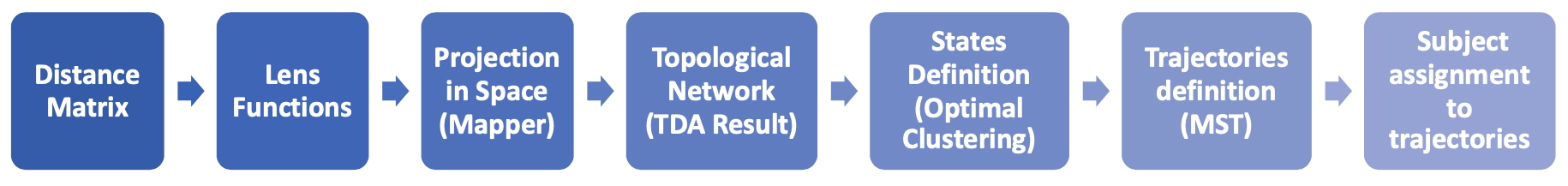
pseudotime TDA
4CE longitudinal lab values
https://github.com/covidclinical/Phase2.1TDAPseudotimeRPackage
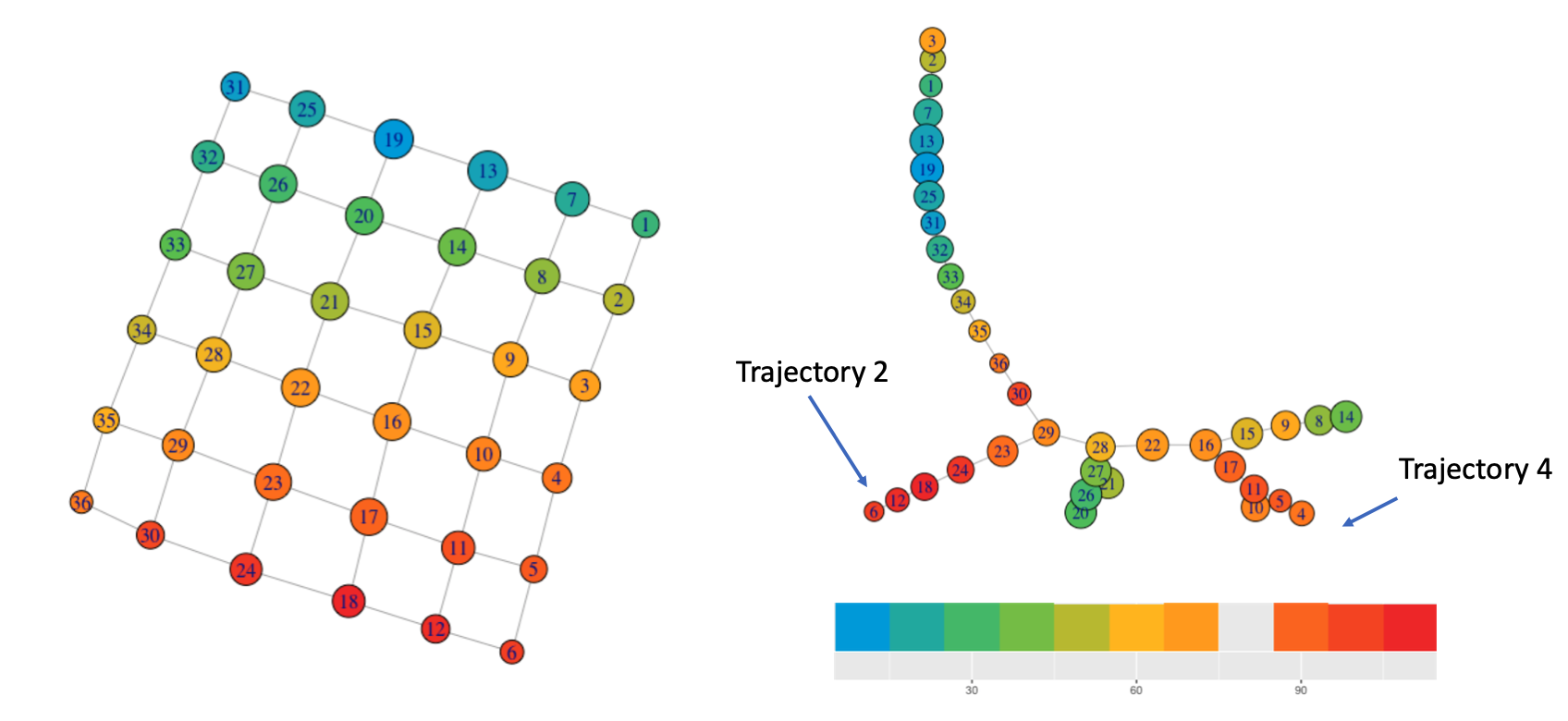
CRP enrichment
- Topological Data Analysis (TDA): mine “connected” disease states via data shape
- Pseudo Time: infer progression among states to reconstruct disease progression trajectories
Challenges
- Missing data
- Not at random
- Slow imputation
- Aggregate result from different sites
- Consistency?
- Parameter tuning?
- Train/test site
- Assumptions
- Number of trajectory clusters
- All patients start out at a similar state
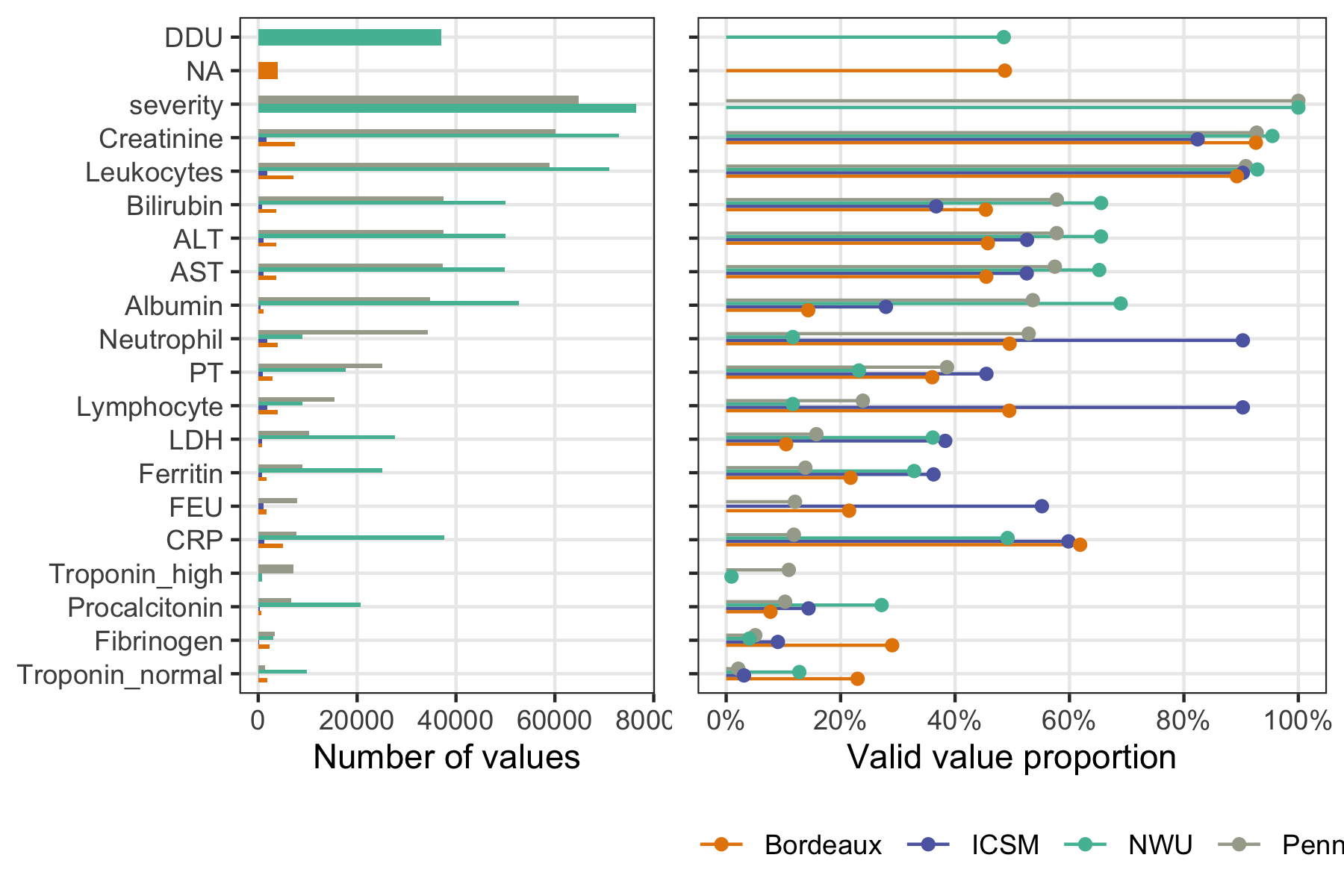
missing data
4CE longitudinal lab values
Does the missingness pattern reflect the healthcare dynamic? (doctor's worry)
workflow proposal
Step 0: Drop variable with high missing rate (> 80%: troponin_high, procalcitonin, fibrinogen, troponin_nomal)
Step 1: Naively impute missing data points of each variable using functional PCA {fdapace}
Step 2: Drop rows with the most (originally) missing values, record the proportion of rows dropped for each patient (pdrop)
Step 3: Put NAs back in the CRP variable where it was missing.
Step 4: Train CRP on Leukocytes, Albumin and pdrop (mixed effect model, XGBoost, Amelia II) with available data
Step 5: Use the fitted model to predict the missing CRP values.
Step 6: Repeat Steps 3–5 separately for each variable that has missing data (Leukocytes and Albumin).
CRP, Albumin, Leukocytes
for each cycle:
evaluation
patient \(p\)
lab \(a\)
time index \(i\)
mask one extra value per lab per patient
Amelia II
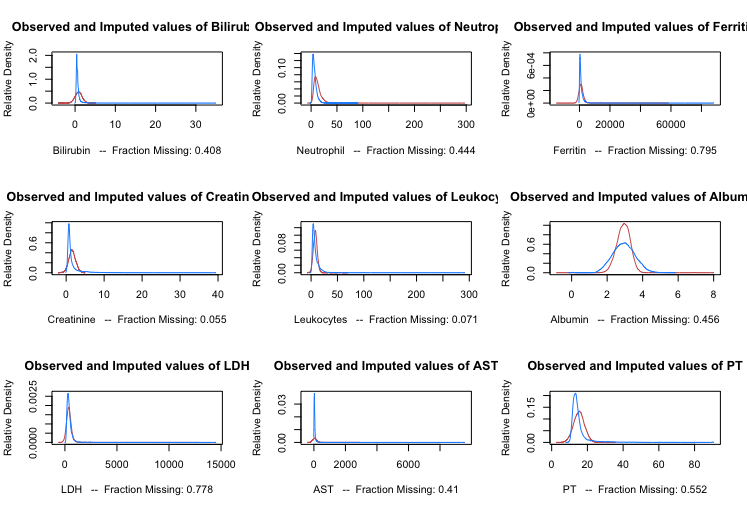
MICE
Limitations
- a proportion of patients would excluded
- missing at random assumption
pattern of missingness?
other details
normalization
Gaussian processes
\(f(t_i)\) have a joint Gaussian distribution
locality constraint
closer time points have more similar measurement values
Step 1: extract separate univariate time series for each patient and variable
Step 2: GPfit: MLE over \(\alpha\) and \(l\)
Step 3: infer values at unobserved time points