Quantum Annealing for Protein Design

Computational Protein Design
Proteins
Proteins are sequences of amino acids that are essential to many bodily functions.
Protein structure is closely related to protein function.
Individual amino acids attract/repel one another.
The protein tends to fold into conformations that minimize its free energy.

What is Computational Protein Design?
Computational Protein Design (CPD) tries to, given a protein structure, find an amino acid sequence that will likely fold into that structure. It is the inverse of the problem of protein folding.
After we know a sequence, we can make that protein in the lab.
CPD has already been used to design proteins for important medical applications (like detecting HIV) and holds promise to treat diseases that involve complex protein-protein interactions (Alzheimer's, cancer, etc.).
Quantum Annealing
What is Quantum Annealing?
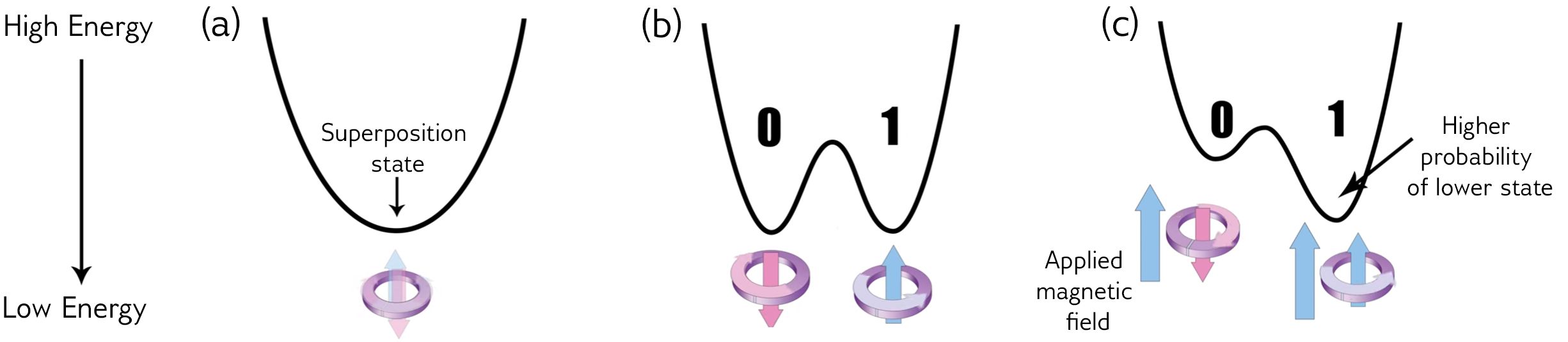
Quantum annealing is a type of quantum computation that does one job: optimizing functions.
By representing the objective function with a system of qubits, quantum annealing takes advantage of quantum effects like entanglement and tunneling to potentially find better solutions than classical methods would.
Note that this is a probabilistic (not exact) method.
The D-Wave Quantum Computer
The D-Wave machine only solves quadratic unconstrained binary optimization (QUBO) problems:
$$\min_{x \in \{0,1\}^n} x^T Q x$$
where Q is the matrix that defines the problem, and x is a binary solution vector.
Entries of the matrix Q are represented by coupling strengths between qubits, and the solution vector x is encoded in the spin (up - 0 or down - 1) of the qubits. So as the annealer evolves to the lowest energy state of the system, it also solves the optimization problem.
Solving a CPD Problem on the
D-Wave
Motivation
This is an NP-hard discrete optimization problem.
The dominant approach (used by Rosetta) is simulated annealing, but this struggles with >100 residues and requires huge computational resources.
Scientists want a low-energy sampling distribution rather than one optimal solution - and the D-Wave is essentially an efficient low-energy probabilistic sampler.
Currently, protein design requires lots of intuition even with computational methods, but by improving these methods, we are hoping to take a lot of the guesswork out of design.
As a QUBO
We use the CPD program Rosetta to calculate an energy matrix for different amino acid values.
We introduce penalties to this matrix to ensure we assign one amino acid per position.
The binary solution vector represents an amino acid sequence.
So we solve for the lowest energy amino acid sequence
$$\min_{x \in \{0,1\}^n} x^T Q x$$
Solving Larger Problems
While the D-Wave 2000 series has 2000 qubits, they are so sparsely connected that we can only fit a 100 variable CPD problem on a single chip.
To solve larger problems, we use a hybrid strategy.
This iterative solver alternates between two methods as it attempts to converge to a global minimum:
- classical heuristic local search (ex. simulated annealing)
- quantum annealing subproblem solver
With this method we get good results on 2000-3000 variable (20-30 residue) problems.
What We Did
Collaborators' Work
We collaborated with two scientists who have been working on the problem for about a year already.
In that time, they developed and implemented the mapping from CPD problem to QUBO matrix, and generated ~6,000 CPD problems of various sizes to test on the D-Wave.
They took one of these design problems, solved it on the D-Wave, and are currently expressing it in a wet lab to experimentally verify if the quantum annealing approach worked.
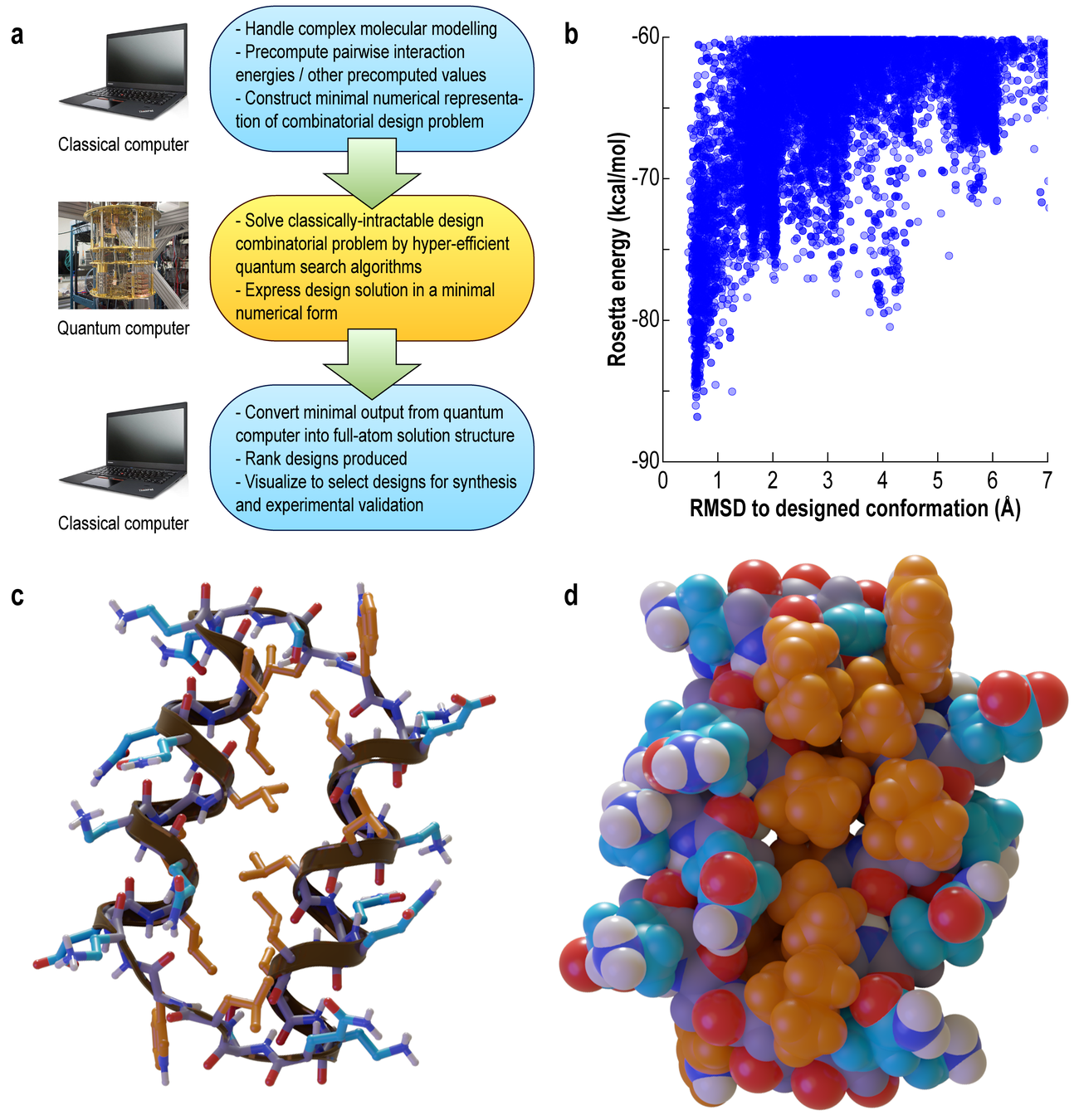
33 residue
quantum-designed protein
Our Work (Ongoing)
Characterizing scaling
Quantum annealing is not yet competitive with classical state-of-the-art solvers. But QA hardware continues improve rapidly.
To make the argument that QA will be a better method once hardware improves, we need to show that our approach scales well as a function of problem size.
We will evaluate scaling on several metrics (energy difference from optimal solution, total annealing time, total iterations).
Our Work (Ongoing)
Improving algorithm for large problems
D-Wave provides an out-of-the-box hybrid solver called QBSolv. It is usable for 20-30 residue design problems, but scales poorly beyond that.
The largest proteins designed to date have about 200 residues.
Since QBSolv uses no domain-specific knowledge to solve CPD problems, we are modifying it to leverage certain assumptions and improve its scaling behavior.
Conclusions
Computational protein design is one of quantum annealing's most convincing near-term applications.
If D-Wave's machines continue to improve at the current rate, expect quantum CPD to be state-of-the-art in 10 years or less.
D-Wave has made quantum annealing very accessible, but the technique it is not a silver bullet.