JOURNAL CLUB
September 12, 2019
and some comments on figures...
Some terminology
- DOL: day of life
- ontogeny: study of ind. dev. from early stage to maturity
- "indexing": adjusted for baseline/DOL0
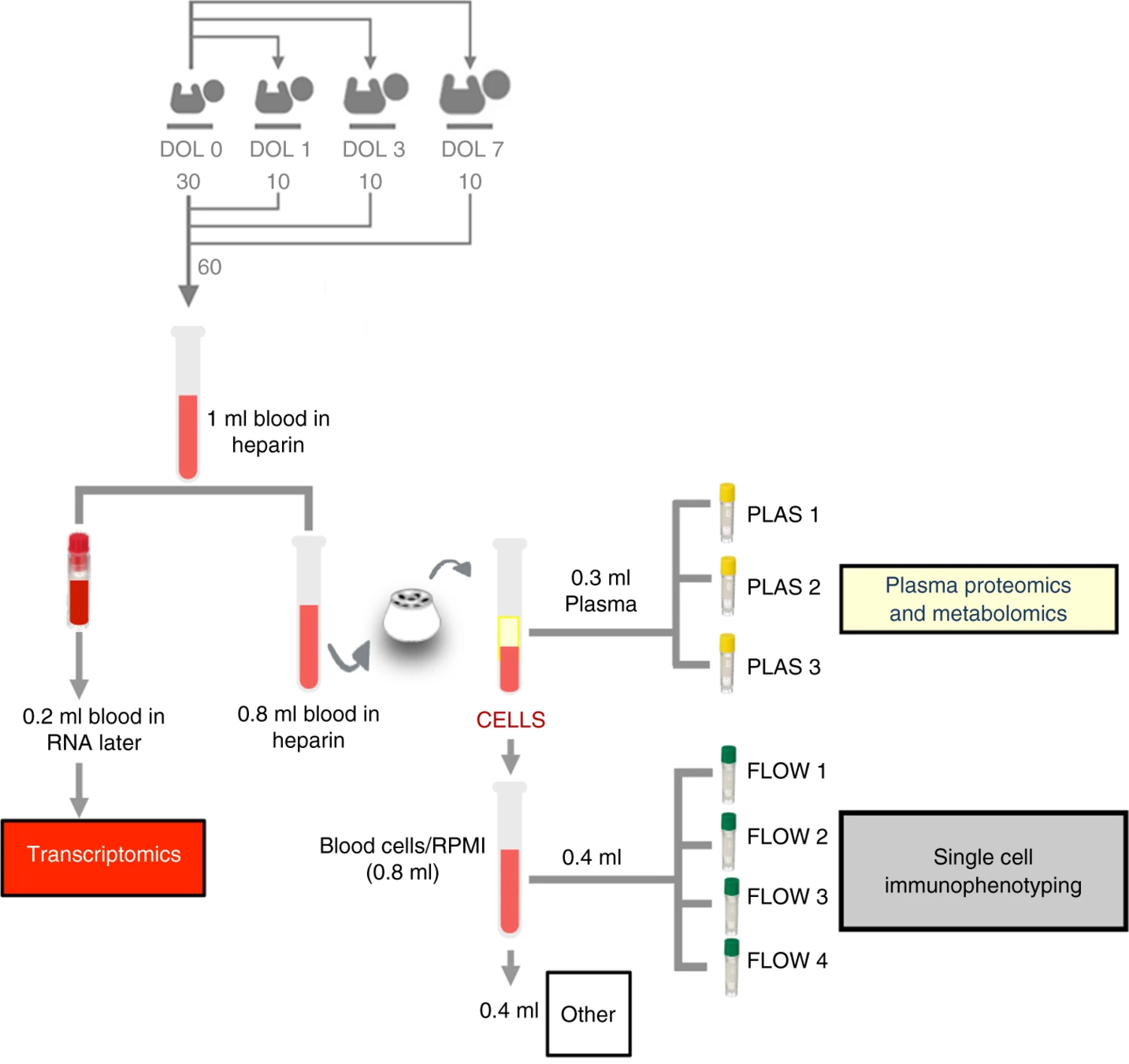
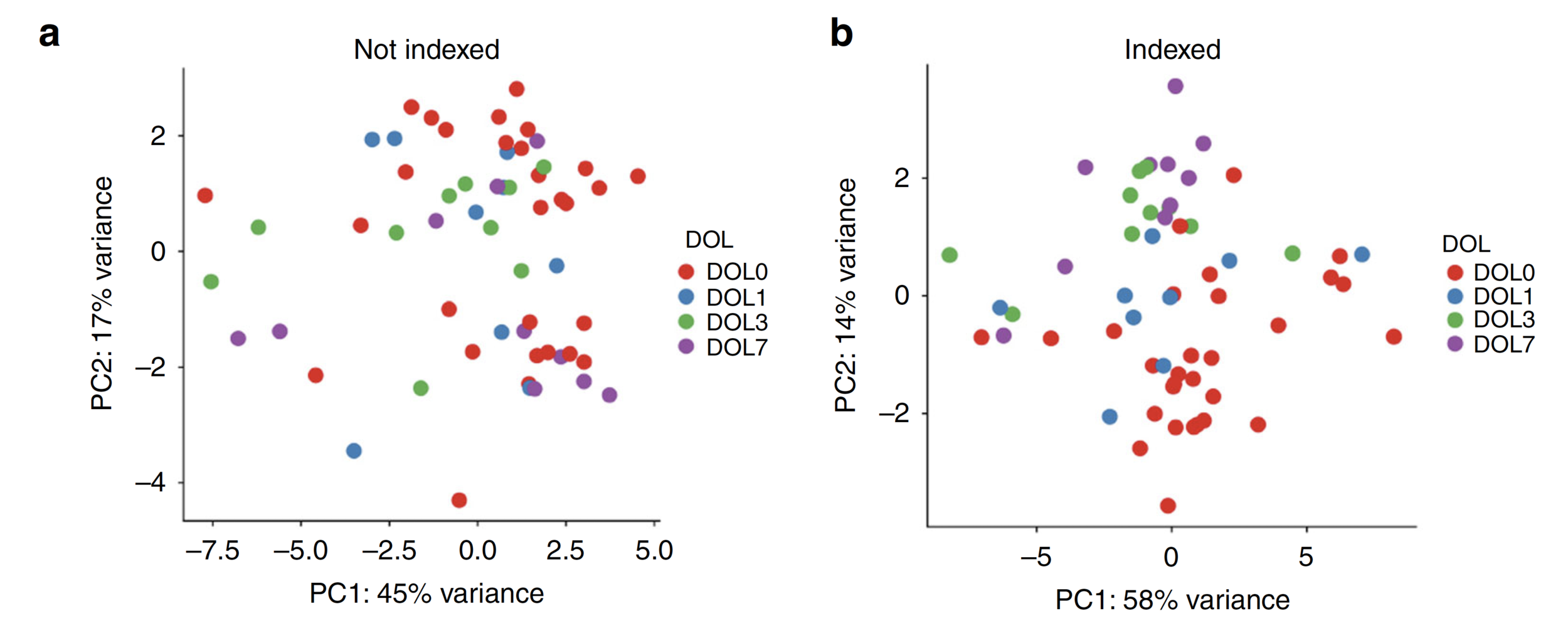
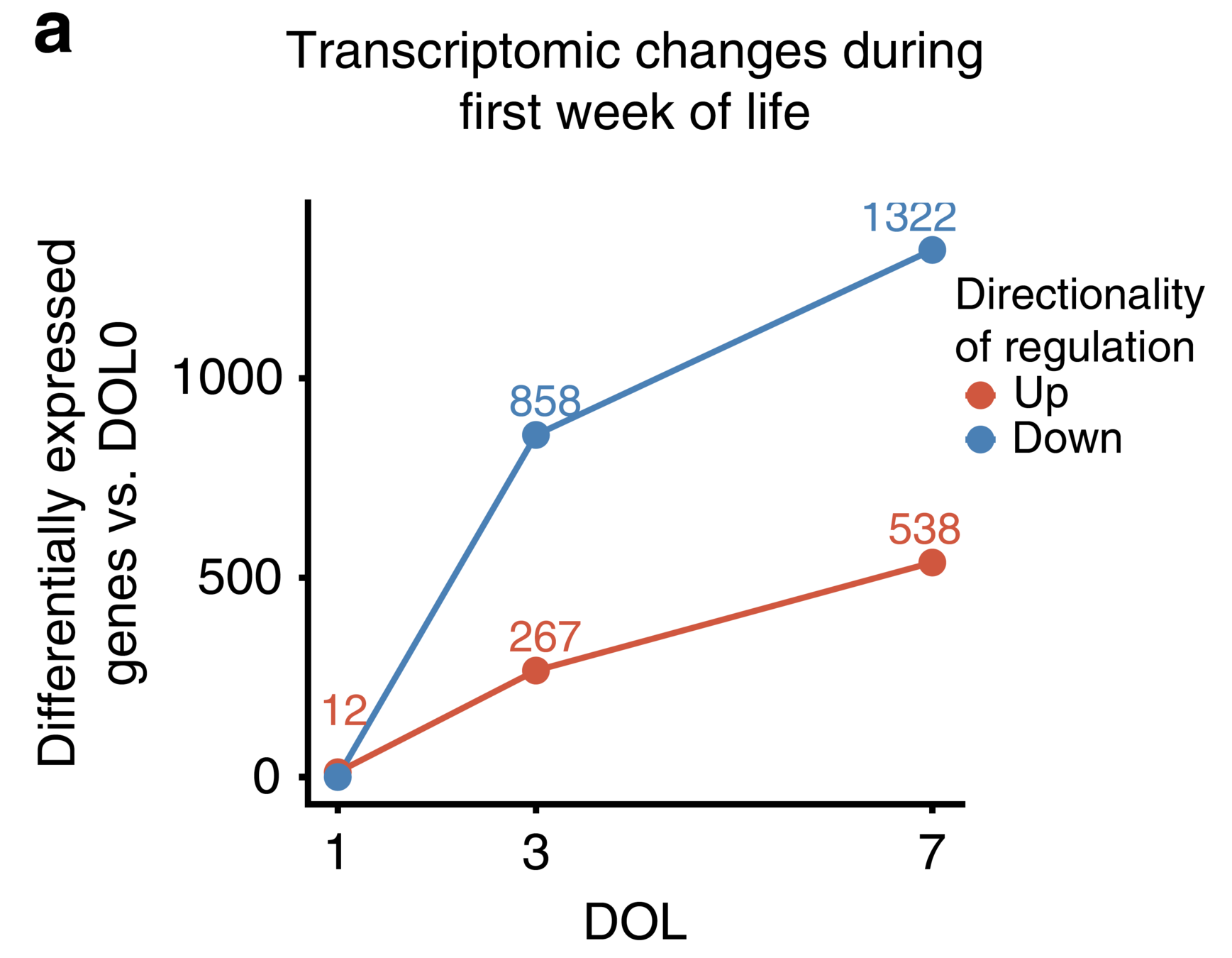
Indexing cell composition to baseline reveals
sample clustering by DOL.
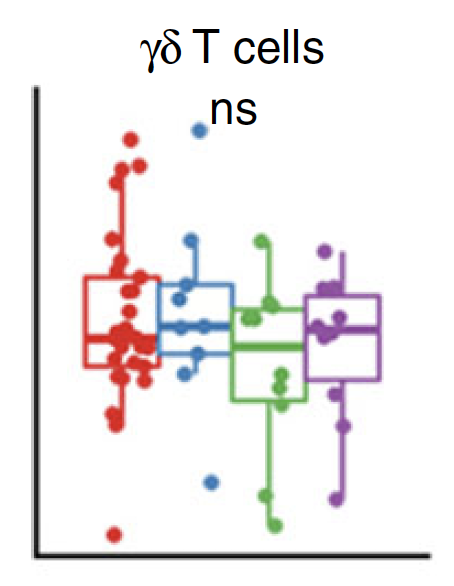
Similar for plasma cytokines/chemokine concentration
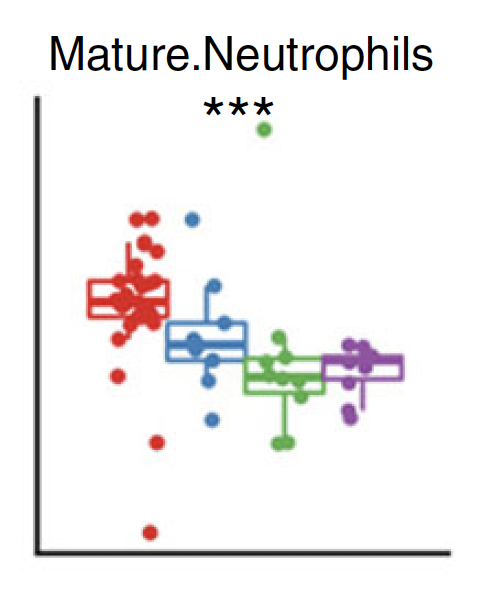

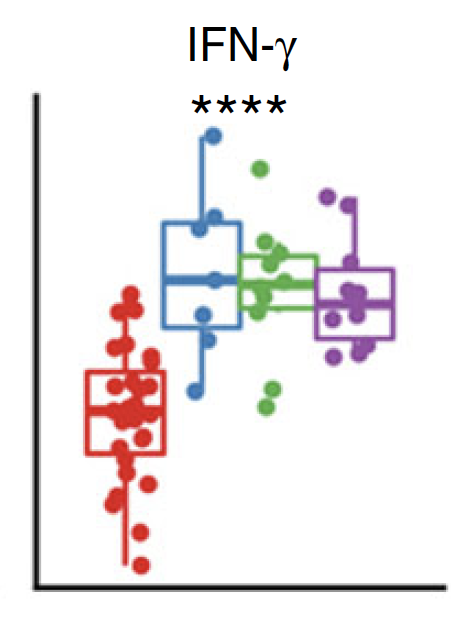
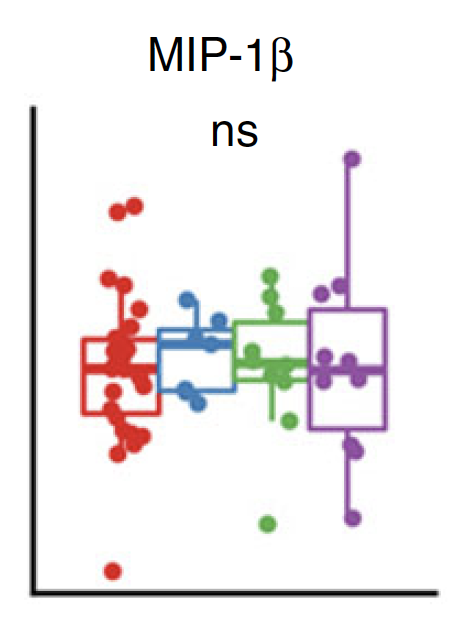
Significant
Non-significant
x 2
x 2
x 8
x 8
Normalized cell counts
Plasma concentration
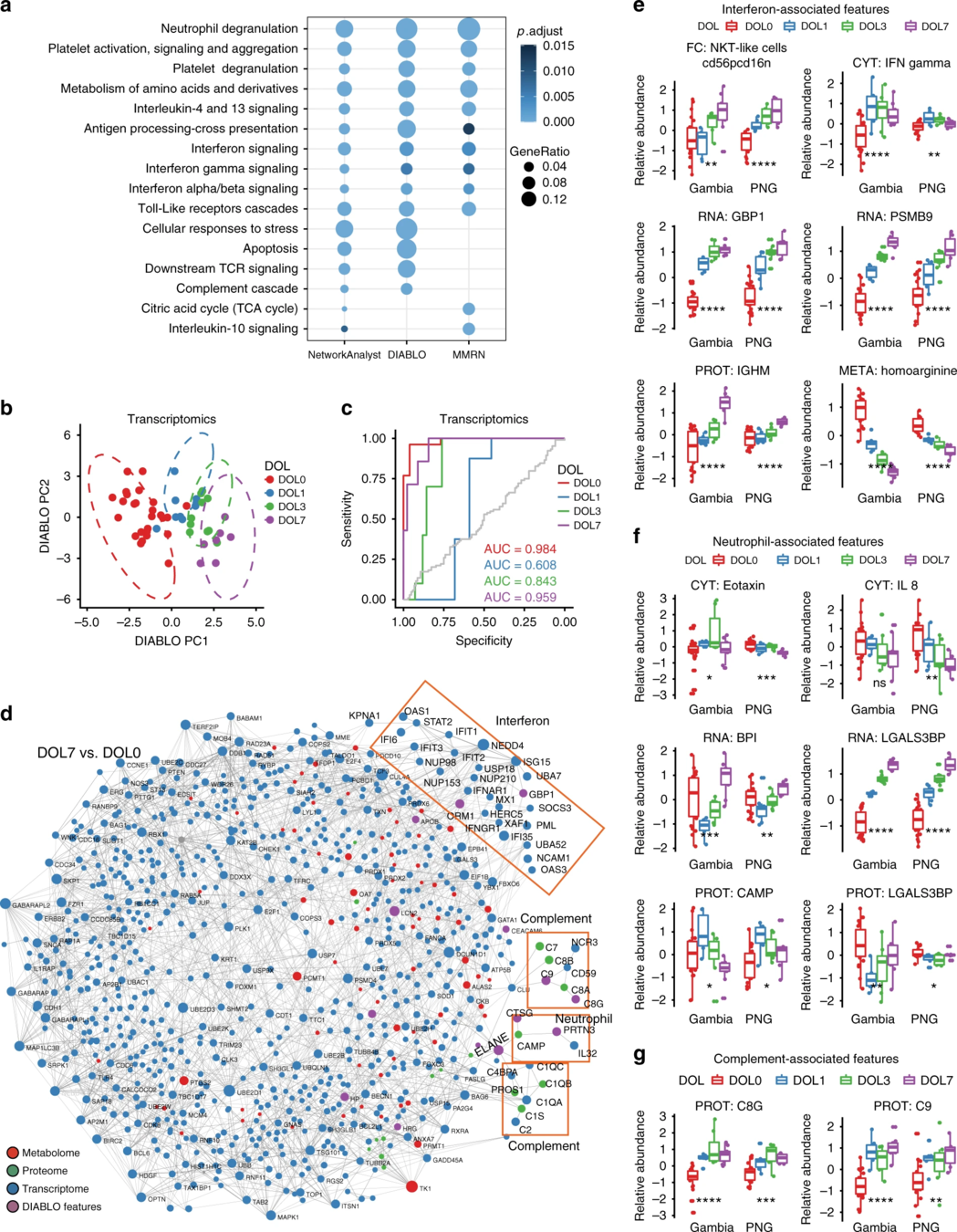
- cellular responses to stress
- detoxification of reactive oxygen
- heme biosynthesis and iron uptake
- interferon signaling
- Toll-like-receptor (TLR)
- negative regulation of Retinoic Acid Inducible Gene I (RIG-I) and complement activation
Similar for proteins and metabolites:
differences in protein composition ↑ as time ↑
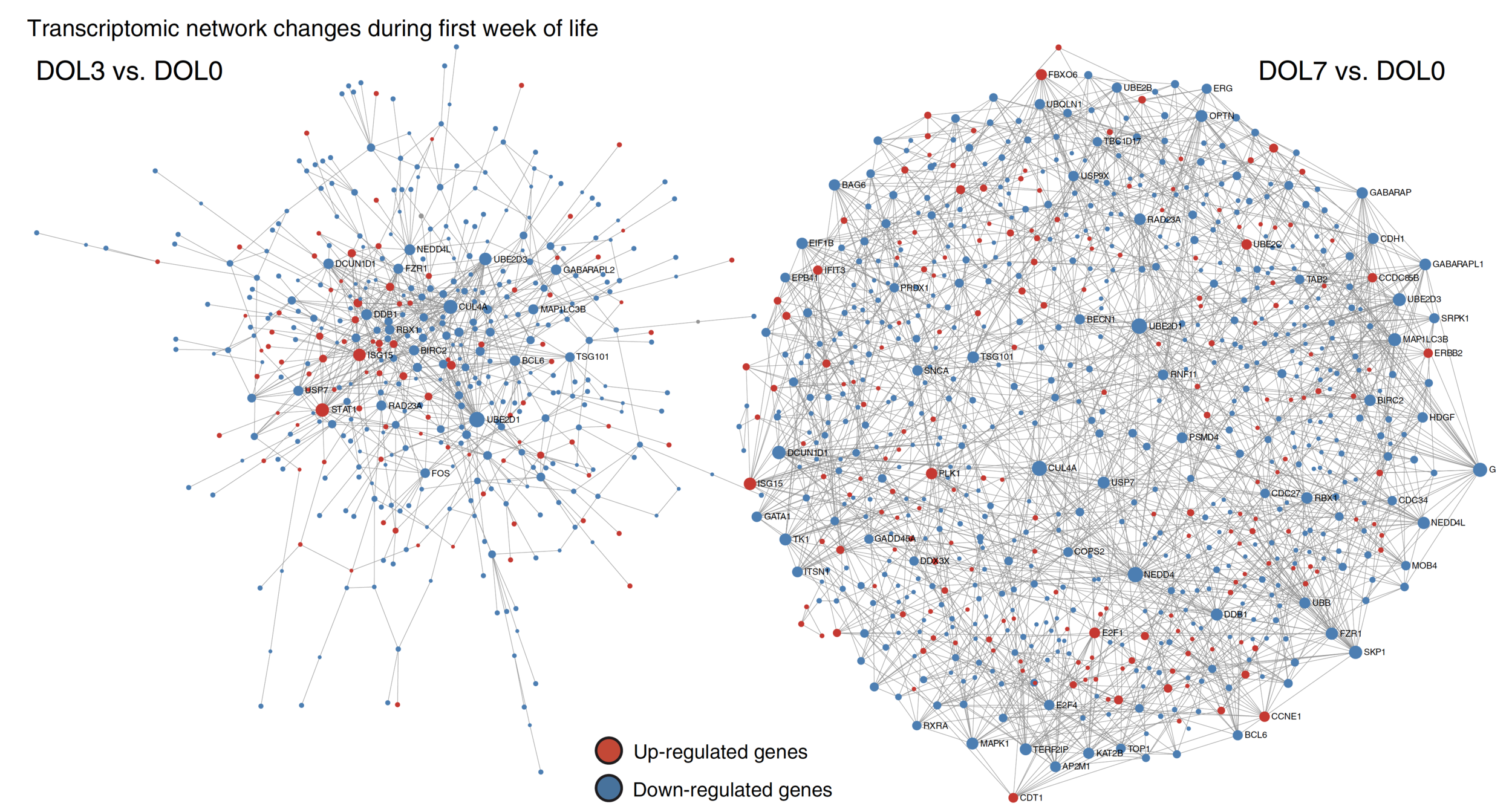
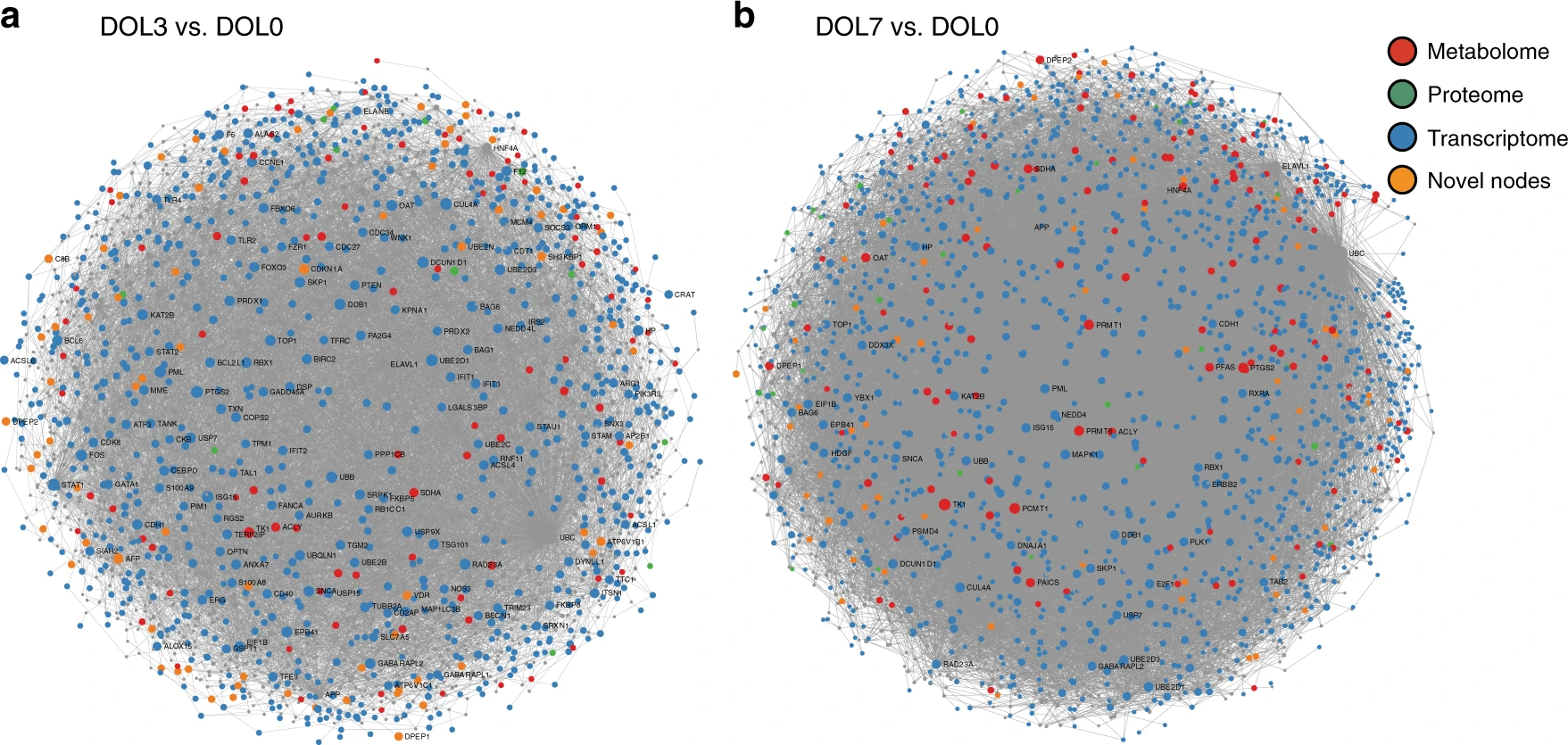
Figure 4 is not very informative. I would use histograms or stacked bar charts for each comparison. Caveat: node sizes not shown.
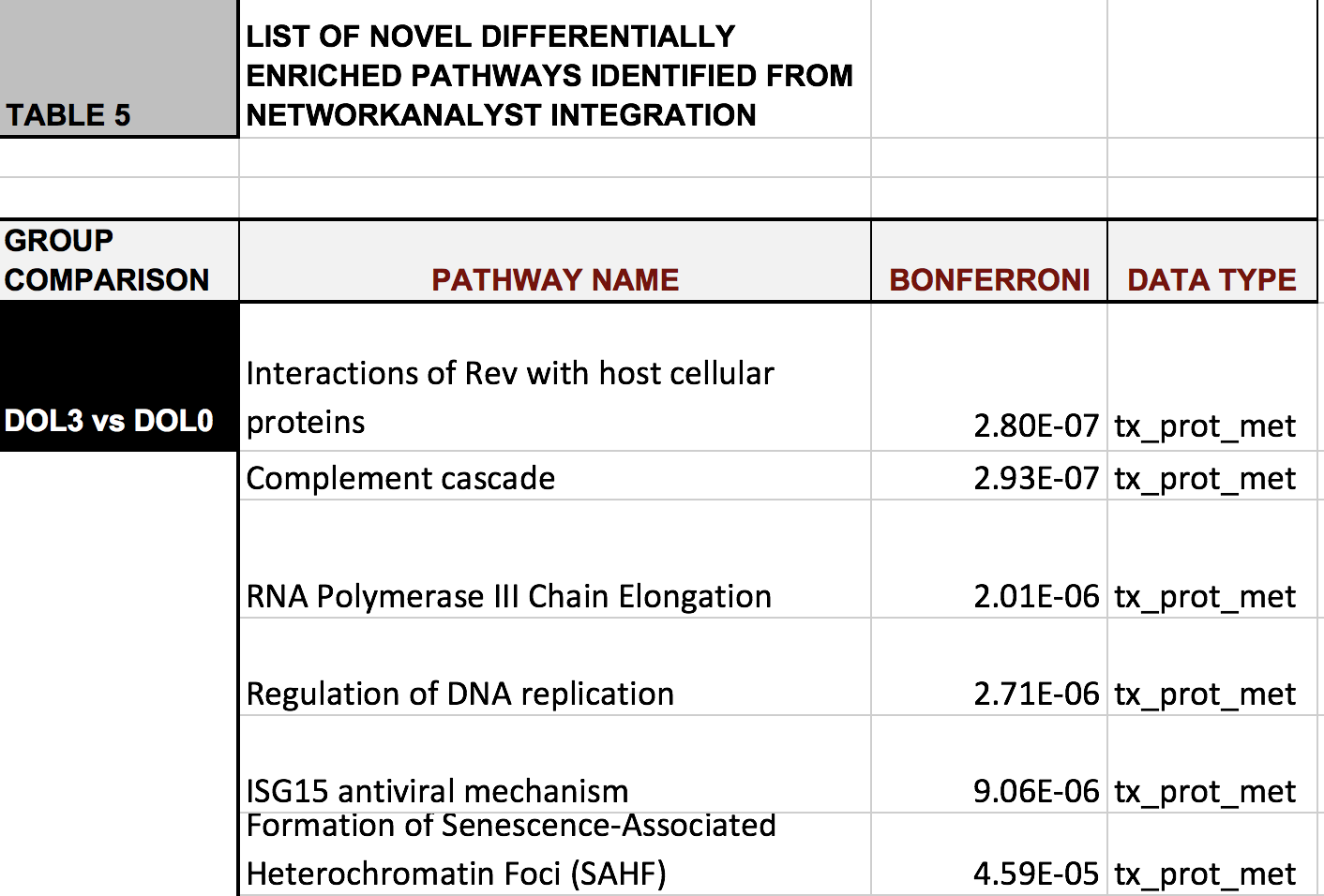
DOL3 vs. DOL0
DOL7 vs. DOL0
Metabolome
Proteome
Transcriptome
Novel nodes
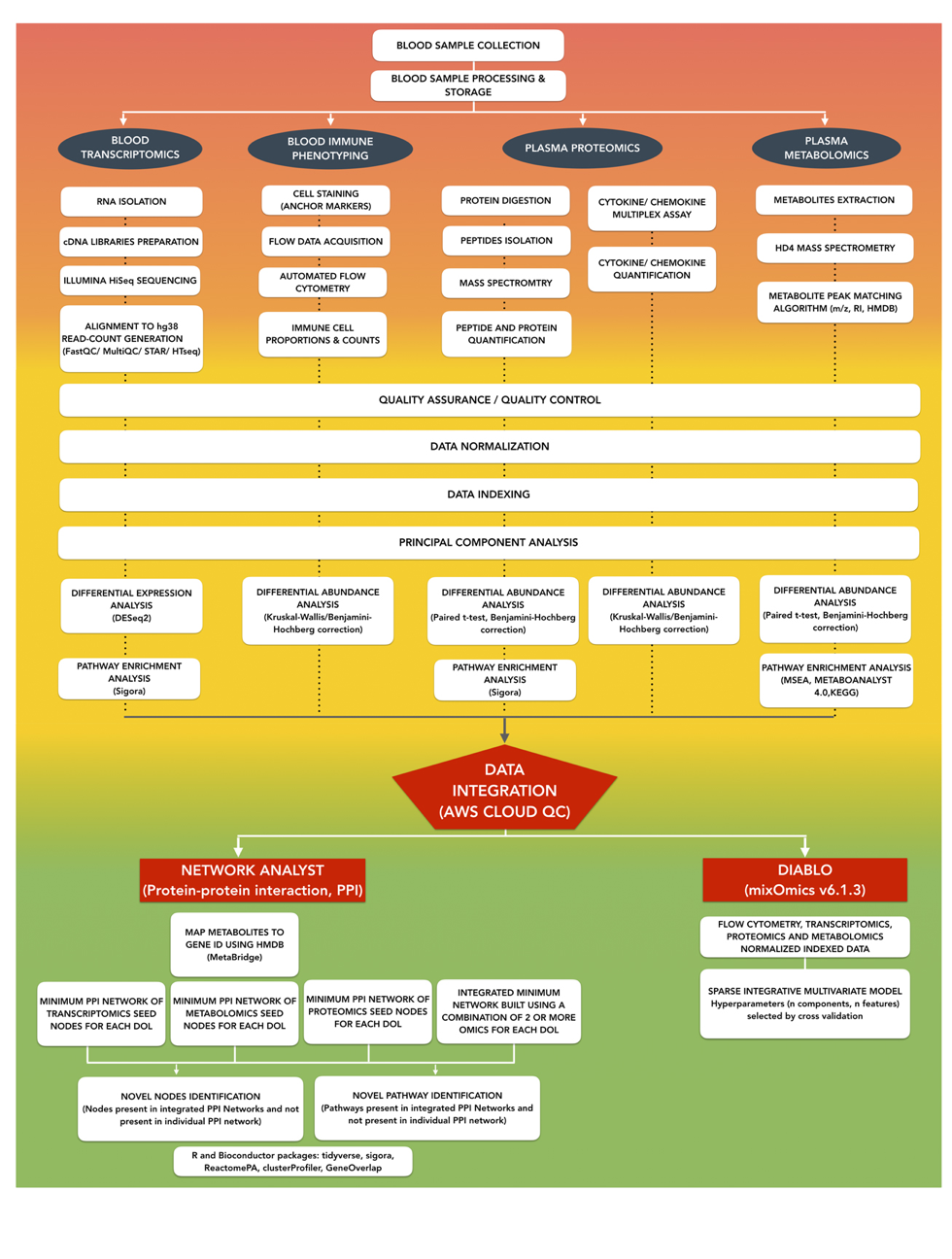
NetworkAnalyst
- creates networks based on public PPI database InnateDB/IMeX
- seed nodes: differential genes/proteins + interactors;
enzymes driving differential metabolites (up to 3 data types) - validates many findings on individual datasets
- new biological insights
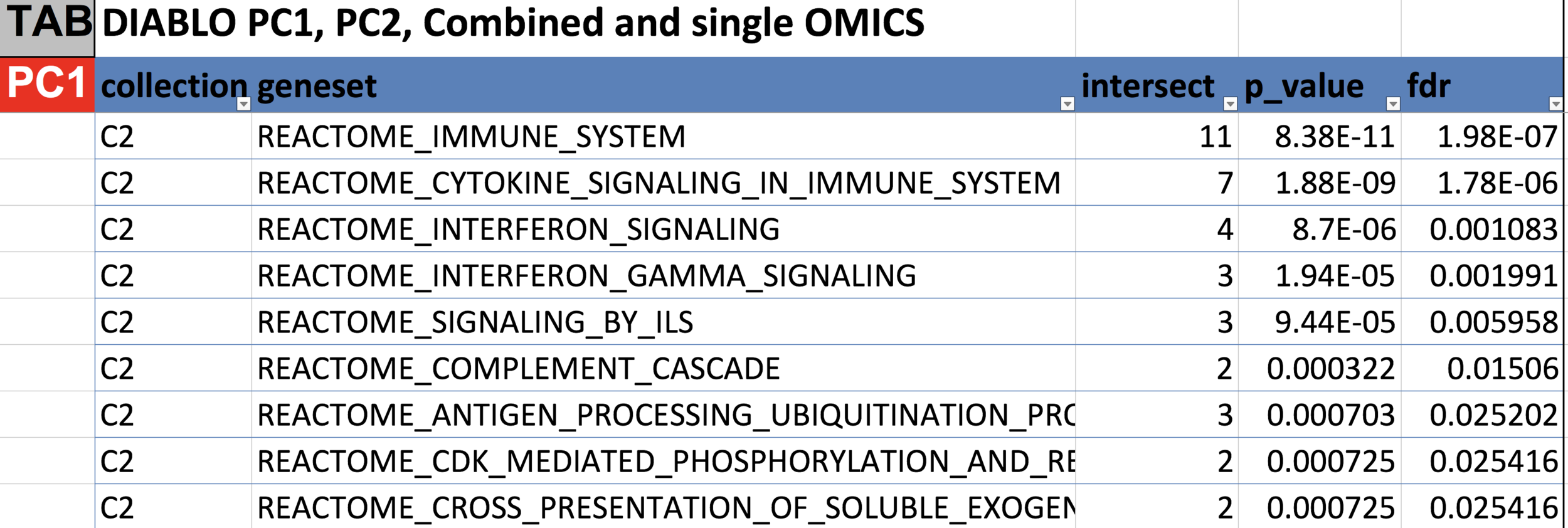
DIABLO
- addresses: complexity (n << p) & heterogeneity (scales, platforms, etc.)
- 5 data types: cells, cyt, tx, met, prot
- produces linear combinations of original data that most explain DOL + identify subset of features associated with DOL (similar to PCR?)
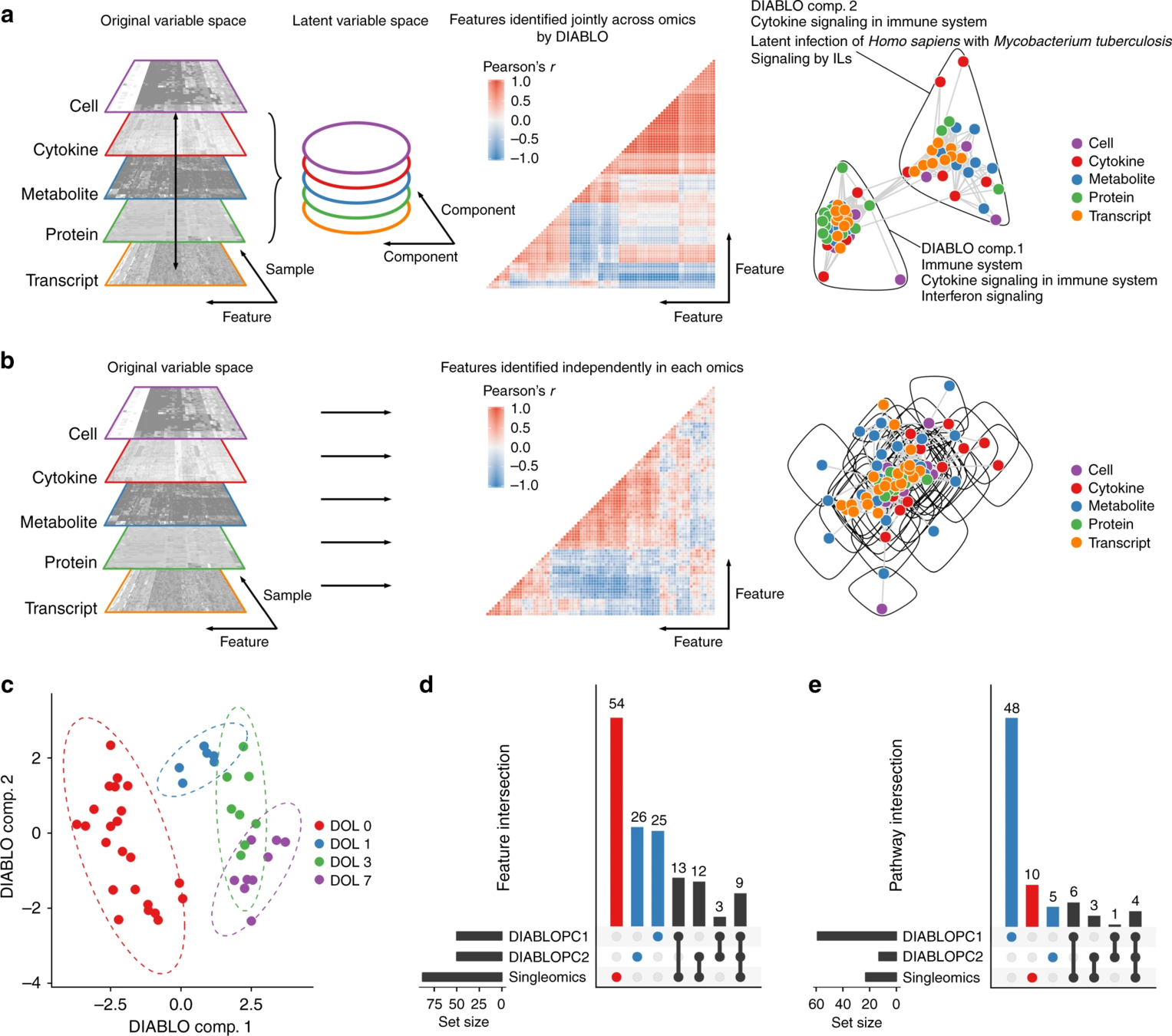
strongly associated across data types
poorly associated across data types

Features identified by DIABLO were more strongly enriched for known biological pathways.
expected
UpSet plots
MMRN
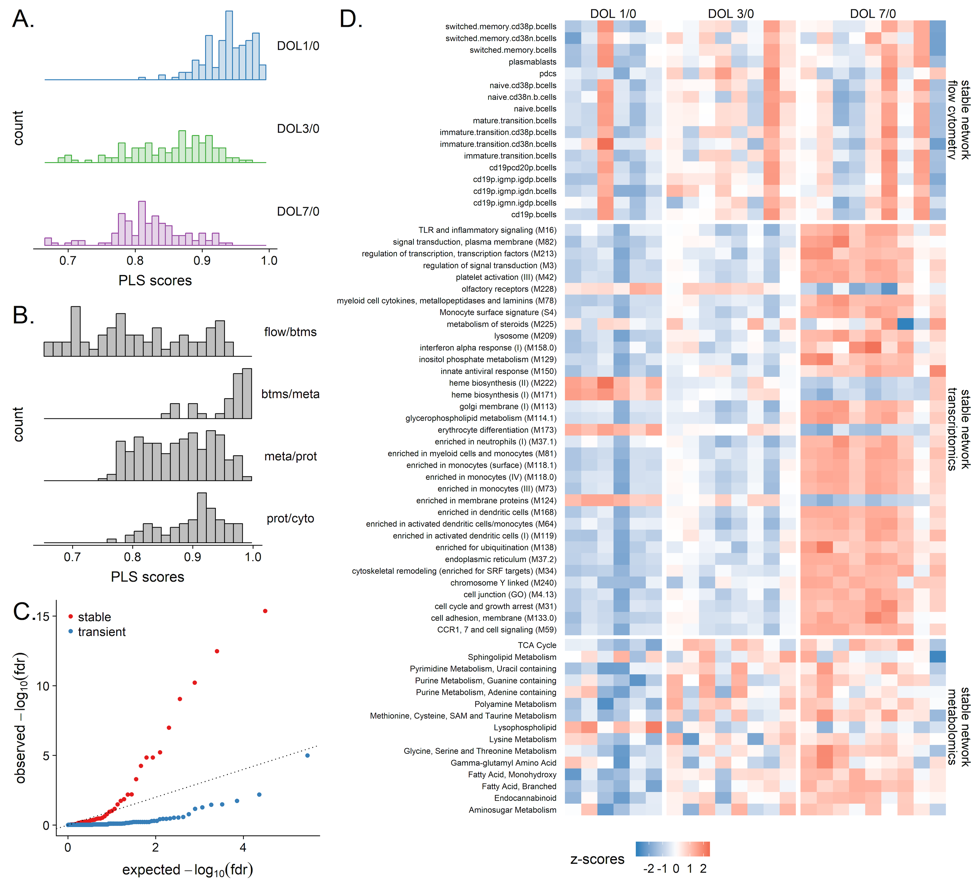
Associations between data types were strongest at DOL1.
- multiscale, multifactorial response network
- estimate correlation network across data types
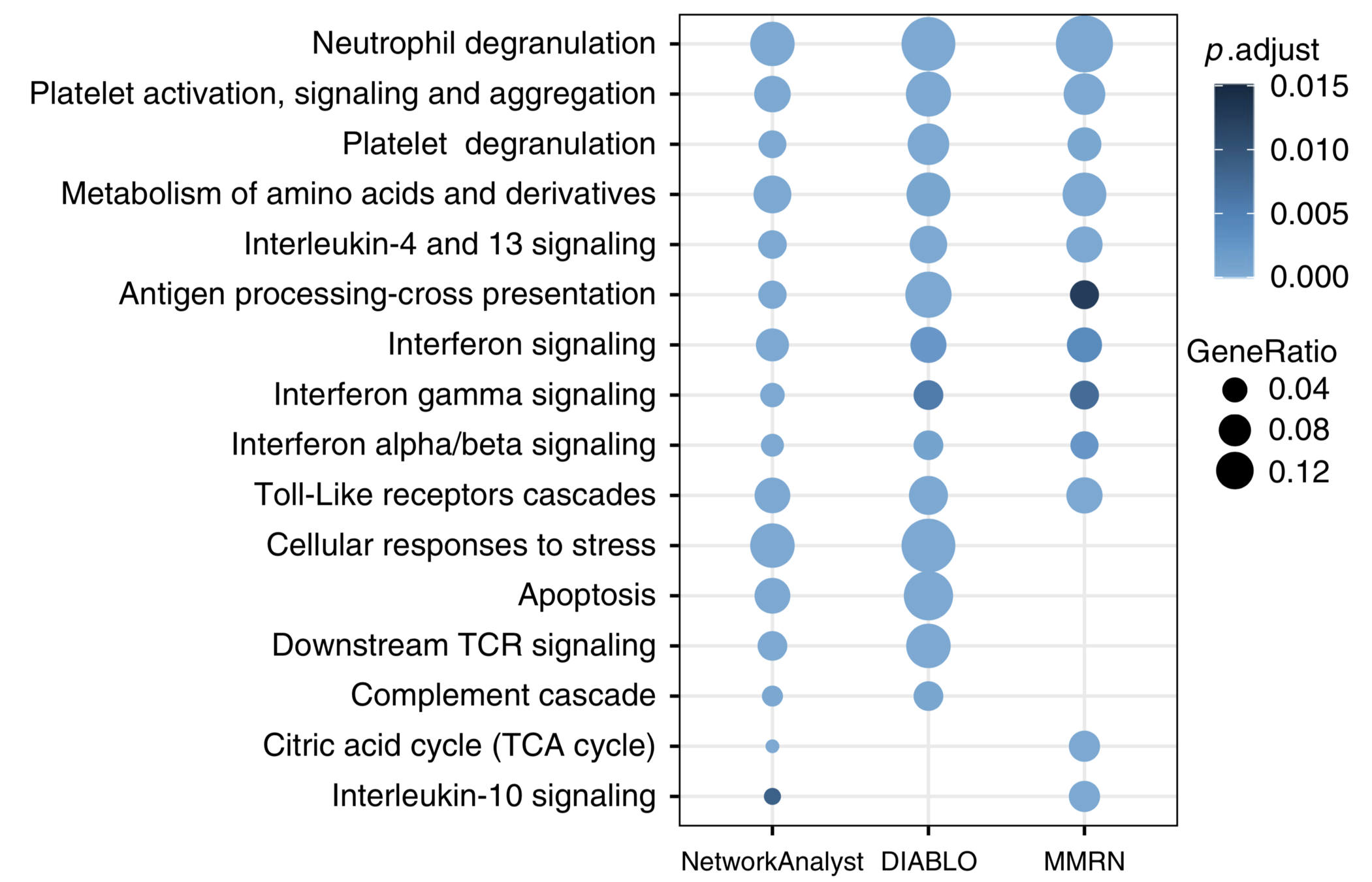
Meta integration shows significant congruence between conclusions from different platforms.
- NetworkAnalyst: 3195 nodes of minimum-connected network
- DIABLO: 428 nodes from components 1 + 2
- MMRN: 675 nodes comprising stable network
In PNG validation set, DIABLO predicted DLO well.
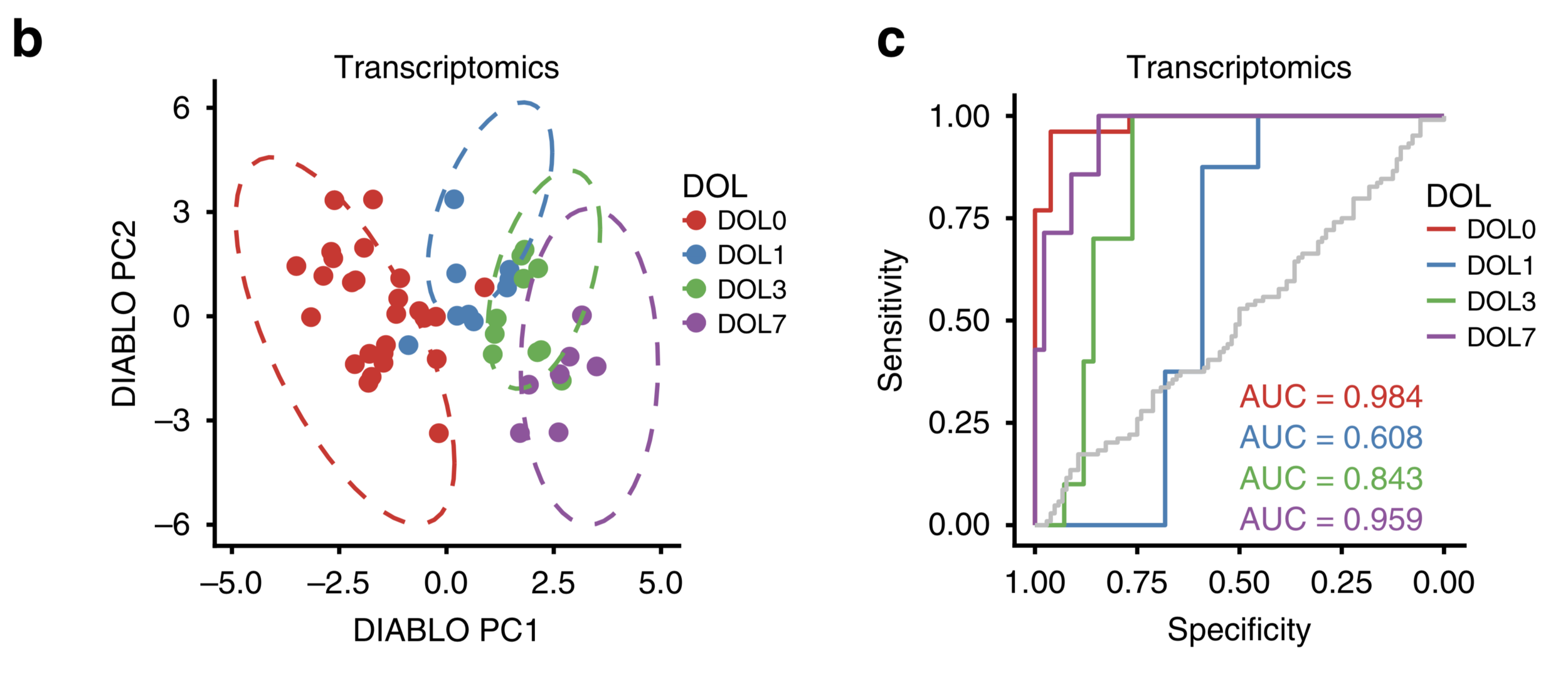
Functional network of DIABLO-selected features highlighted pathways lated to inteferon signaling, complement and neutrophil, as in Gambian cohort.
Take-home
- Developmental trajectory (trend) is consistent across the first week of life.
- Contributors: cell autonomous, immune functions
- subset selection
- MMRN
- DIABLO
- molecular information
- NetworkAnalyst
Potential ethical issues
- 1 cc of blood is quite a lot to extract from newborns. This was done twice within the first week of life. Is this standard?
The transcriptomics data presented in this publication were submitted to the NCBI Gene Expression Omnibus under accession numbers GSE111404 and GSE123070. All other datasets including immune phenotyping, Luminex, metabolomics and proteomics data were archived on ImmPort (https://immport.niaid.nih.gov/home) under accession numbers SDY1256 and SDY1412.
All data are publicly available.