Multilocus risk scores:
rethinking genetic risk scores
to account for epistasis
Trang Lê
University of Pennsylvania
BIOSTEC, Bioinformatics, Valletta, Malta
2020-02-25
- environmental exposures
- lifestyle factors
-
genetic susceptibility ← heritability
-
single nucleotide polymorphisms
(SNPs)
-
single nucleotide polymorphisms
What do we mean when we say disease risk
Individuals with high genetic risk scores for a disease are more susceptible to that disease
and may benefit from prioritized interventions.

Polygenic Risk Score (PRS) aggregates genetic risk factors.
\[PRS(i)=\sum_{j=1}^{k} \beta_j \times SNP_{ij}\]
a
effect size of \(SNP_j\) in discovery sample from OLS or logistic reg.
a
subject
number of minor alleles at \(SNP_j\)
for subject \(i\)
a
Probabilistic susceptibility
→ identify groups of individuals who need prioritized interventions and screenings
→ life planning
Current PRS model does not account for the effect of SNP-SNP interactions.
but...
\[PRS(i)=\sum_{j=1}^{k} \beta_j \times SNP_{ij}\]
Our goal is to expand risk score models
beyond main effect of individual variants on disease risk.

Polygenic Risk Scores
- individual effects
- manual encodings
Multilocus Risk Scores
- interaction effects
- automatic encodings
MRS method utilizes
model-based multifactor dimensionality reduction (MB-MDR).

HLO matrix
High = 1
Low = -1
O = 0
\(\gamma\)
\[MRS_d(i) = \sum_{j = 1}^{k_d} \gamma_j \times \textrm{HLO}_j(X_{ij})\]
\[PRS(i)=\sum_{j=1}^{k} \beta_j \times SNP_{ij}\]
a
interaction dimension
a
SNP combination
a
\(j^{th}\) HLO matrix
a
MB-MDR test statistic
a
subject
\[MRS_d(i) = \sum_{j = 1}^{k_d} \gamma_j \times \textrm{HLO}_j(X_{ij})\]

\[MRS_2(Alice) = 0.8\times 1 + \cdots\]
Bob has the combination (aa, aa) for these two SNPs.
\(\gamma = 0.8\)
\[MRS_2(Bob) = 0.8\times (-1) + \cdots\]
Suppose Alice has the combination (AA, Aa) and
450 datasets: 1000 individuals and 10 SNPs
For an individual, each genotype was randomly assigned with:
- 1/2 probability heterozygous Aa (coded as 1)
- 1/4 probability homozygous major AA (coded as 0)
- 1/4 probability homozygous minor aa (coded as 2).
How do we simulate data with interaction effects?
Evolutionary-based method: Heuristic Identification of Biological Architectures for simulating Complex Hierarchical Interactions (HIBACHI)
-
80% training: run MB-MDR to obtain
- the \(\gamma\) coefficients
- the HLO matrix
-
20% holdout:
- calculate MRS for each individual
- assess the performance of the MRS by comparing the area under the Receiving Operator Characteristic curve (auROC) with that of the standard PRS method.
How do we test our method?
MRS produces improved auROC in 335 of 450 simulated datasets.

~50% auROC increase at the second peak
\(MRS = MRS_1 + MRS_2\) increasingly outperforms standard PRS
as dataset contains more main and interaction effects.

\[ME = \sum_{j} I(SNP_j; Y) = \sum_{j} \left(H(Y) - H(Y|SNP_j)\right).\]
\[SE = \sum_{j} IG(X_j; Y) = \sum_{j} \left(I(SNP_{j_1}, SNP_{j_2}; Y) - I(SNP_{j_1}; Y) - I(SNP_{j_2}; Y)\right)\]
Next steps
- Apply MRS to real-world data
- increase number of variants in simulation
- pre-select variants
- increase computational efficiency
- Aggregate other risk factors
Hoyt Gong
Elisabetta Manduchi
Patryk Orzechowski
Jason H. Moore
Acknowledgements

funded by the
National Institutes of Health
BIOSTEC organizers
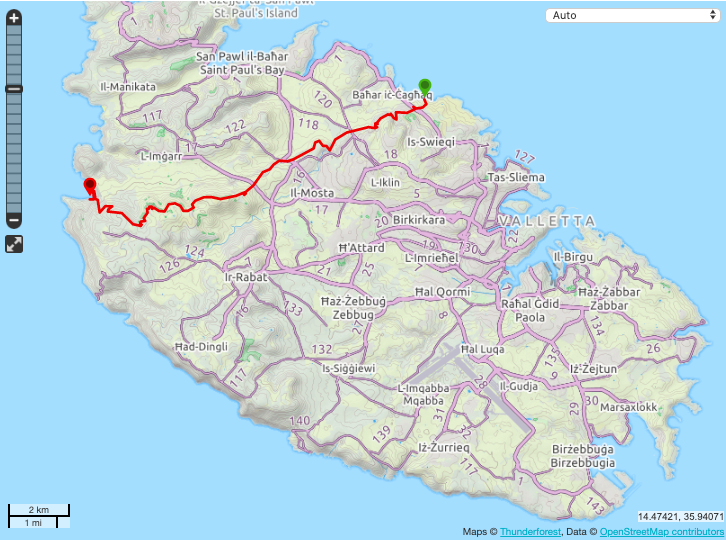
Want to run the Victoria Lines?
Meet me at Excelsior Level 1, 5:45 AM tomorrow (Wednesday).
My (optimistic) estimate: ~ 3 hours, 18 km, 600 m vertical gain