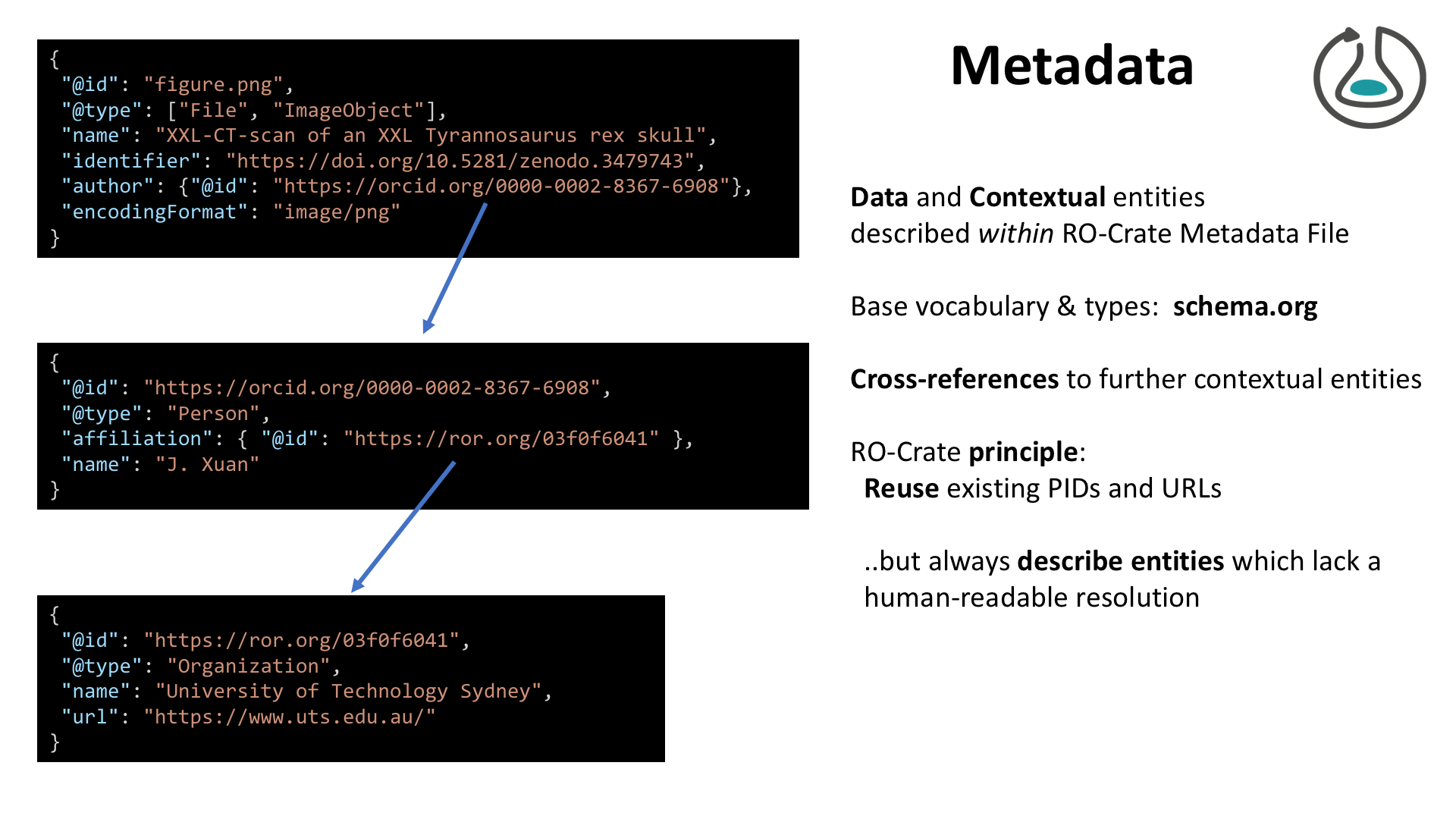
{
"@context": "https://w3id.org/ro/crate/1.1/context",
"@graph": [
{
"@id": "ro-crate-metadata.json",
"@type": "CreativeWork",
"about": {
"@id": "./"
},
"conformsTo": [
{
"@id": "https://w3id.org/ro/crate/1.1"
},
{
"@id": "https://about.workflowhub.eu/Workflow-RO-Crate/"
}
]
},
{
"@id": "ro-crate-preview.html",
"@type": "CreativeWork",
"about": {
"@id": "./"
}
},
{
"@id": "./",
"@type": "Dataset",
"mainEntity": {
"@id": "Galaxy-Workflow-Purge_duplicates_from_hifiasm_assembly_v1.0.ga"
},
"hasPart": [
{
"@id": "Galaxy-Workflow-Purge_duplicates_from_hifiasm_assembly_v1.0.ga"
},
{
"@id": "purge_image.png"
}
],
"identifier": "https://workflowhub.eu/workflows/237?version=1",
"url": "https://workflowhub.eu/workflows/237/ro_crate?version=1",
"name": "Research Object Crate for Purge duplicates from hifiasm assembly v1.0 (HiFi genome assembly stage 3)",
"description": "Optional workflow to purge duplicates from the contig assembly.\r\n\r\nInfrastructure_deployment_metadata: Galaxy Australia (Galaxy)",
"sdDatePublished": "2021-11-23 10:44:32 +0000",
"creativeWorkStatus": "Stable"
},
{
"@id": "Galaxy-Workflow-Purge_duplicates_from_hifiasm_assembly_v1.0.ga",
"@type": [
"File",
"SoftwareSourceCode",
"ComputationalWorkflow"
],
"programmingLanguage": {
"@id": "#galaxy"
},
"image": {
"@id": "purge_image.png"
},
"contentSize": 115723,
"dct:conformsTo": "https://bioschemas.org/profiles/ComputationalWorkflow/1.0-RELEASE/",
"description": "Optional workflow to purge duplicates from the contig assembly.\r\n\r\nInfrastructure_deployment_metadata: Galaxy Australia (Galaxy)",
"name": "Purge duplicates from hifiasm assembly v1.0 (HiFi genome assembly stage 3)",
"url": "https://workflowhub.eu/workflows/237?version=1",
"keywords": "Assembly, purge_dups, HiFi",
"version": 1,
"license": "https://opensource.org/licenses/GPL-3.0",
"creator": {
"@id": "https://workflowhub.eu/people/139"
},
"producer": [
{
"@id": "https://workflowhub.eu/projects/54"
},
{
"@id": "https://workflowhub.eu/projects/30"
}
],
"dateCreated": "2021-11-15T01:39:22Z",
"dateModified": "2021-11-15T01:40:24Z",
"isPartOf": [
],
"input": [
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFiASM 1o assembly"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFi reads as FASTQ"
}
],
"output": [
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-split_fasta"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-alignment_output"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stat_file"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_cov"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_wig"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-hist"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_log"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_tab"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_log"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_bed"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_hap"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_purged"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stats"
}
],
"sdPublisher": {
"@id": "http://about.workflowhub.eu"
}
},
{
"@id": "purge_image.png",
"@type": [
"File",
"ImageObject",
"WorkflowSketch"
],
"contentSize": 118673
},
{
"@id": "https://about.workflowhub.eu/Workflow-RO-Crate/",
"@type": "CreativeWork",
"name": "Workflow RO-Crate Profile",
"version": "0.2.0"
},
{
"@id": "https://workflowhub.eu/people/139",
"@type": "Person",
"name": "Gareth Price"
},
{
"@id": "https://workflowhub.eu/projects/54",
"@type": [
"Project",
"Organization"
],
"name": "Galaxy Australia"
},
{
"@id": "https://workflowhub.eu/projects/30",
"@type": [
"Project",
"Organization"
],
"name": "Australian BioCommons"
},
{
"@id": "#galaxy",
"@type": "ComputerLanguage",
"name": "Galaxy",
"identifier": {
"@id": "https://galaxyproject.org/"
},
"url": {
"@id": "https://galaxyproject.org/"
}
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFiASM%201o%20assembly",
"@type": "FormalParameter",
"name": "HiFiASM 1o assembly",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFi%20reads%20as%20FASTQ",
"@type": "FormalParameter",
"name": "HiFi reads as FASTQ",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-split_fasta",
"@type": "FormalParameter",
"name": "split_fasta",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-alignment_output",
"@type": "FormalParameter",
"name": "alignment_output",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stat_file",
"@type": "FormalParameter",
"name": "stat_file",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_cov",
"@type": "FormalParameter",
"name": "pbcstat_cov",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_wig",
"@type": "FormalParameter",
"name": "pbcstat_wig",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-hist",
"@type": "FormalParameter",
"name": "hist",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_log",
"@type": "FormalParameter",
"name": "calcuts_log",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_tab",
"@type": "FormalParameter",
"name": "calcuts_tab",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_log",
"@type": "FormalParameter",
"name": "purge_dups_log",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_bed",
"@type": "FormalParameter",
"name": "purge_dups_bed",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_hap",
"@type": "FormalParameter",
"name": "get_seqs_hap",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_purged",
"@type": "FormalParameter",
"name": "get_seqs_purged",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stats",
"@type": "FormalParameter",
"name": "stats",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "http://about.workflowhub.eu",
"@type": "Organization",
"name": "WorkflowHub",
"url": "http://about.workflowhub.eu"
}
]
}
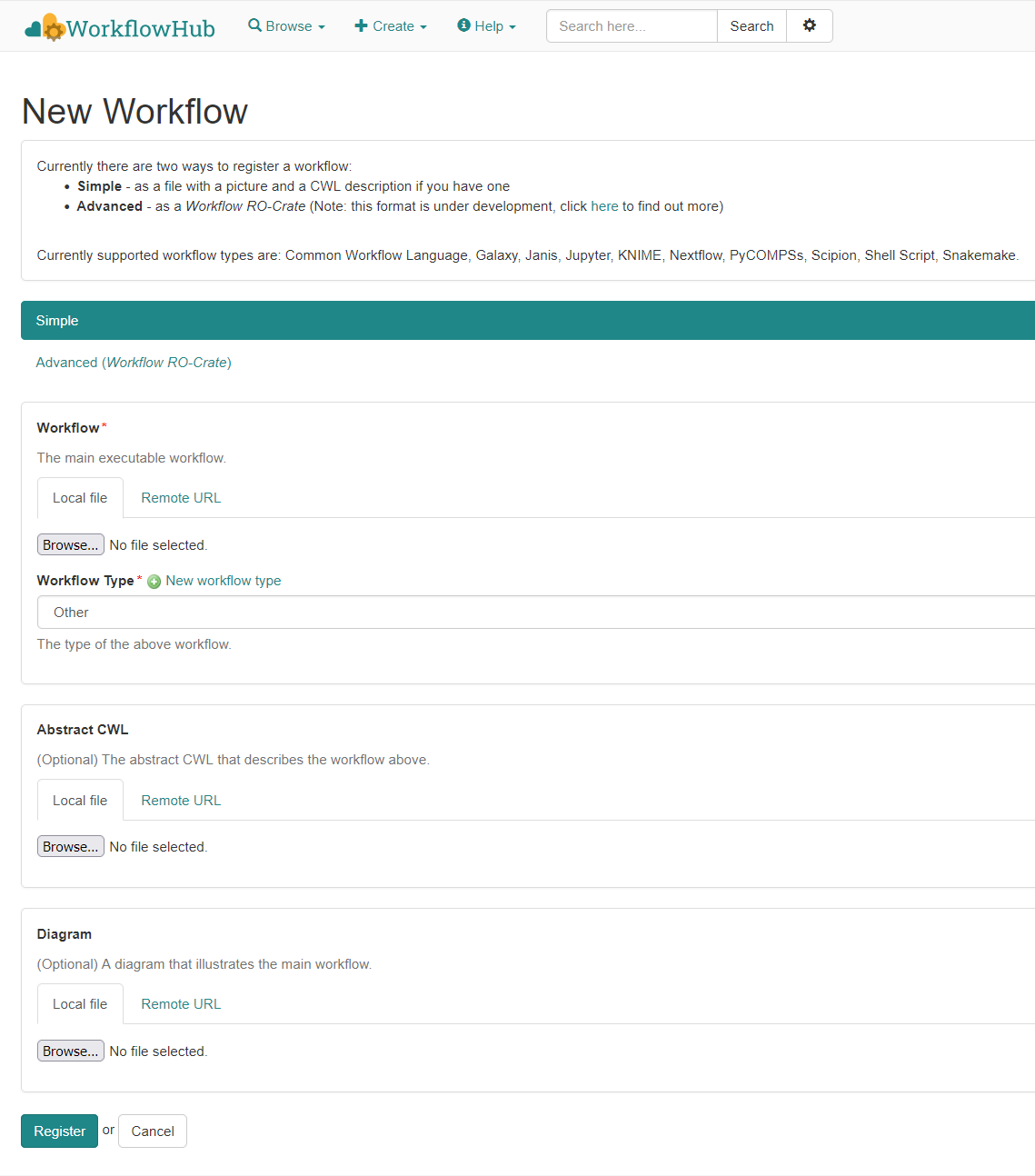
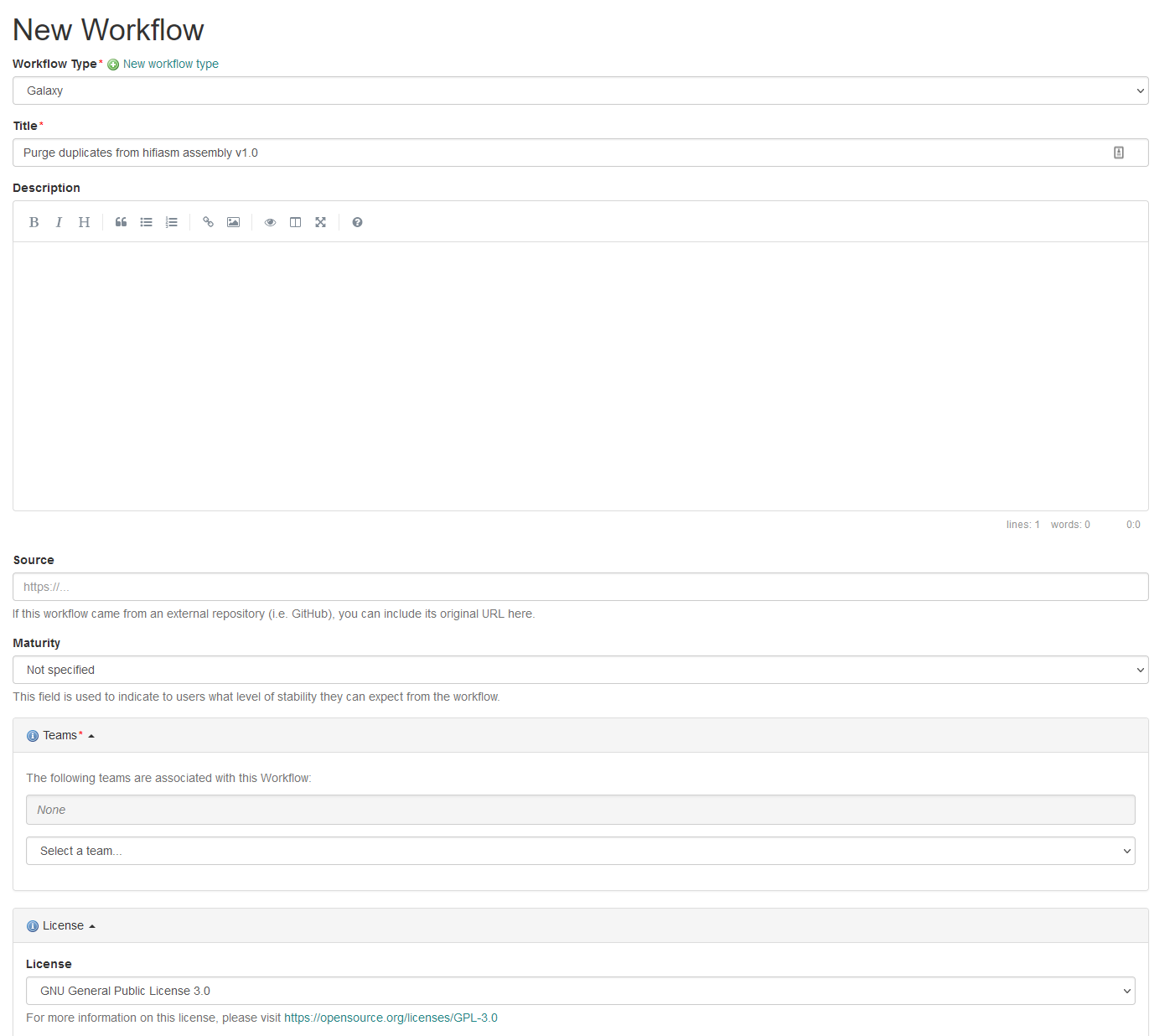
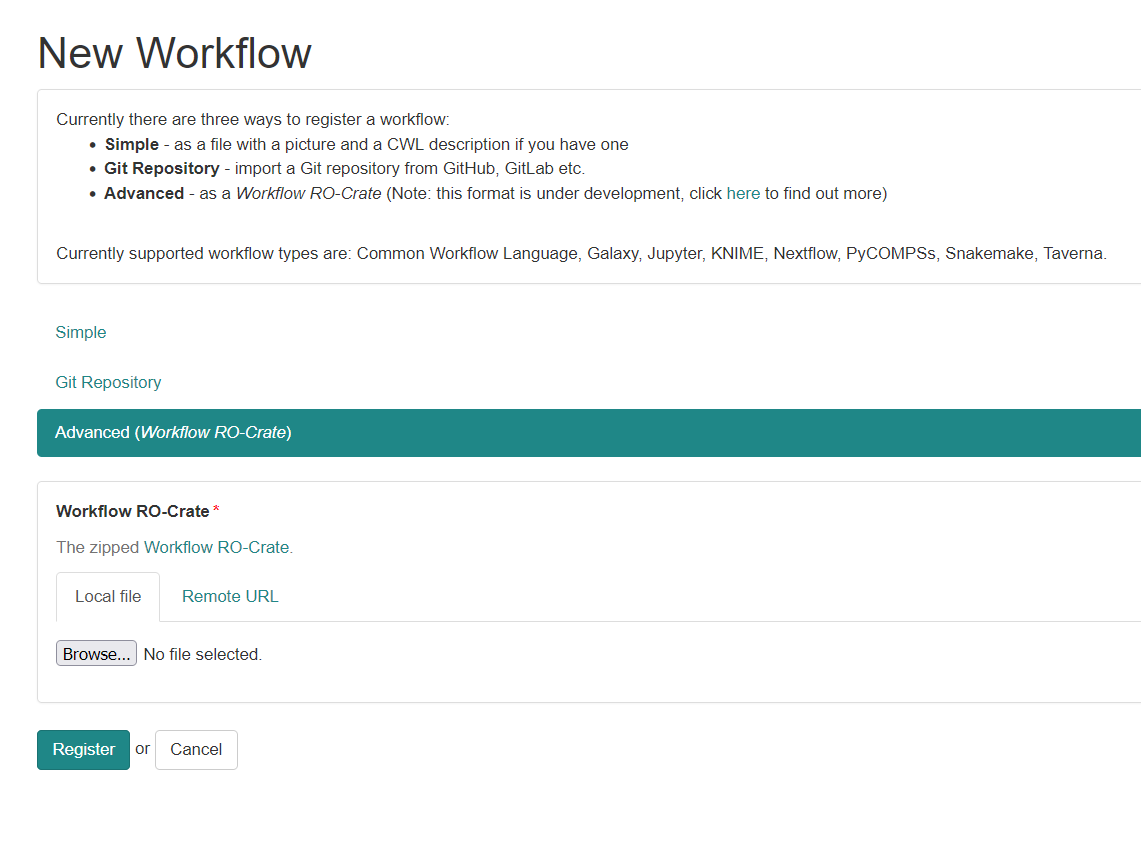
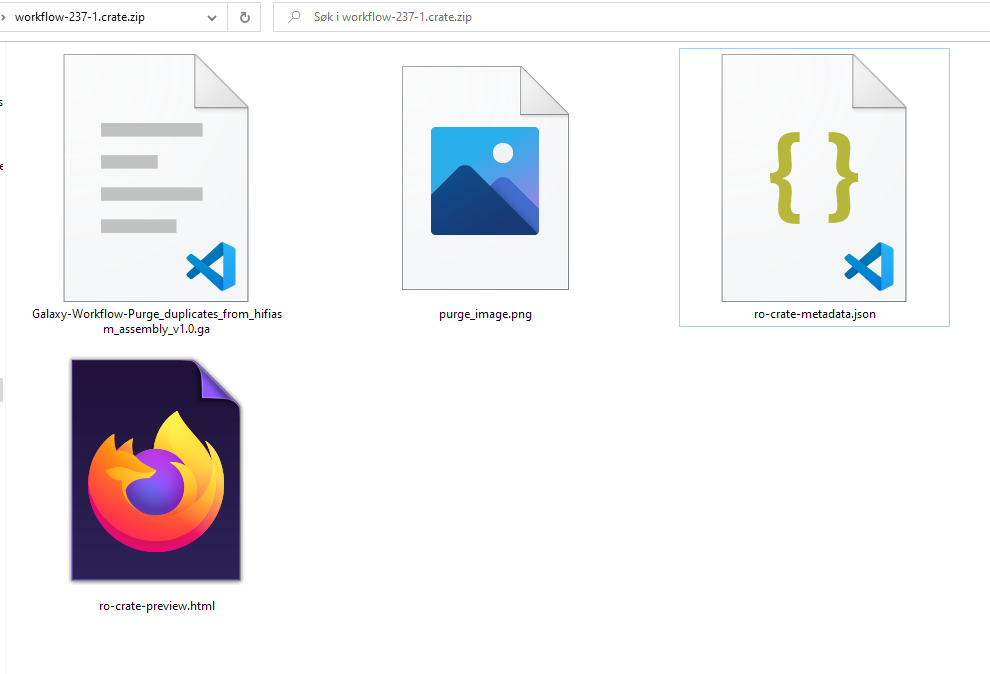
ro-crate-metadata.json


Text
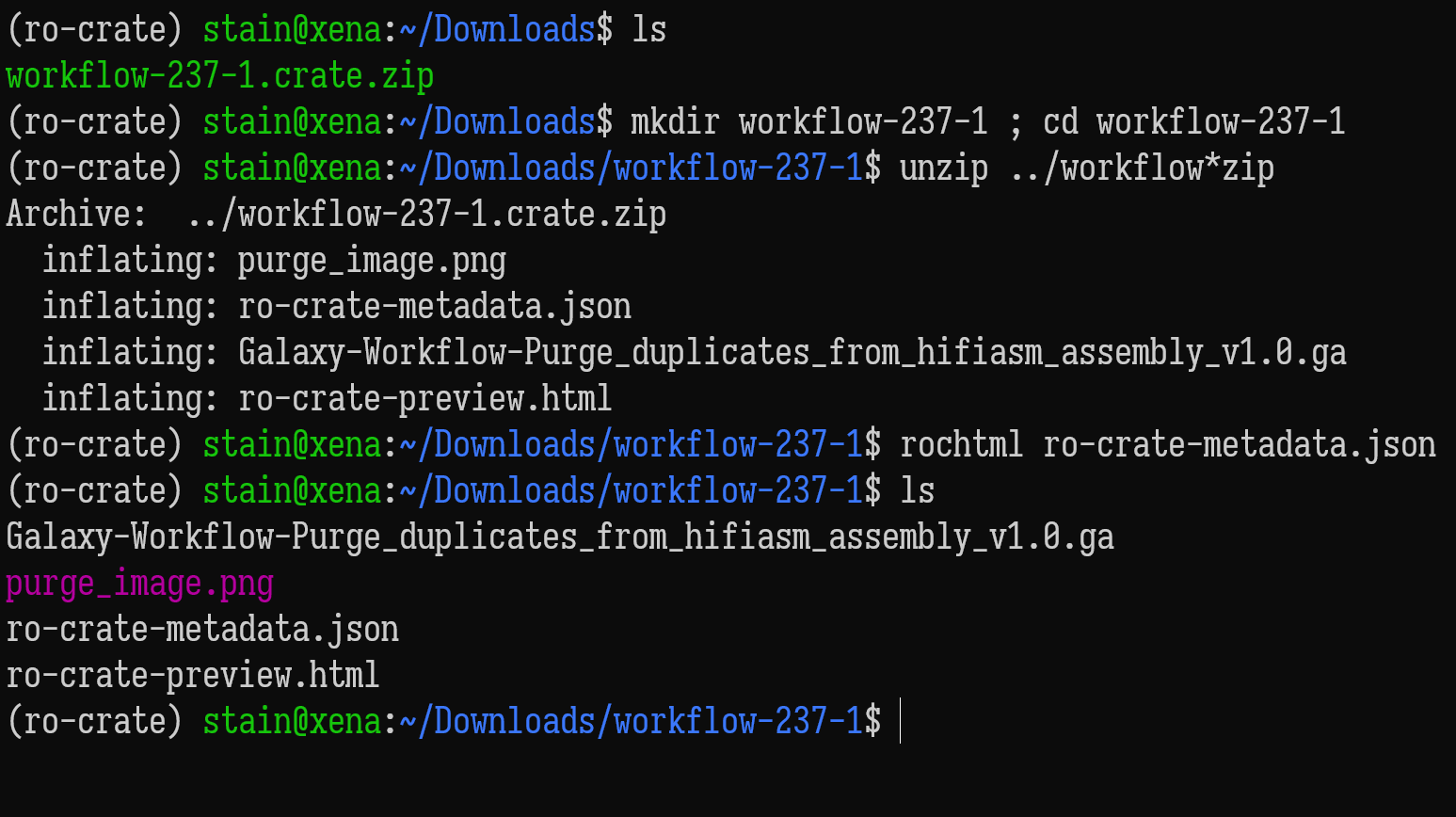
Generate a HTML preview from JSON-LD





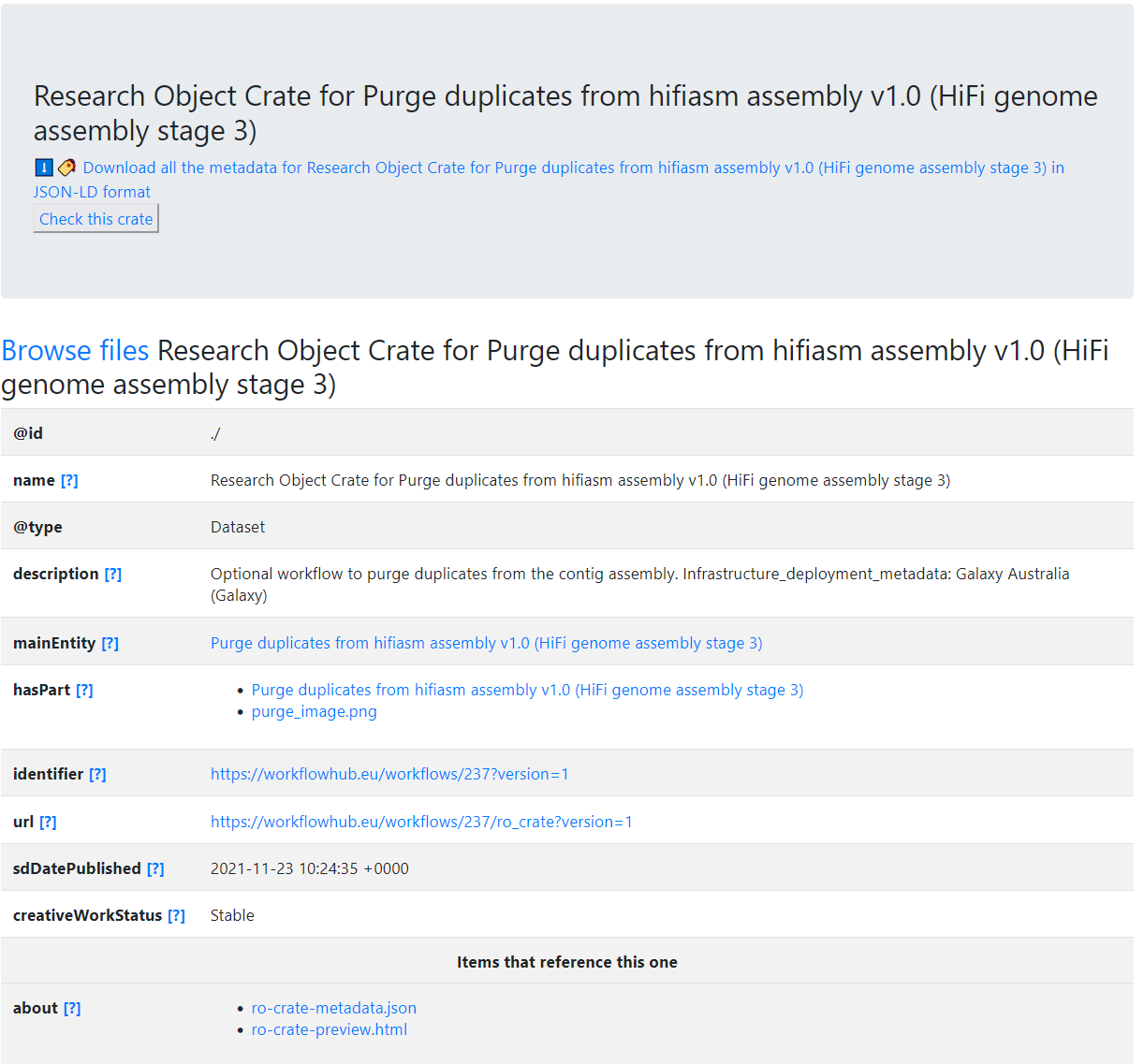
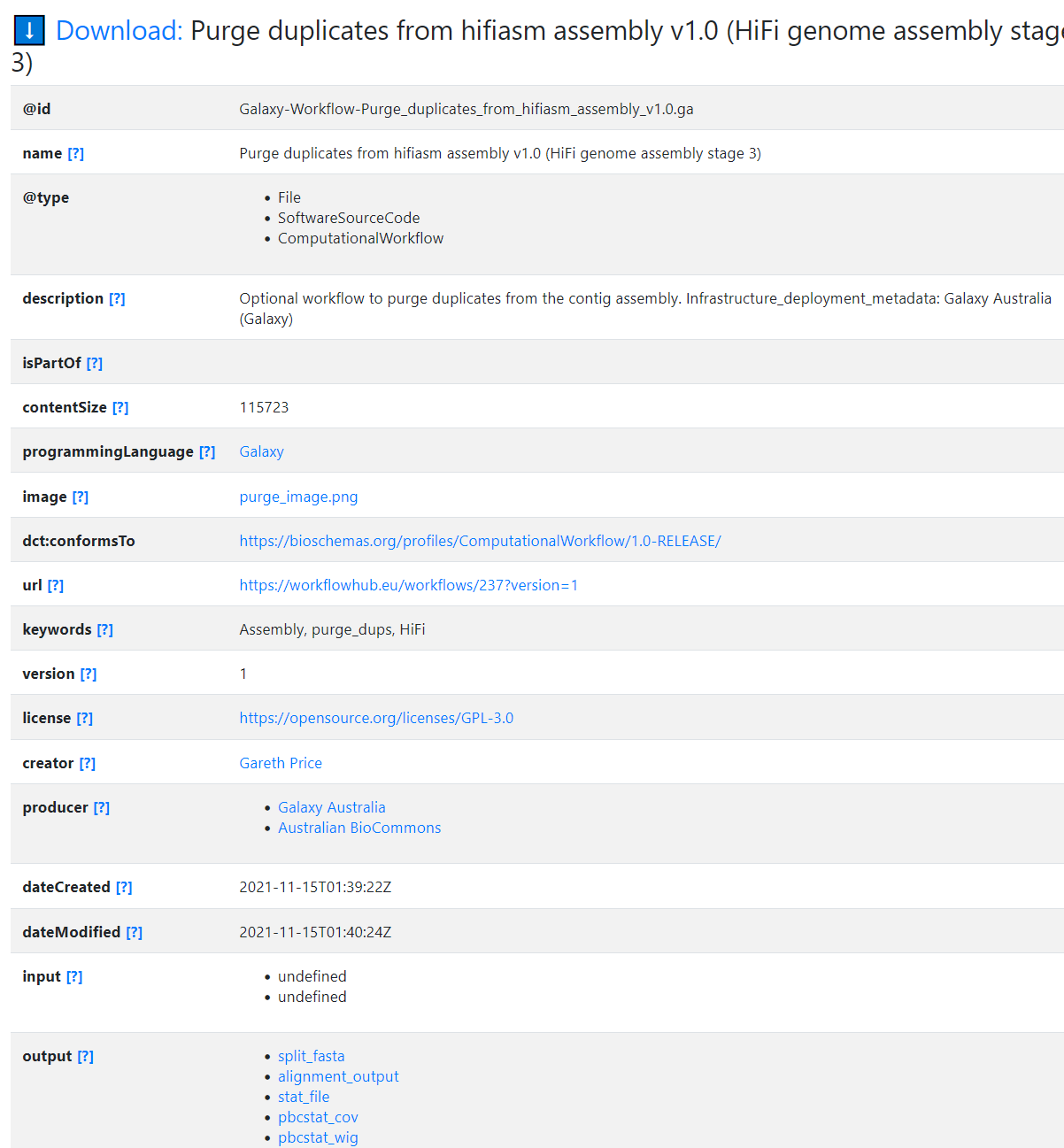

Techie deep-dive!
Warning: JSON ahead














RO-Crate tutorials


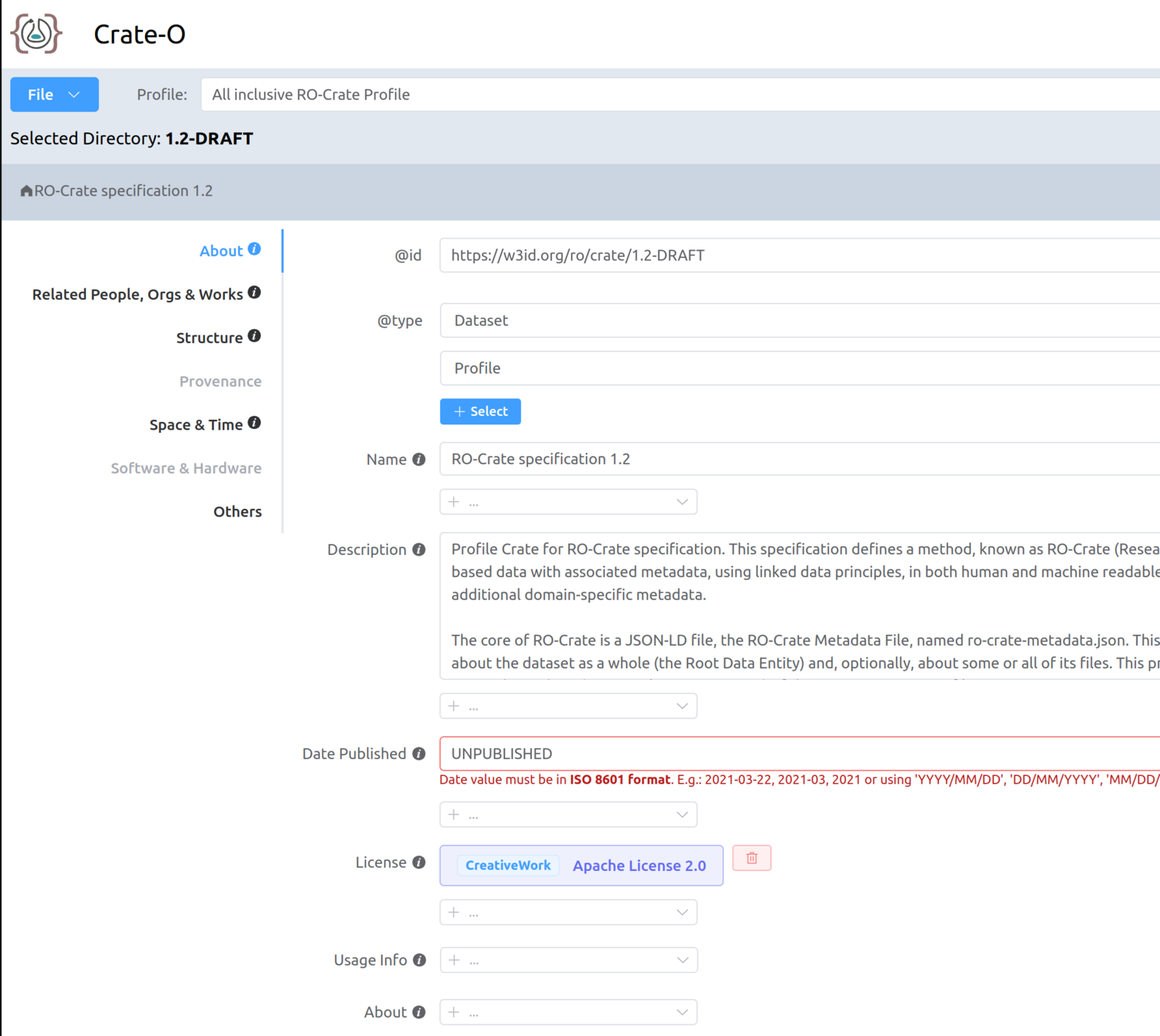
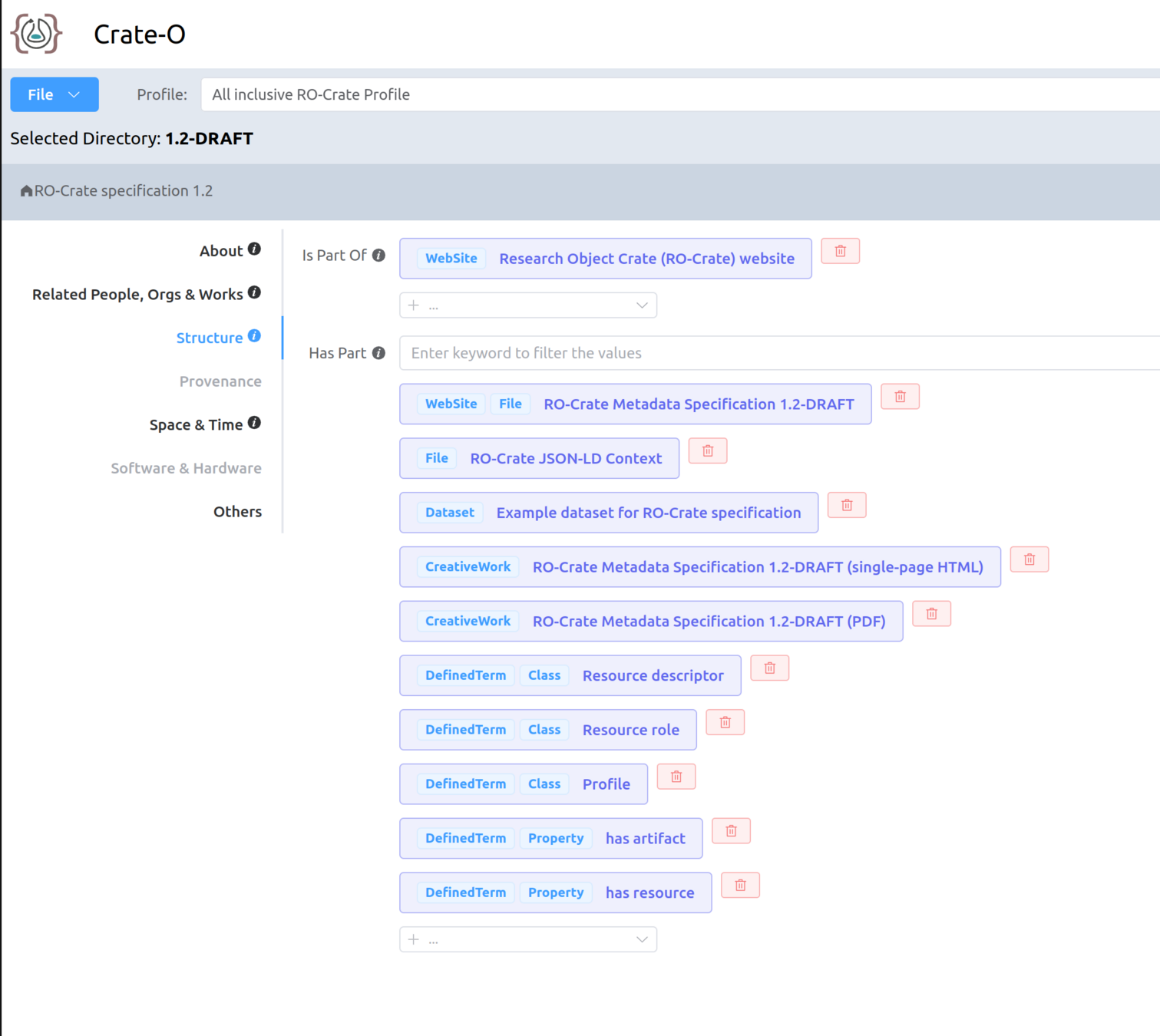
Making your own RO-Crate with Crate-O
Provenance of computational processes
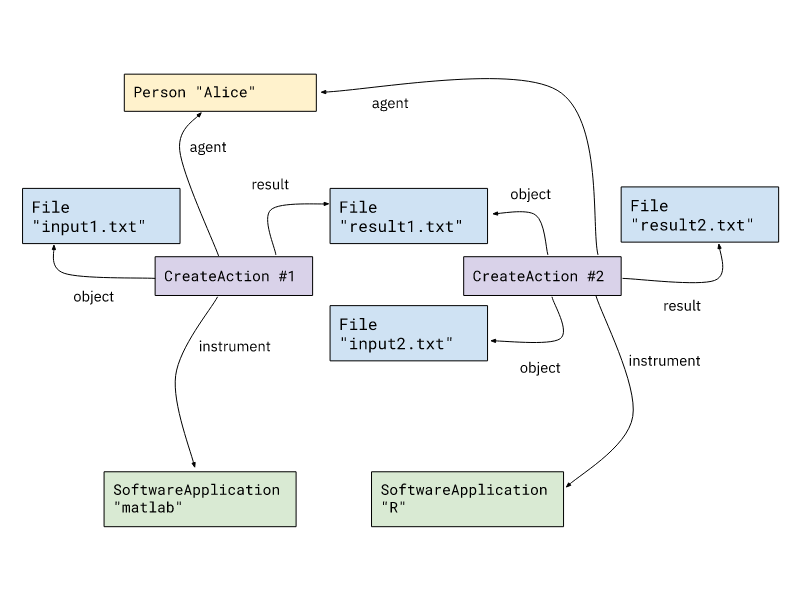
Workflow definition is optional,
but recommended
{
"@id": "#wfrun-5a5970ab-4375-444d-9a87-a764a66e3a47",
"@type": "CreateAction",
"name": "Galaxy workflow run 5a5970ab-4375-444d-9a87-a764a66e3a47",
"endTime": "2018-09-19T17:01:07+10:00",
"instrument": {"@id": "Galaxy-Workflow-Hello_World.ga"},
"subjectOf": {"@id": "https://usegalaxy.eu/u/5dbf7f05329e49c98b31243b5f35045c/p/invocation-report-a3a1d27edb703e5c"},
"object": [
{"@id": "inputs/abcdef.txt"},
{"@id": "#verbose-pv"}
],
"result": [
{"@id": "outputs/Select_first_on_data_1_2.txt"},
{"@id": "outputs/tac_on_data_360_1.txt"}
]
},
{
"@id": "Galaxy-Workflow-Hello_World.ga",
"@type": ["File", "SoftwareSourceCode", "ComputationalWorkflow"],
"name": "Hello World (Galaxy Workflow)",
"author": {"@id": "https://orcid.org/0000-0001-9842-9718"},
"creator": {"@id": "https://orcid.org/0000-0001-9842-9718"},
"programmingLanguage": {"@id": "https://w3id.org/workflowhub/workflow-ro-crate#galaxy"},
"input": [
{"@id": "#simple_input"},
{"@id": "#verbose-param"}
],
"output": [
{"@id": "#reversed"},
{"@id": "#last_lines"}
]
},Choice of layered profiles:
Process Run Crate – some tool was executed
Workflow Run Crate – the tool was a workflow
Provenance Run Crate – we know which tools the workflow ran
RO-Crate in biodiversity



Jonas Grieb, Claus Weiland
Senckenberg – Leibniz Institution for Biodiversity and Earth System Research
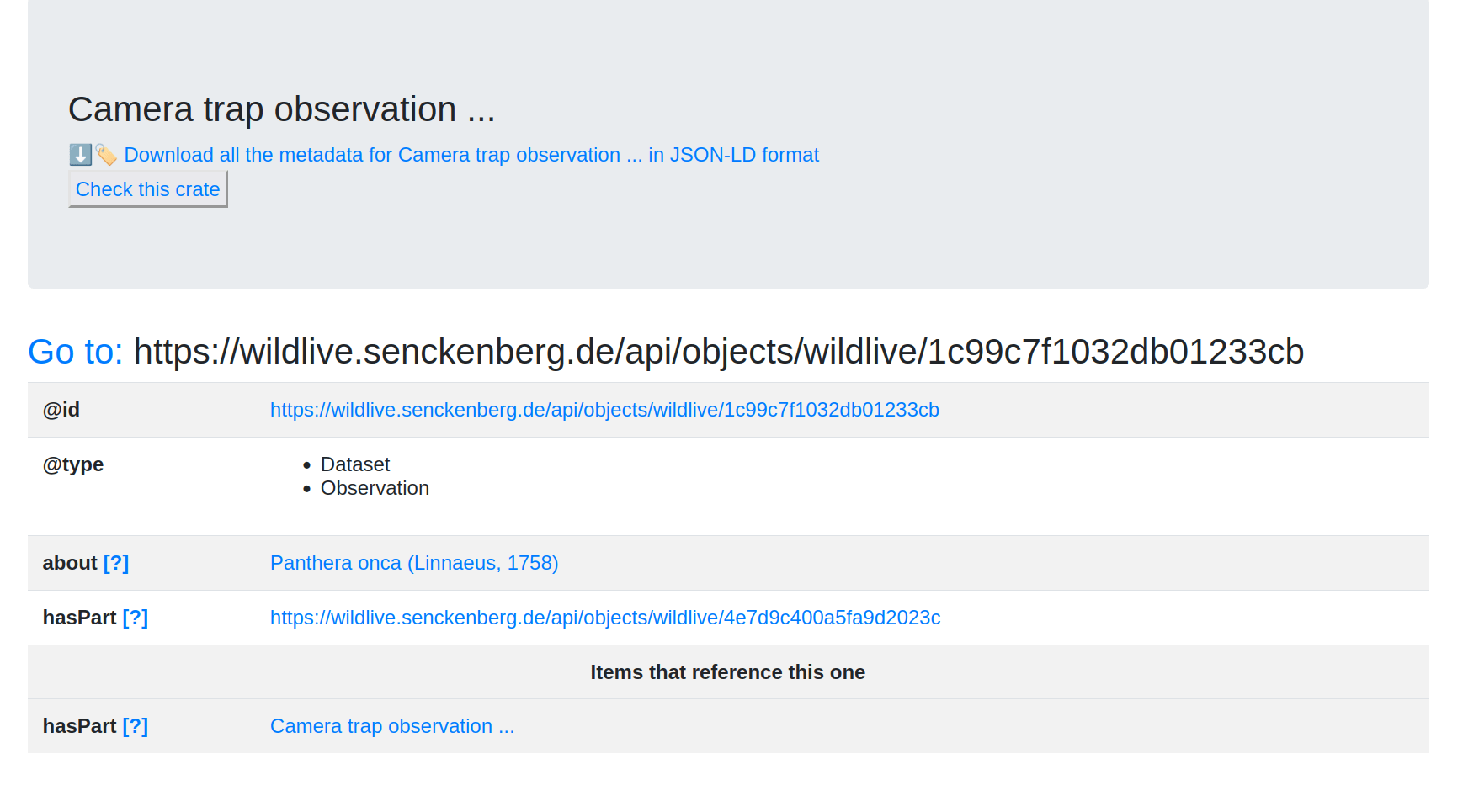
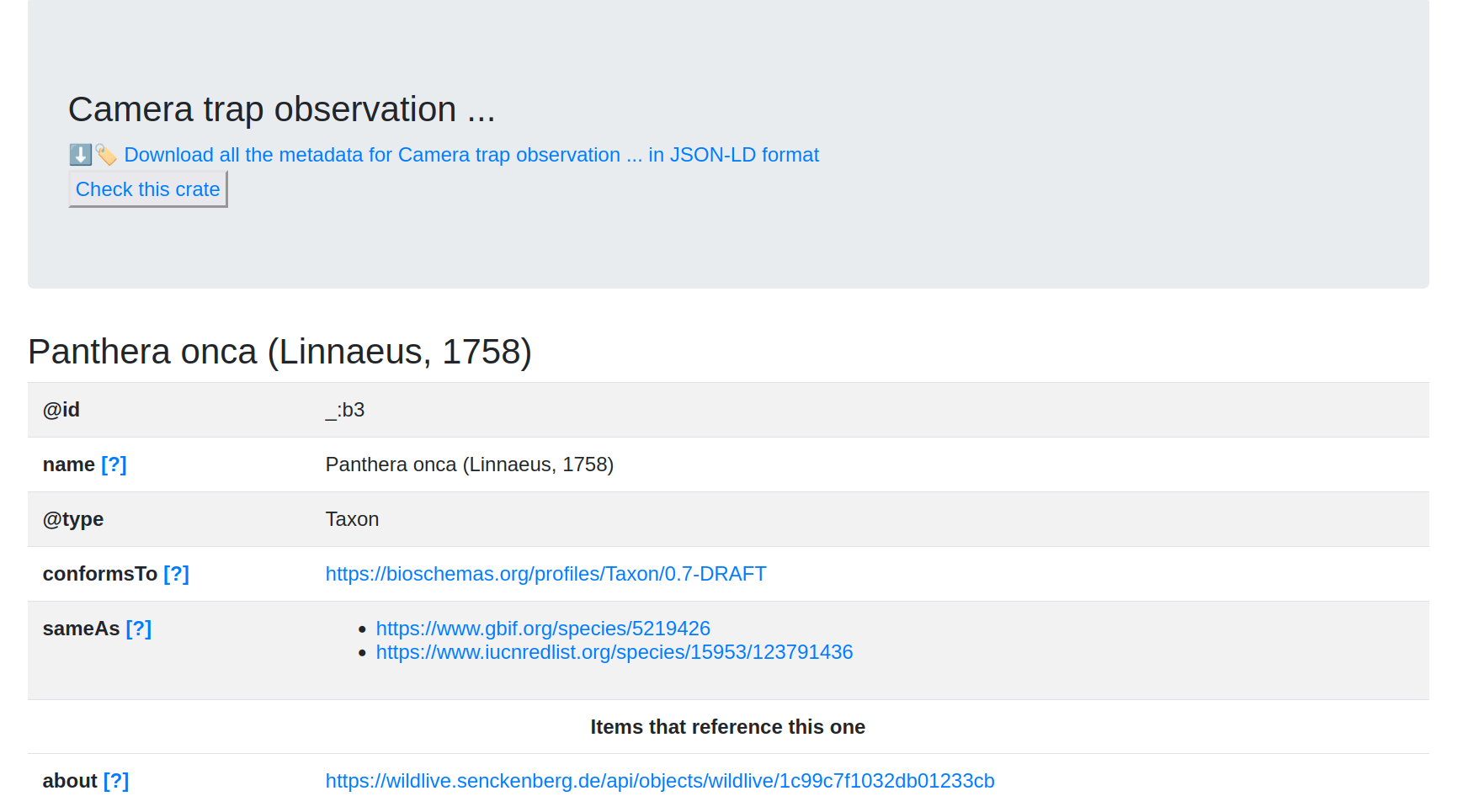
(base) stain@xena:~$ signposting https://wildlive.senckenberg.de/captureevent/wildlive/7df91e6d148a386cc674
Signposting for https://wildlive.senckenberg.de/captureevent/wildlive/7df91e6d148a386cc674
CiteAs: <https://wildlive.senckenberg.de/api/objects/wildlive/7df91e6d148a386cc674>
License: <http://spdx.org/licenses/CC-BY-4.0>
Author: <https://orcid.org/0000-0002-2631-4601>
DescribedBy: <https://wildlive.senckenberg.de/api/call?objectId=wildlive/7df91e6d148a386cc674&method=getAsROCrate> application/ld+json
Item: <https://wildlive.senckenberg.de/api/objects/wildlive/6f6afb2850b946bb9394?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_601.JPG> image/jpeg
<https://wildlive.senckenberg.de/api/objects/wildlive/4e7d9c400a5fa9d2023c?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_607.JPG> image/jpeg
<https://wildlive.senckenberg.de/api/objects/wildlive/dbfaee3660bef31479a3?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_605.JPG> image/jpeg
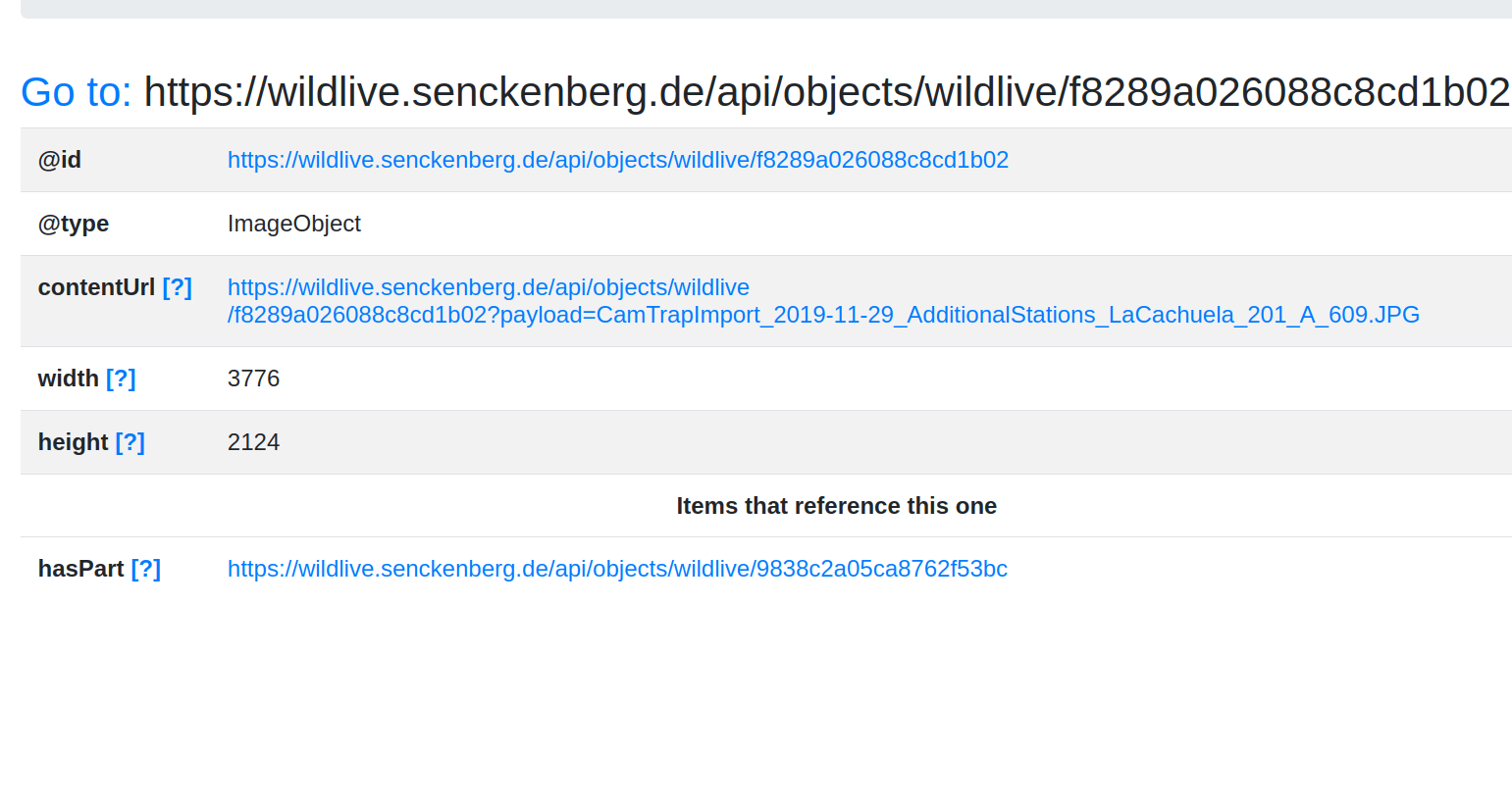
<https://wildlive.senckenberg.de/api/objects/wildlive/f8289a026088c8cd1b02?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_609.JPG> image/jpeg
<https://wildlive.senckenberg.de/api/objects/wildlive/a98330d9df31a8d10847?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_608.JPG> image/jpeg
<https://wildlive.senckenberg.de/api/objects/wildlive/ffafa0893d4a2af6d0ba?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_606.JPG> image/jpeg
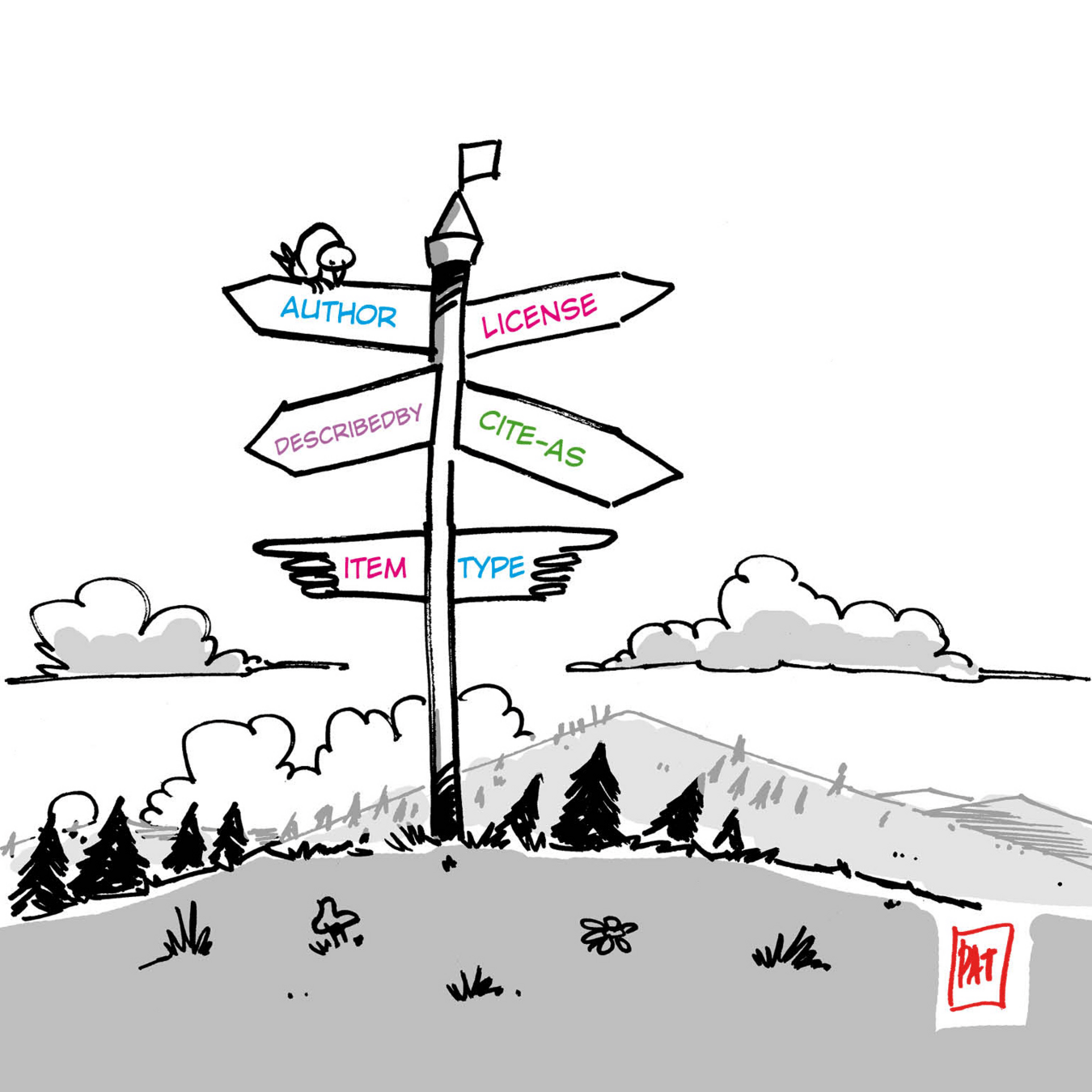
Machine-actionable navigation to an RO-Crate using Signposting
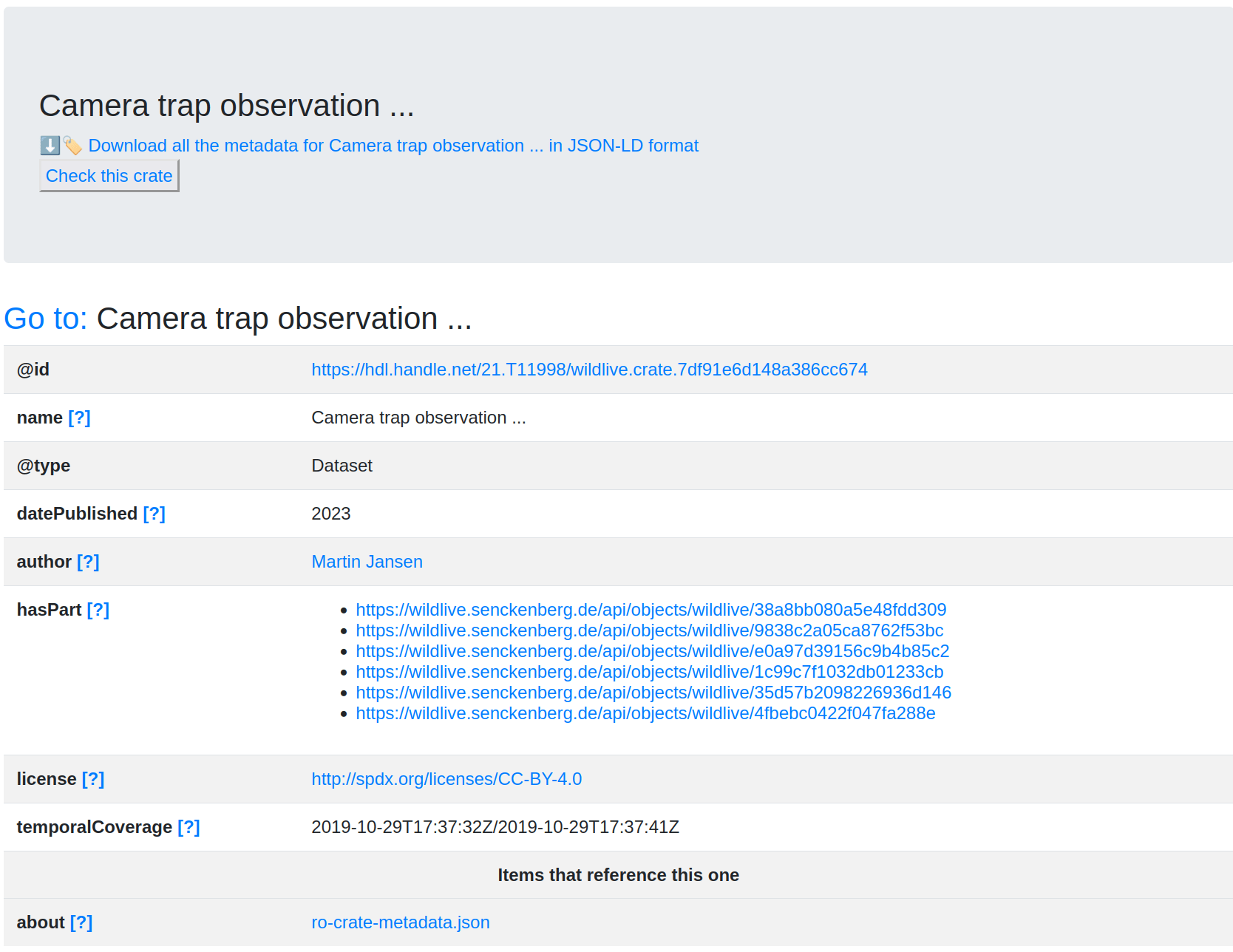


Building BioDT RO-Crate FDO profiles
Kernel Attributes Table
IndexPropertyDescriptionTypeCardinalityCommentsExample
| 1 | profile | The FDO profile that a certain metadata description follows. | URL | 1/1 | For now, we can refer to this repo for the profiles. | |
| 2 | license | License for the FDO the metadata description is about. | URL | 1/1 | The FDO might have a certain license, however, the metadata file should have a CC0 license. | "http://creativecommons.org/licenses/by/4.0/" |
| 3 | type | The nature of the object the metadata describes. For now, this follows the RO-Crate conventions. | Text or URL | 1/many | "Dataset" | |
| 4 | identifier | An identifier for the FDO. Ideally, a globally unique, persistent and resolvable identifier. | Text or URL | 1/1 | "https://doi.org/10.1111/1365-2664.12222" | |
| 5 | name | Name of the FDO. | Text | 1/many | "BEEHAVE model" | |
| 6 | description | Short description of what the FDO is. | Text | 1/many | "BEEHAVE is a computer model to simulate the development of a honeybee colony and its nectar and pollen foraging behavior in different..." | |
| 7 | datePublished | Date of publication of the FDO. | Date | 1/1 | "2014-03-04" | |
| 8 | dateModified | Most recent date where the FDO was modified. | Date | 1/1 | "2014-03-04" | |
| 9 | author | Author of the FDO. | Person or Organization | 1/many | If possible, use an ORCiD, otherwise use full name in natural order. | "https://orcid.org/0000-0003-0791-7164" |

Model-specific Attributes Table
IndexPropertyDescriptionTypeCardinalityCommentsExample
| 201 | codeRepository | Link to the repository or repositories where the un-compiled, human-readable code is located. | URL | 1/many | A link to GitHub, GitLab, or similar. Preferably, the code repository should be under the BioDT GitHub organisation so people in the project have access to it. | "https://github.com/BioDT/biodt-fair" |
| 202 | contributor | Additional people or organisations that contributed to developing the model, despite not having authorship. | Person or Organization | 0/many | If possible, use an ORCiD (or ROR ID, for organizations), otherwise use full name in natural order. | "https://orcid.org/0000-0003-0791-7164" |
| 203 | softwareVersion | The version of the model instance. | Text | 1/1 | It is advised to follow the semantic versioning guidelines. | "2.0.13" |
| 204 | programmingLanguage | The computer programming language the code is written in. | Text or ComputerLanguage | 1/many | "Python" | |
| 205 | runtimePlatform | Runtime platform or script interpreter dependencies. | Text | 1/many | "Python 3.11" | |
| 206 | operatingSystem | Operating systems supported. | Text | 1/many | "Ubuntu 22.04.3" | |
| 207 | softwareRequirements | Component dependency requirements for the model. | Text or URL | 0/many | This includes runtime environments and shared libraries that are not included as part of the model but are required to run it. | ".NET runtime" |
Julian Lopez Gordillo, Naturalis


First prototype: Use the COPO data brokering platform to support the processing, validation, and ingestion of standardised sample, barcode and sequencing metadata into the biodiversity genomics data ecosystem
Next: Consuming as part of larger pipeline
-
Use cases welcome!
-
RO-Crate as submission format

Debby Ku, Aaliyah Providence, Seanna McTaggart, Felix Shaw (Earlham Institute), ++
Attached vs Detached RO-Crate
Attached RO-Crate are used when there is some kind of folder-structure, typically archived as a ZIP file,
or on a traditional website.
Absolute URLs may still be listed as parts, but typically the parts are "files" and "directories" using relative paths.
{ "@context": "https://w3id.org/ro/crate/1.2-DRAFT/context",
"@graph": [
{
"@type": "CreativeWork",
"@id": "ro-crate-metadata.json",
"conformsTo": {"@id": "https://w3id.org/ro/crate/1.2-DRAFT"},
"about": {"@id": "./"}
},
{
"@id": "./",
"@type": [
"Dataset"
],
"hasPart": [
{
"@id": "cp7glop.ai"
},
{
"@id": "lots_of_little_files/"
}
]
},
{
"@id": "cp7glop.ai",
"@type": "File",
"name": "Diagram showing trend to increase",
"contentSize": "383766",
"description": "Illustrator file for Glop Pot",
"encodingFormat": "application/pdf"
},
{
"@id": "lots_of_little_files/",
"@type": "Dataset",
"name": "Too many files",
"description": "This directory contains many small files, that we're not going to describe in detail."
}
]
}Detached RO-Crate has a standalone JSON-LD metadata file, e.g. returned by an API.
All the resources have absolute URIs
This style may also be suitable for a repository exposing the metadata from an RO-Crate ZIP file without exposing its individual files.
{ "@context": "https://w3id.org/ro/crate/1.2-DRAFT/context",
"@graph": [
{
"@type": "CreativeWork",
"@id": "ro-crate-metadata.json",
"conformsTo": {"@id": "https://w3id.org/ro/crate/1.2-DRAFT"},
"about": {"@id": "https://example.com/item/15?crate"}
},
{
"@id": "https://example.com/item/15?crate",
"@type": [
"Dataset"
],
"hasPart": [
{
"@id": "https://example.com/data/12312/cp7glop.ai"
}
]
},
{
"@id": "http://example.com/data/12312/cp7glop.ai",
"@type": "File",
"name": "Diagram showing trend to increase",
"contentSize": "383766",
"description": "Illustrator file for Glop Pot",
"encodingFormat": "application/pdf"
}
]
}