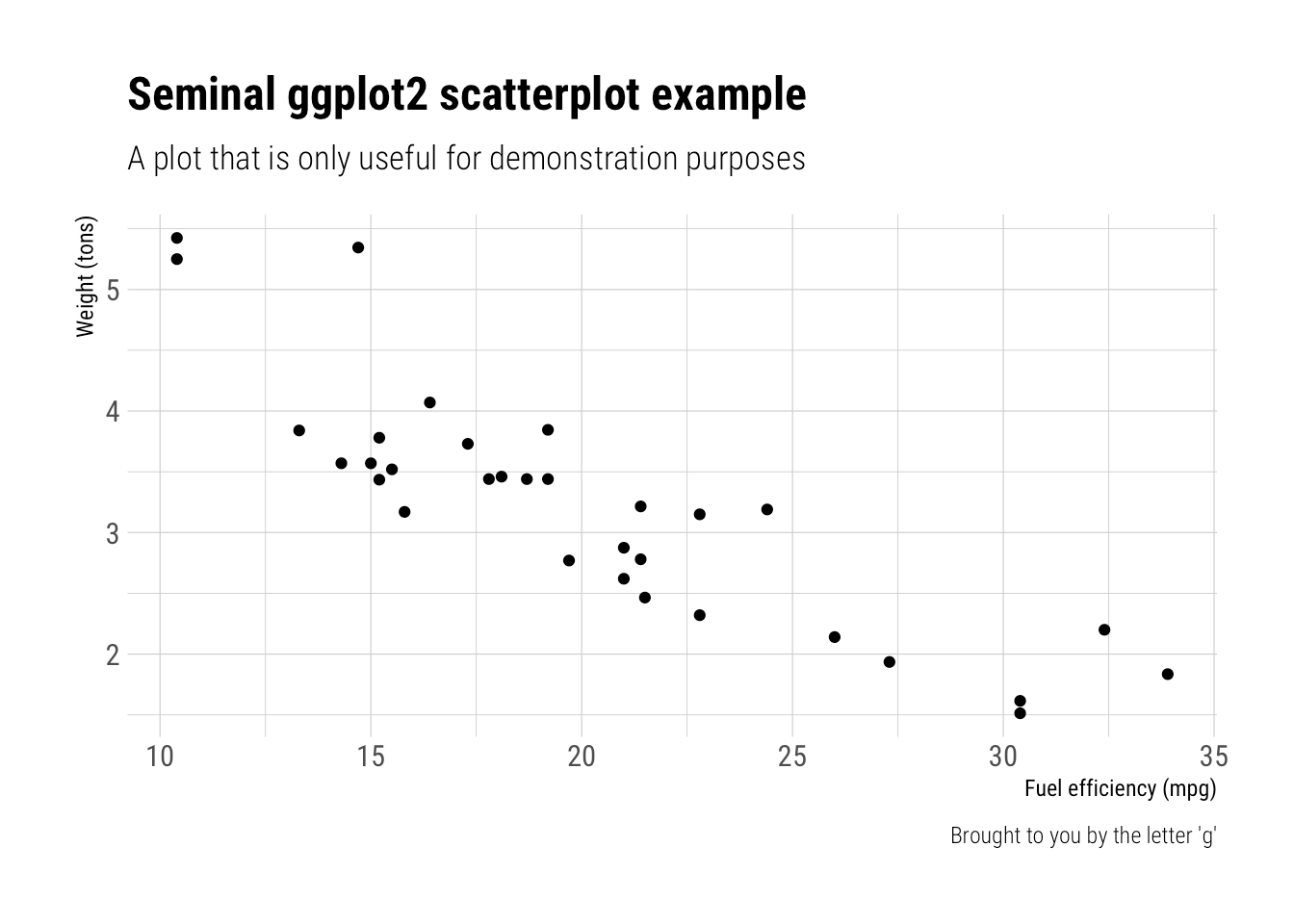
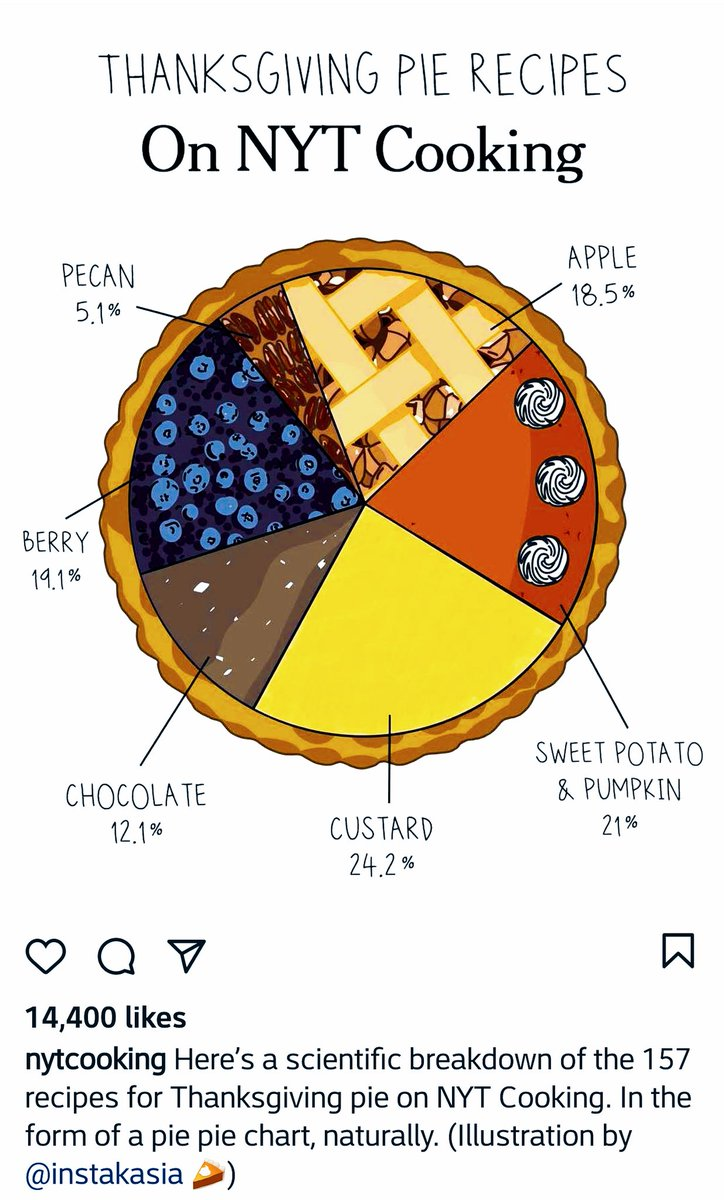
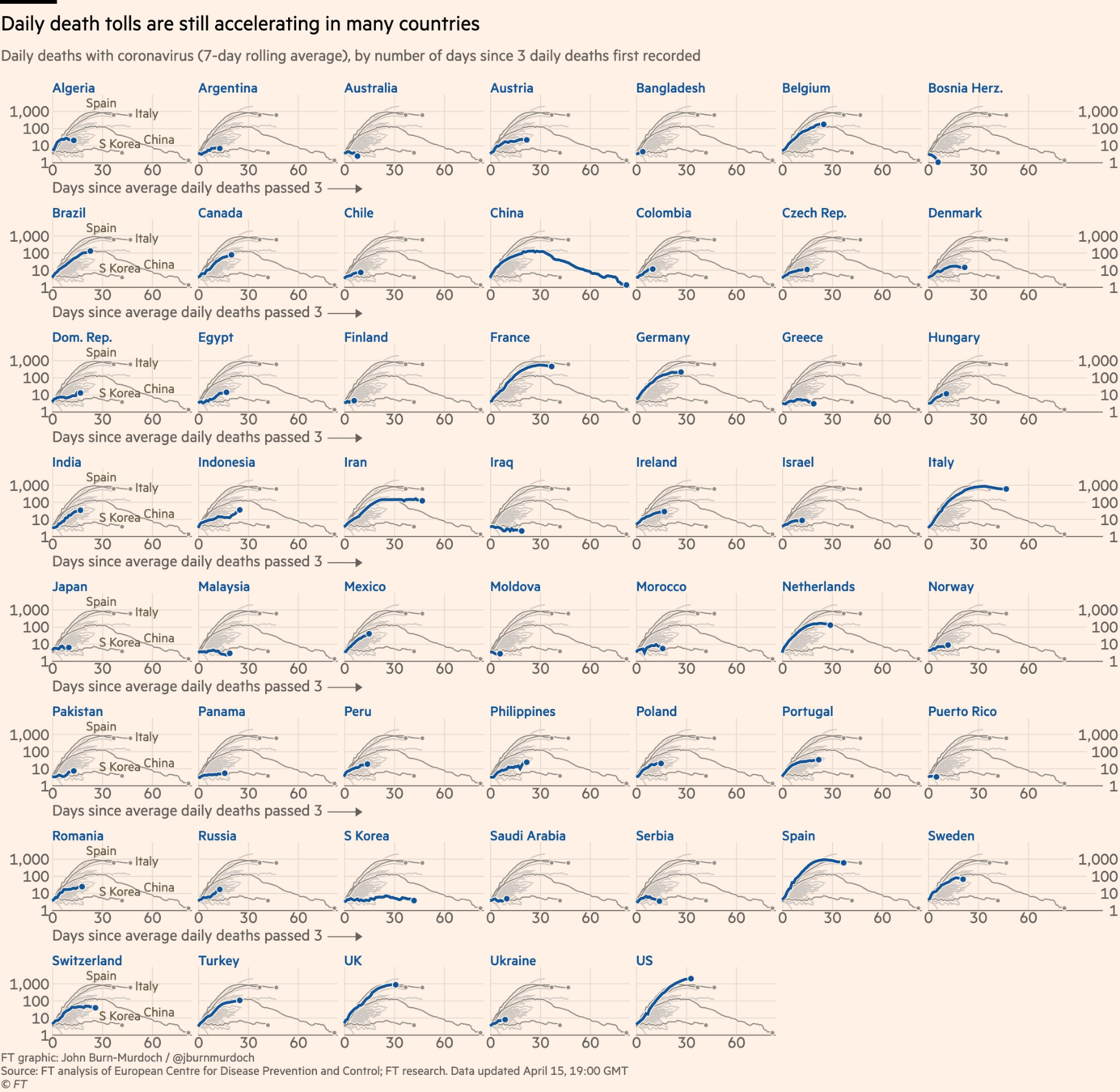
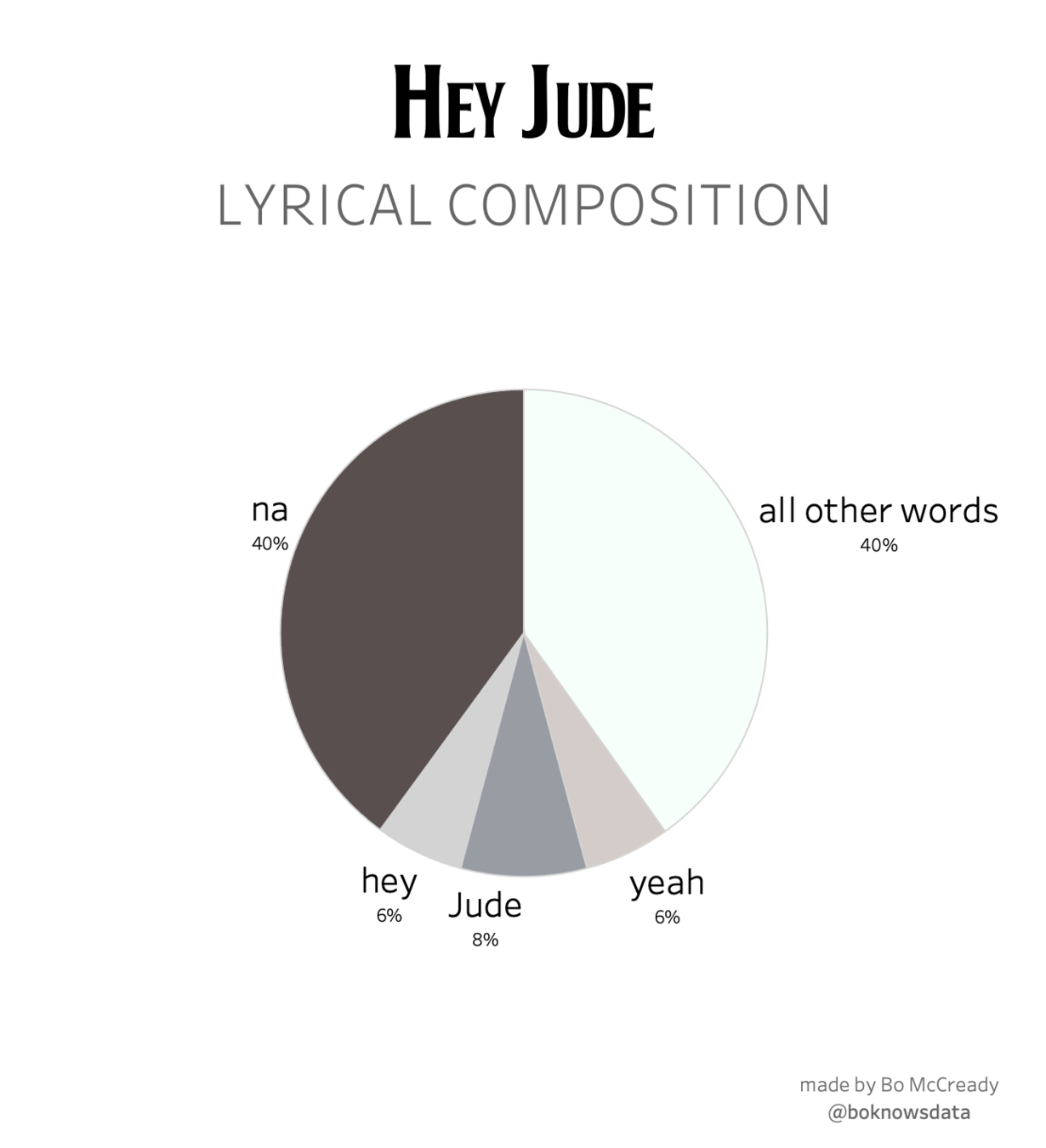
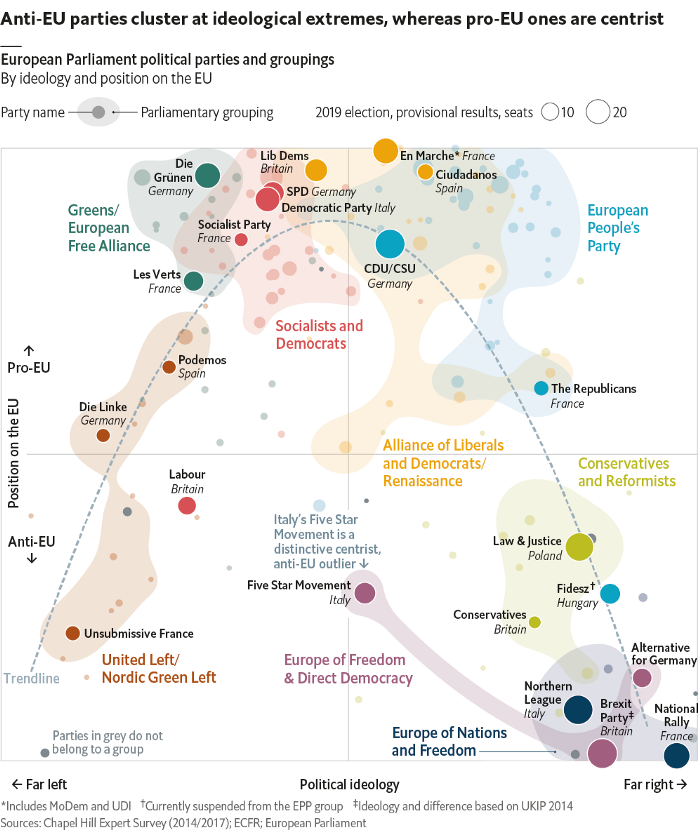
Take a sad chart and make it better
with ggplot2 and other tools
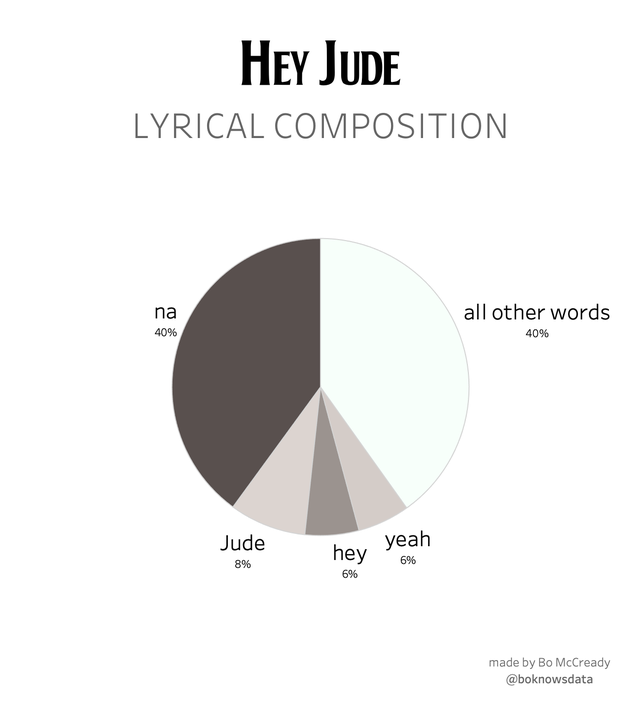


@instakasia
Rules don't always apply.
Never use 3D.
Never use 3D.
@tylermorganwall
Use < 7 colors.
Use < 7 colors.
@trang1618



I'm Trang Le.
I have made some (very) bad charts.
https://trang.page
What's your favorite R package?
Let's learn a bit about you!
reduce cognitive burden
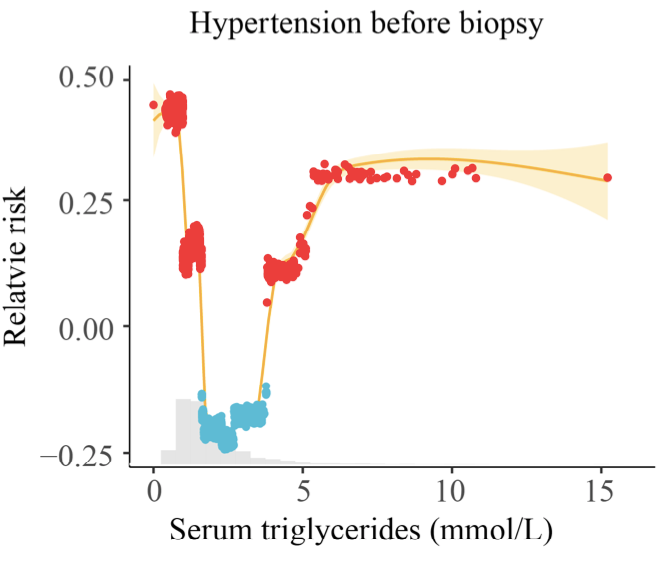
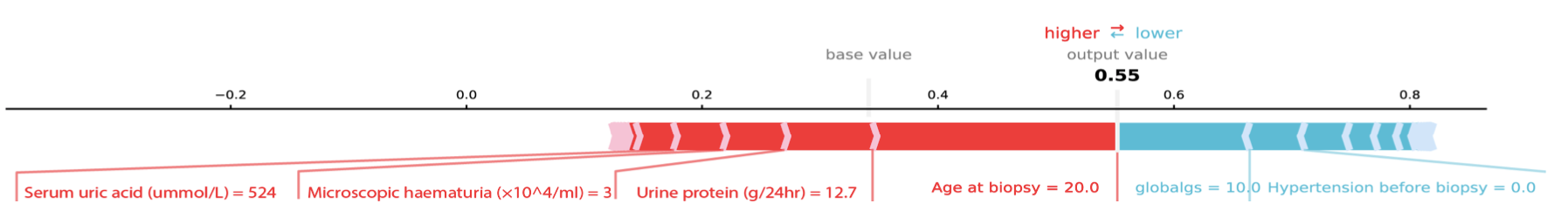
- direct labeling
- reduce number of labels
- highlighting
- consistent color scheme
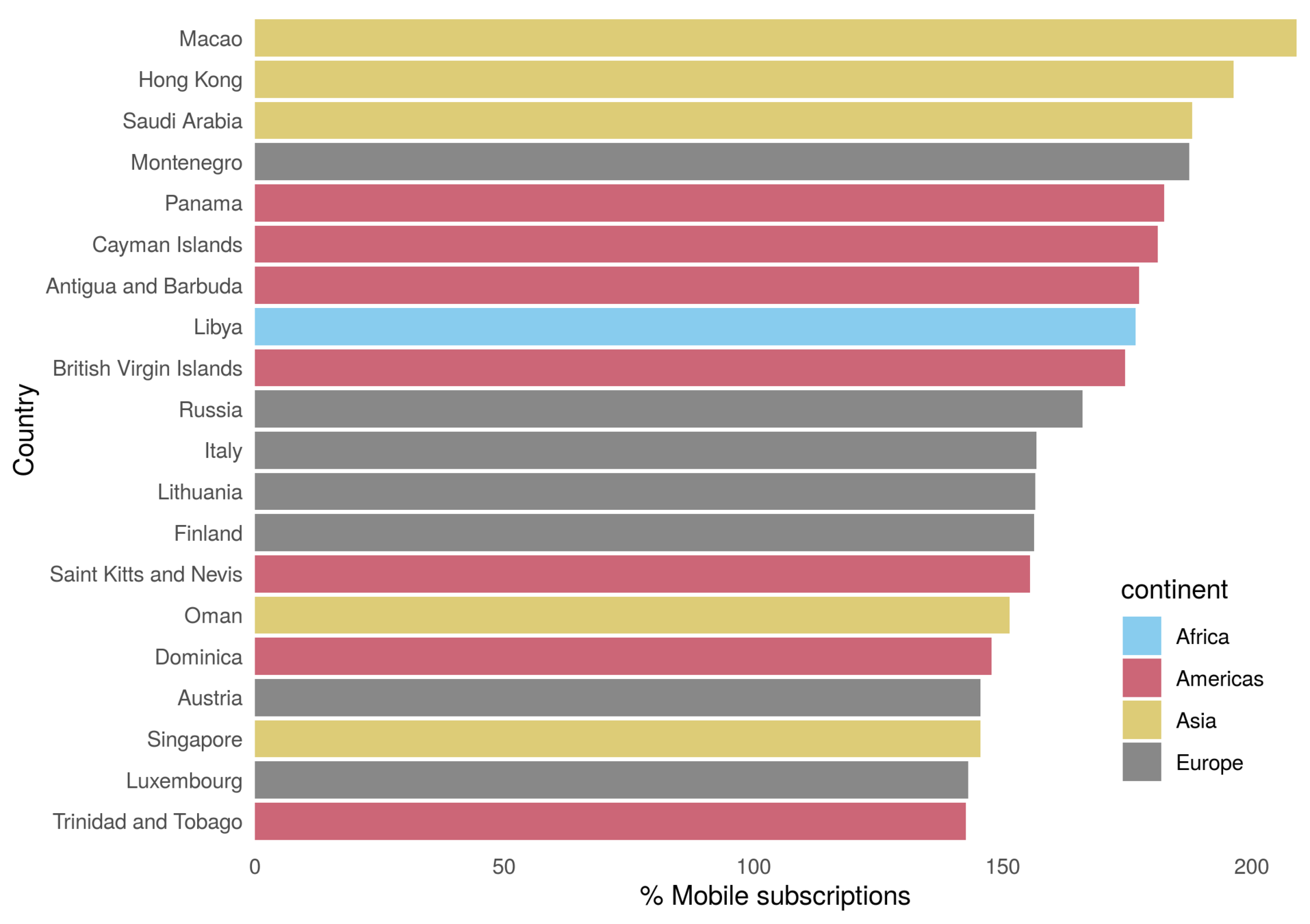
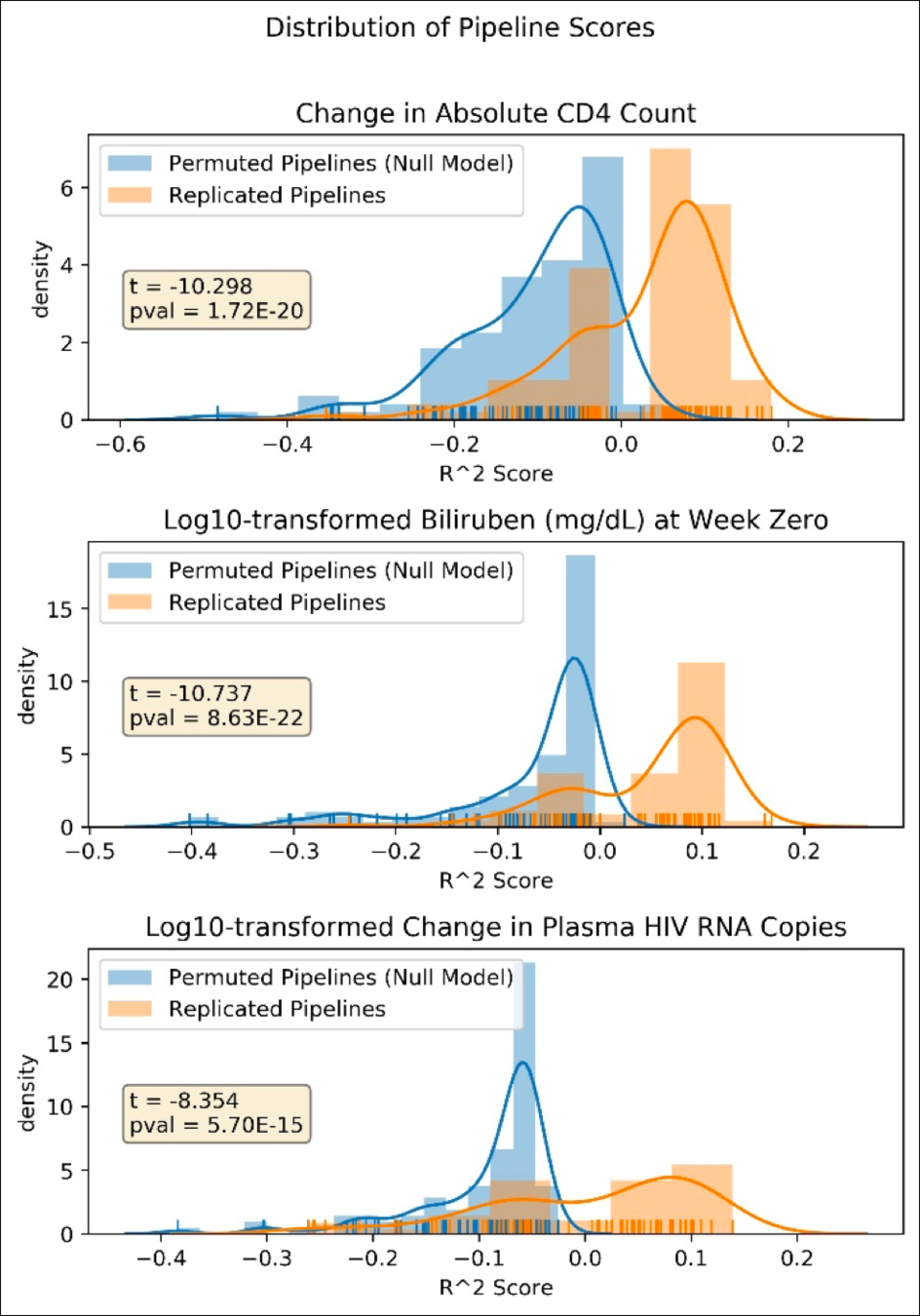
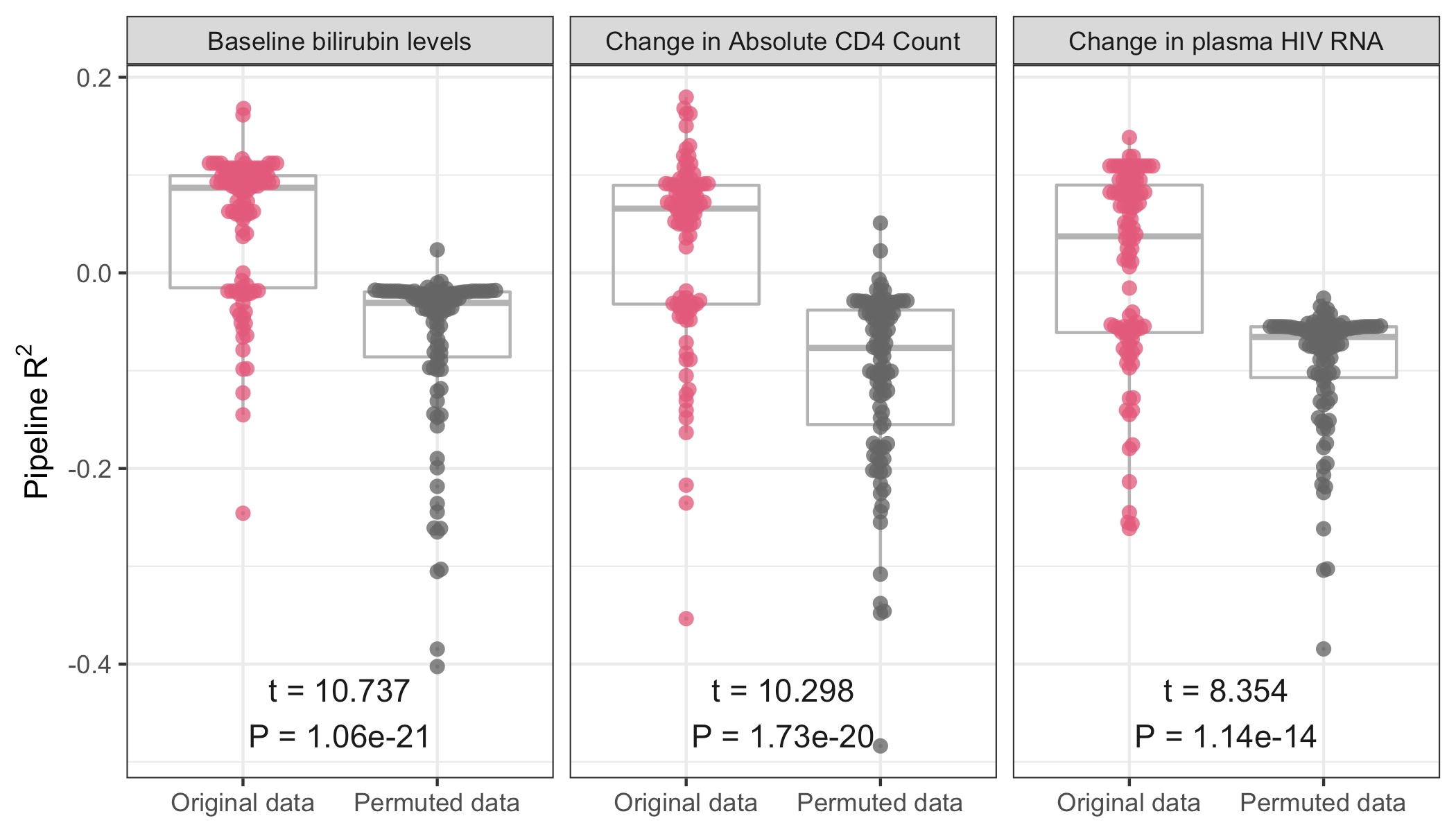
Risk of hypertension
What is the ggplot term for
facet_grid(
cols = vars(...),
scales = 'free',
space = 'free')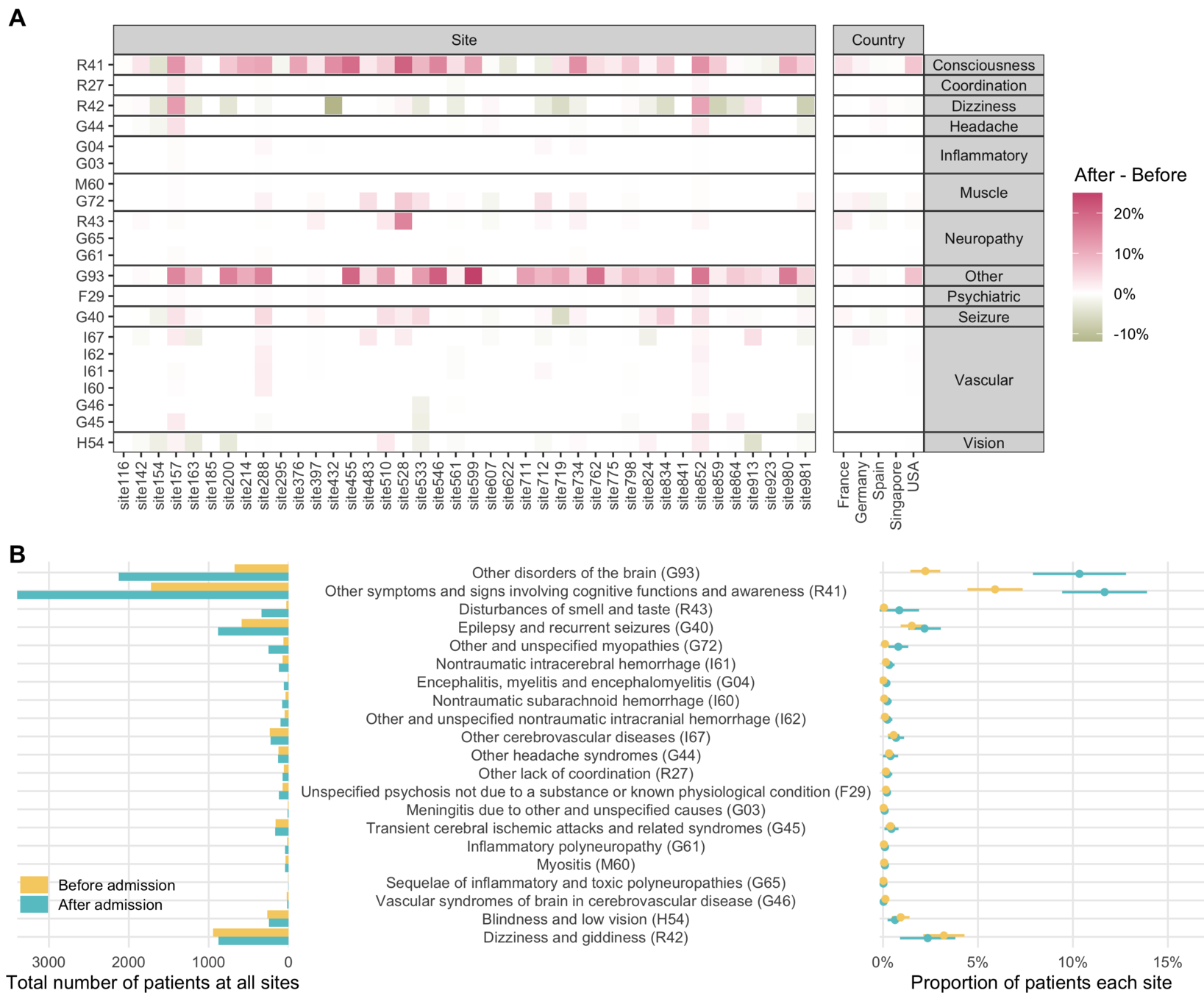
scales = 'free'
space = 'free'
"strips"
facets are extremely useful to improve simplicity!
identify key idea

select chart type
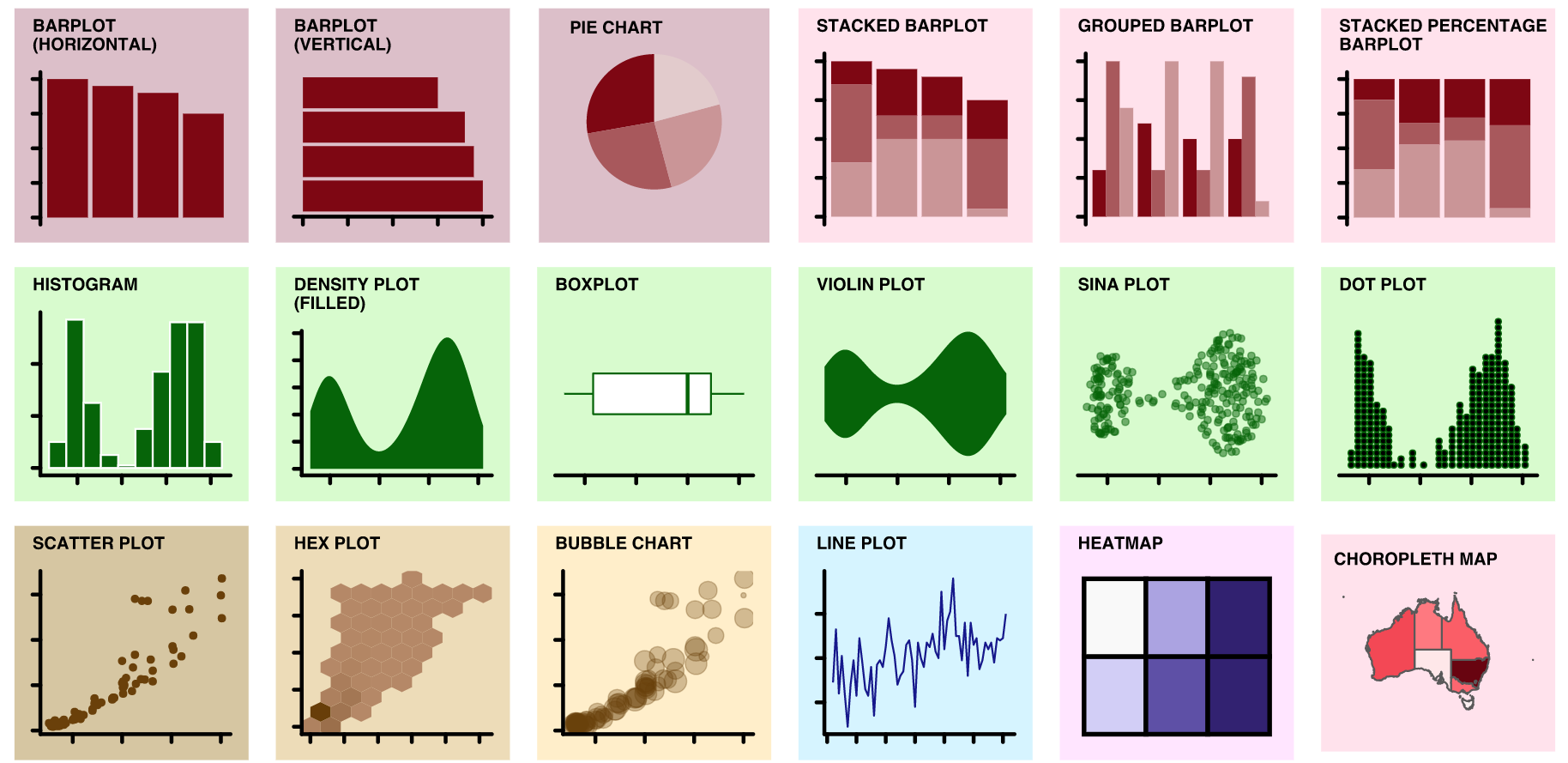
some examples
I would use histograms or stacked bar charts for each comparison.
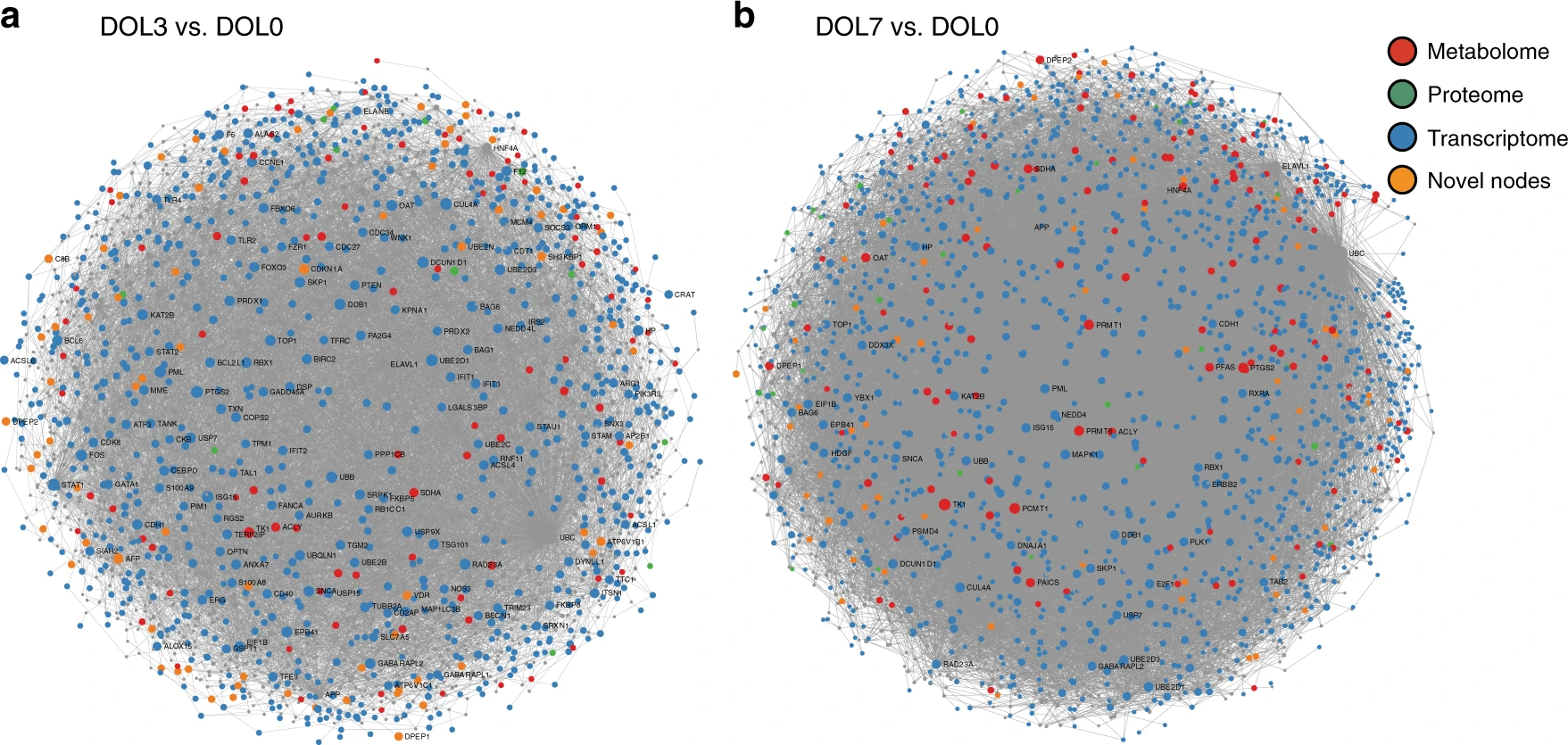
Trade-off: node sizes, edges not shown.
Day 3 vs. Day 0
Day 7 vs. Day 0
Metabolome
Proteome
Transcriptome
Novel nodes
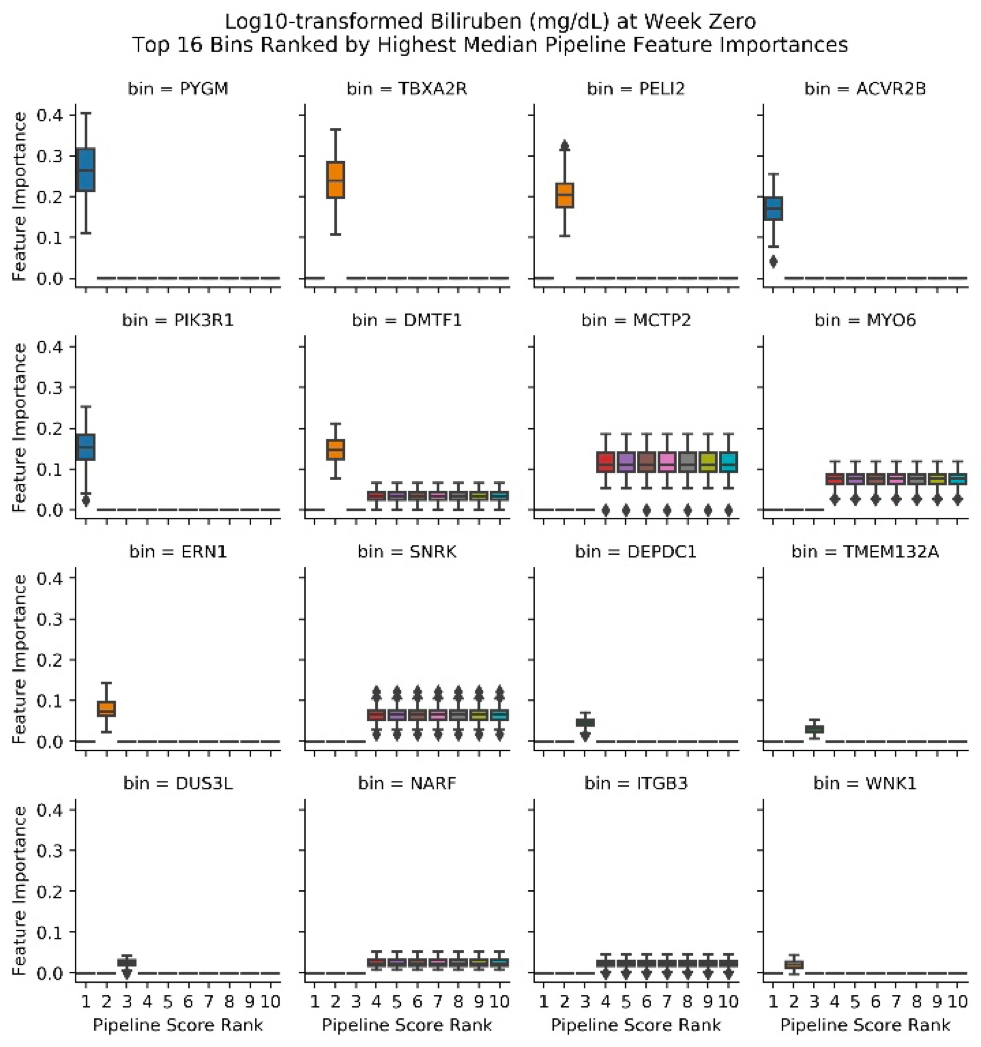
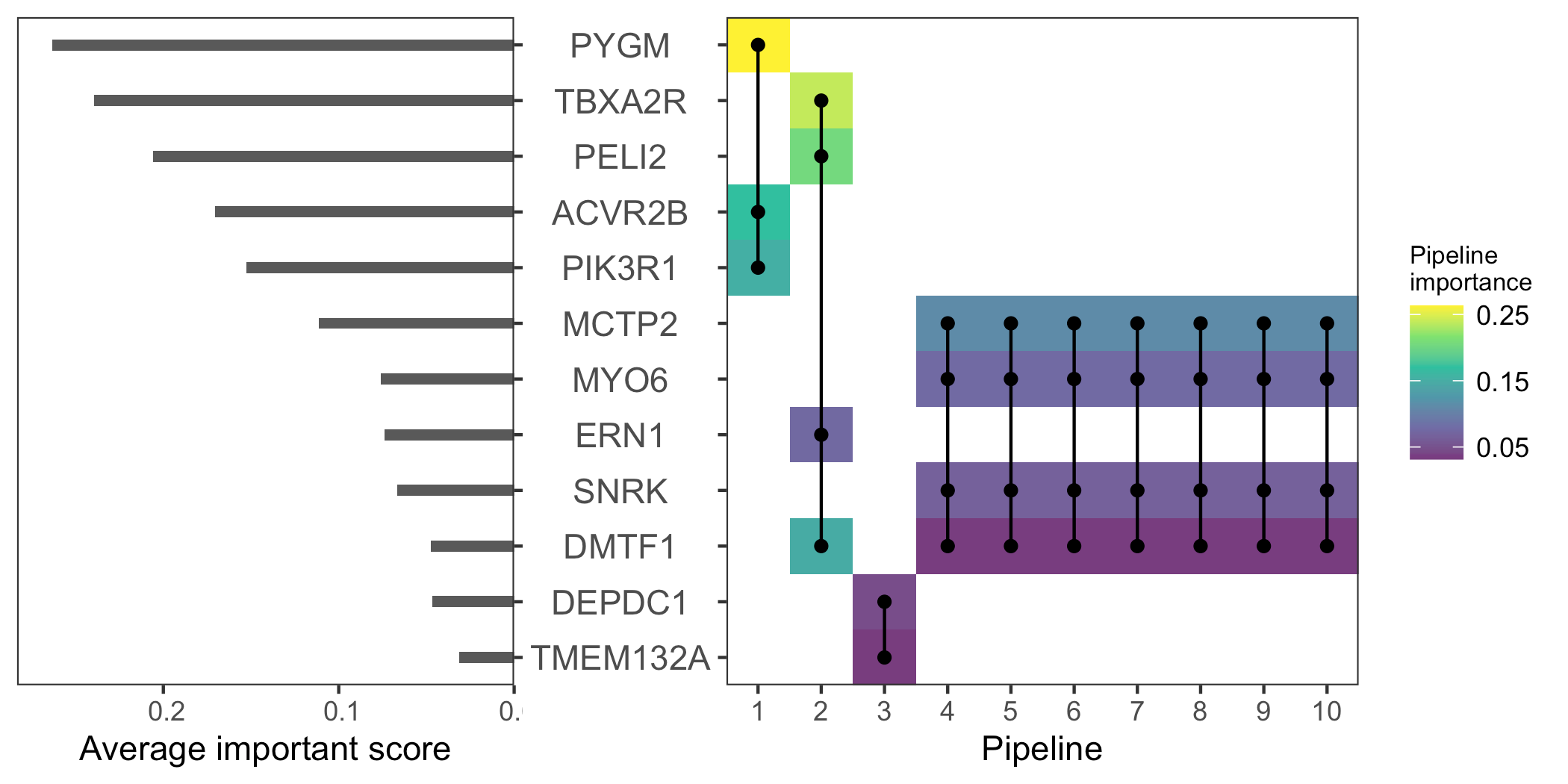
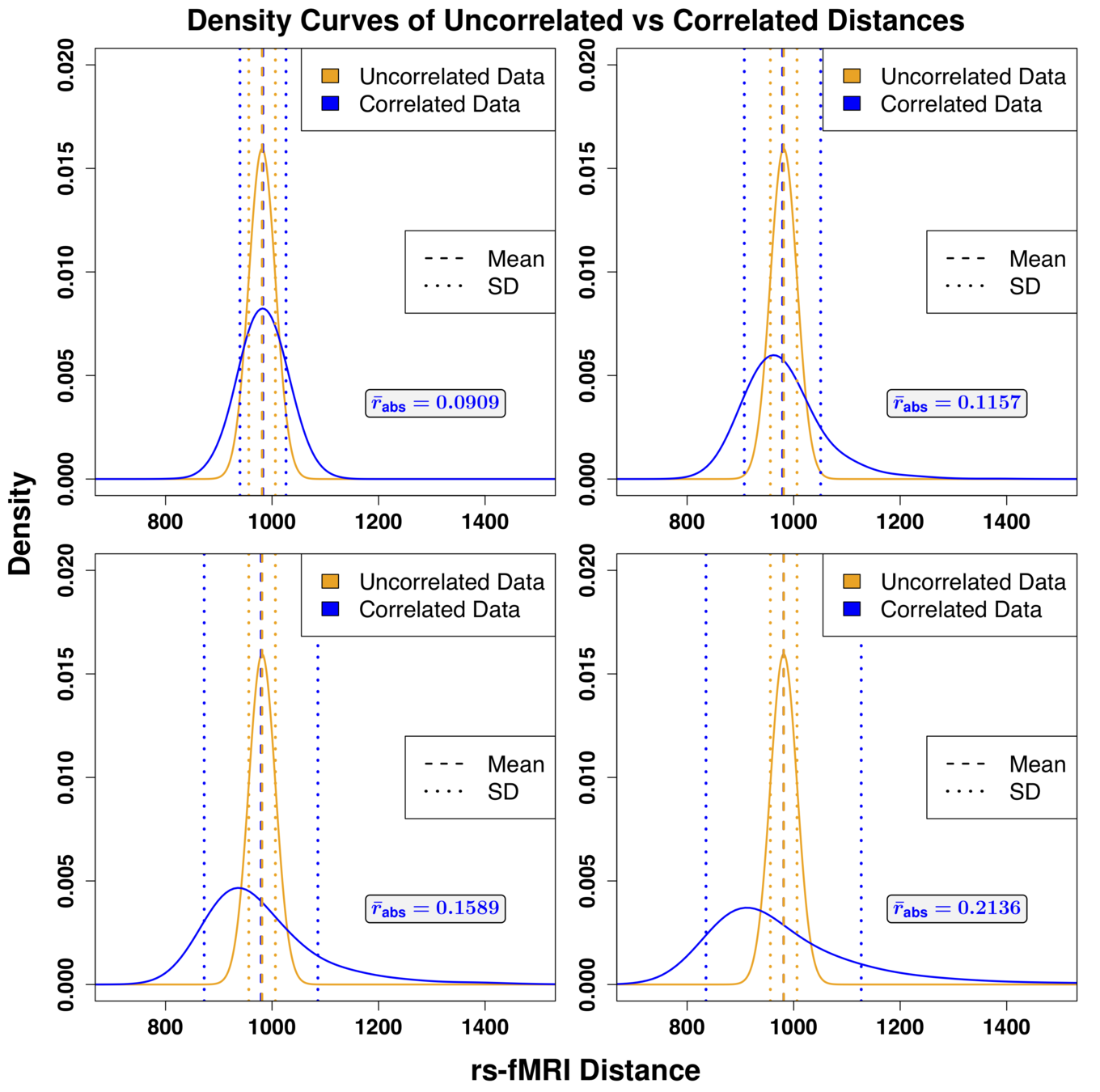
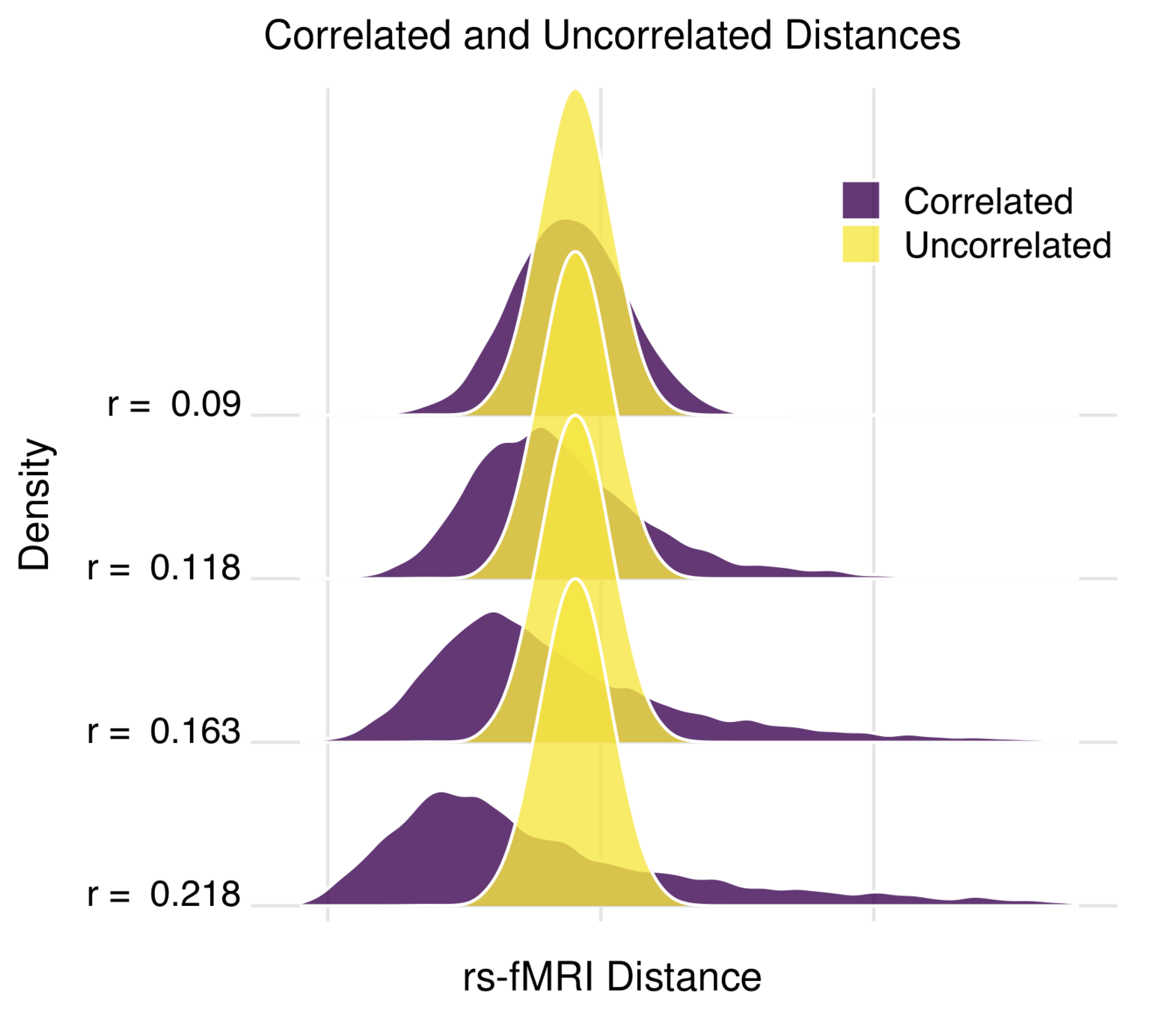

performance
Model B – Model A
Model B
Model A
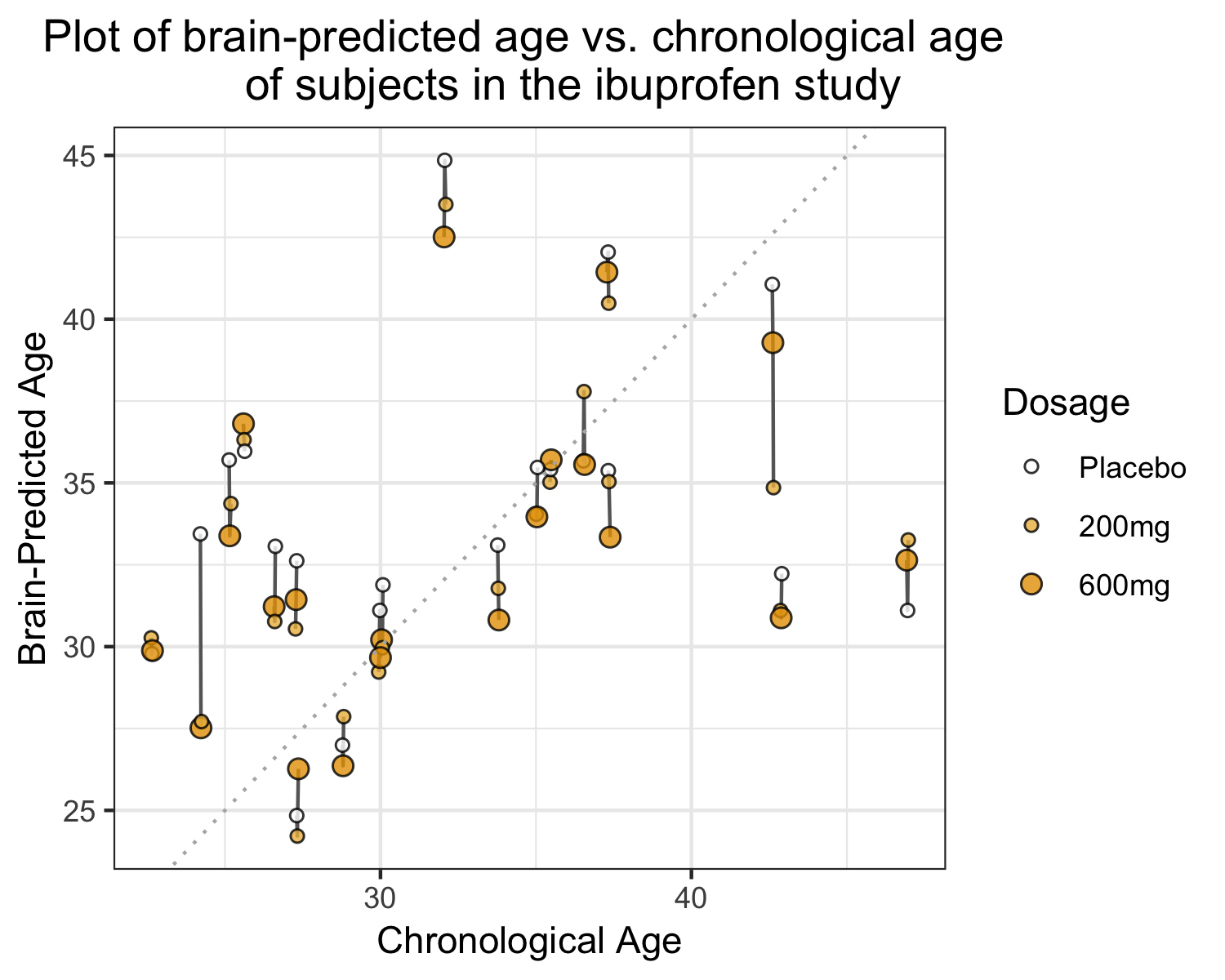
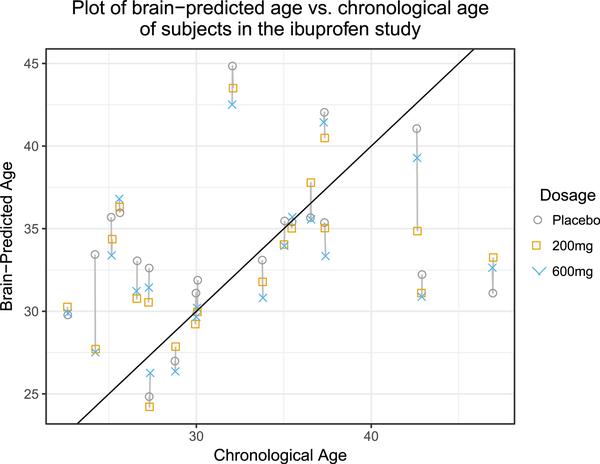
how much information
should I show?
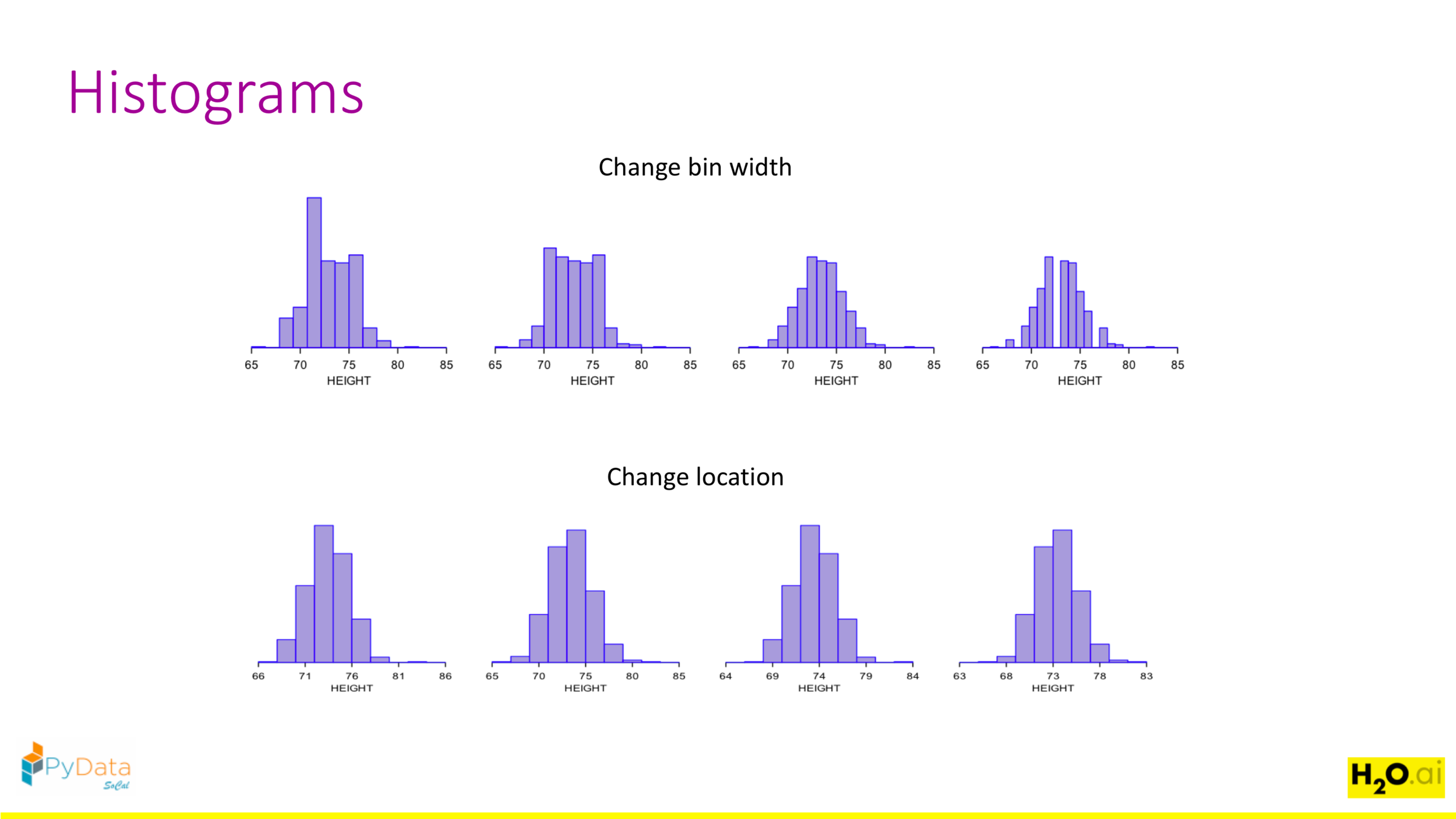
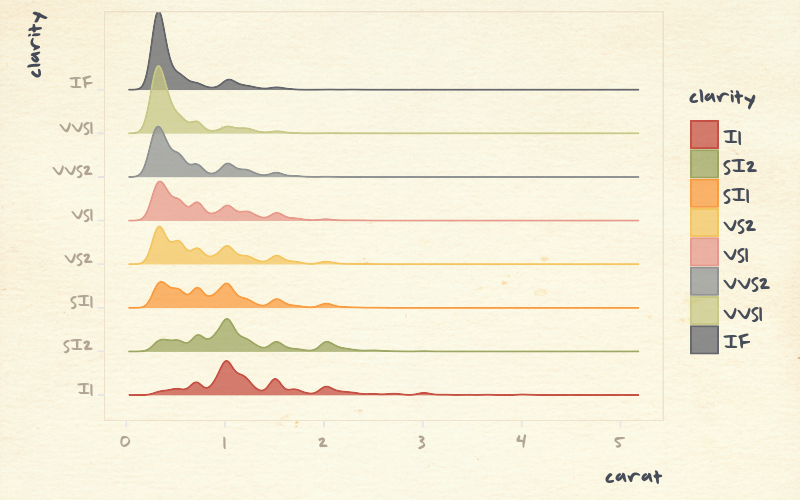
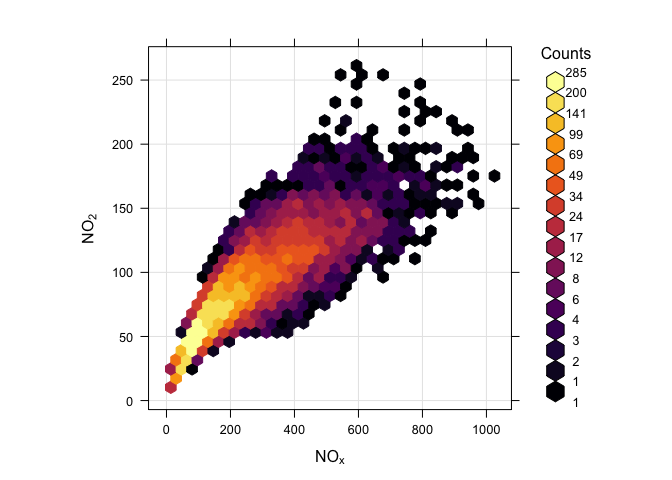
Histograms heavily depend on bin width and location
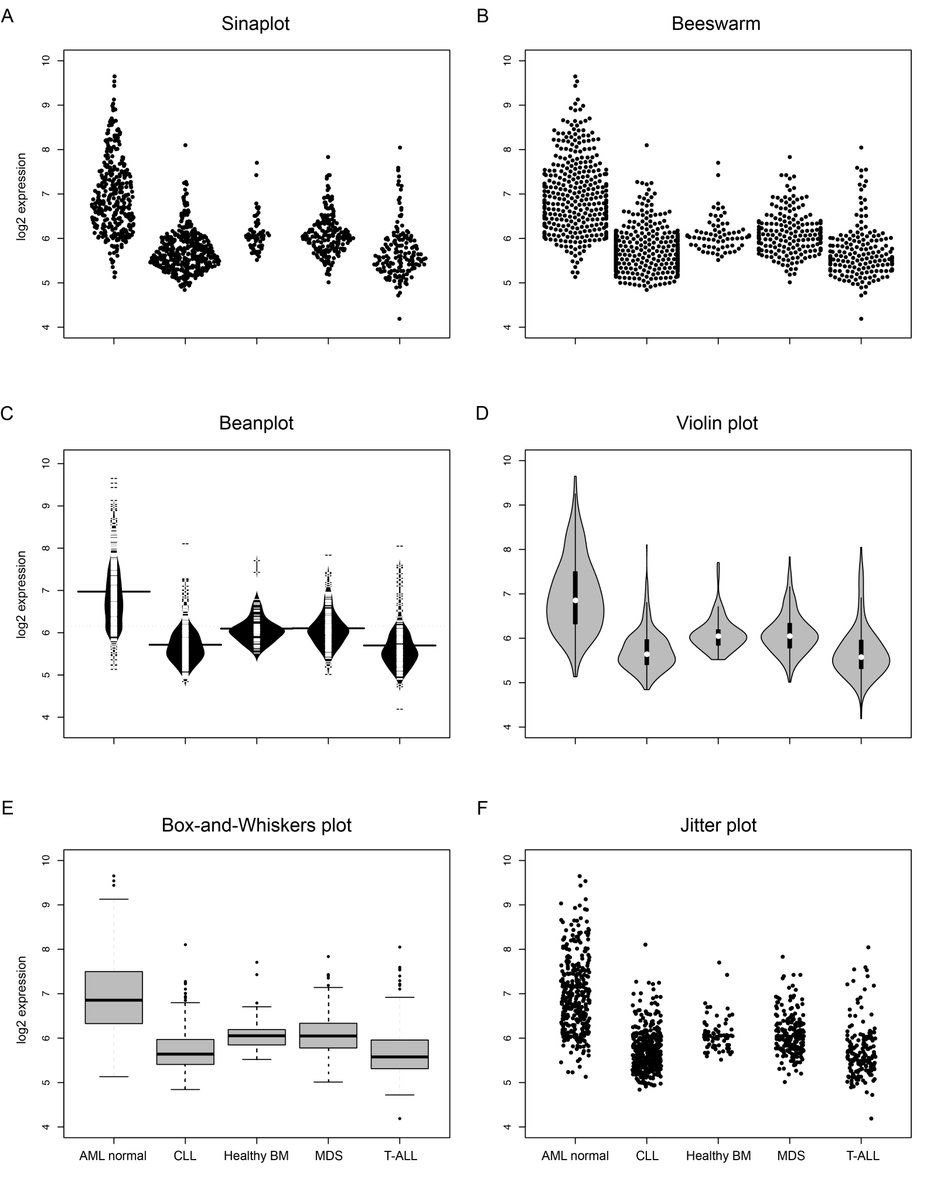
but sometimes binning is helpful...
e.g. to solve overcrowding problem
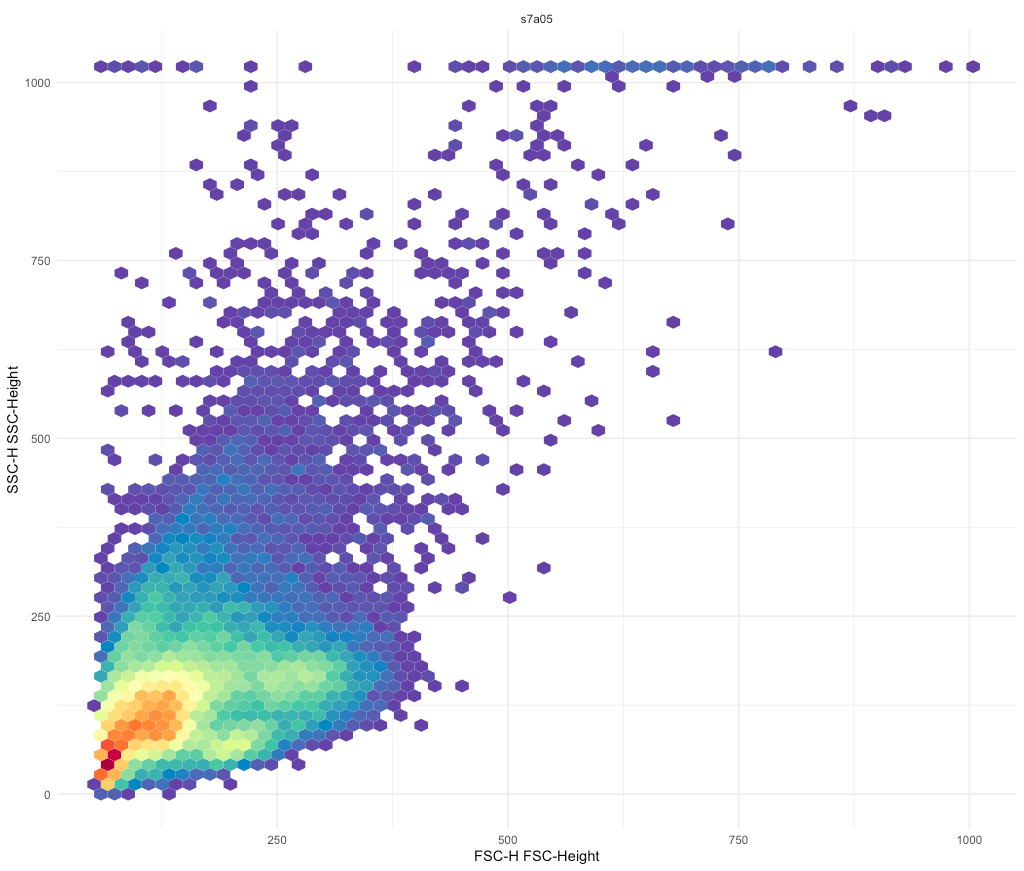
ggcyto::ggcyto() + geom_hex(bins = 64)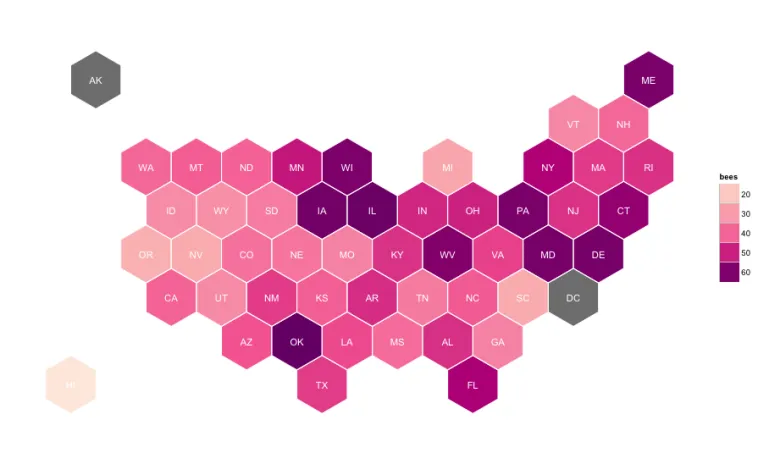
paper ≠ presentation
- amount of information
- annotation
- highlights
- abbreviations
- builds
- text sizes
- ...
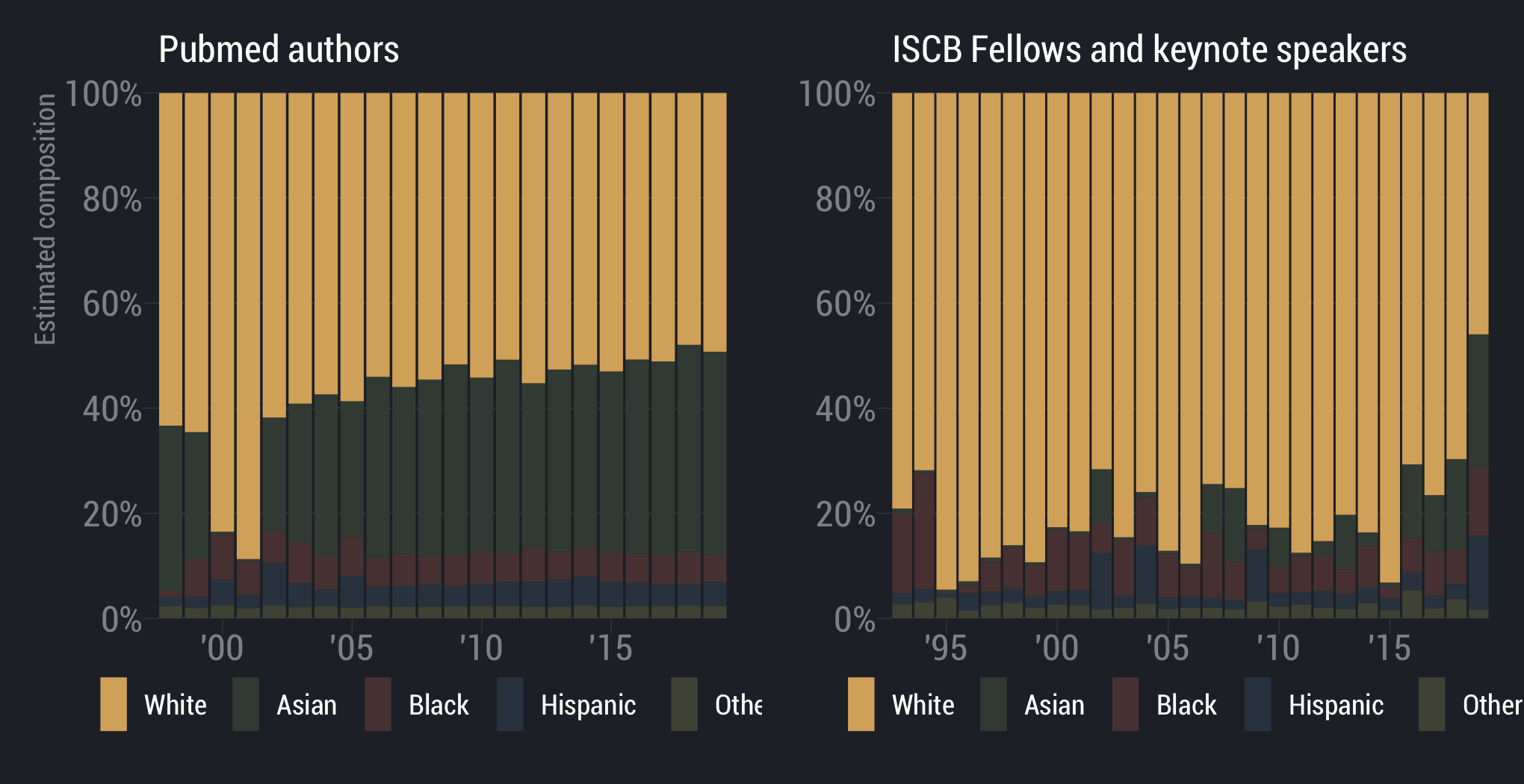
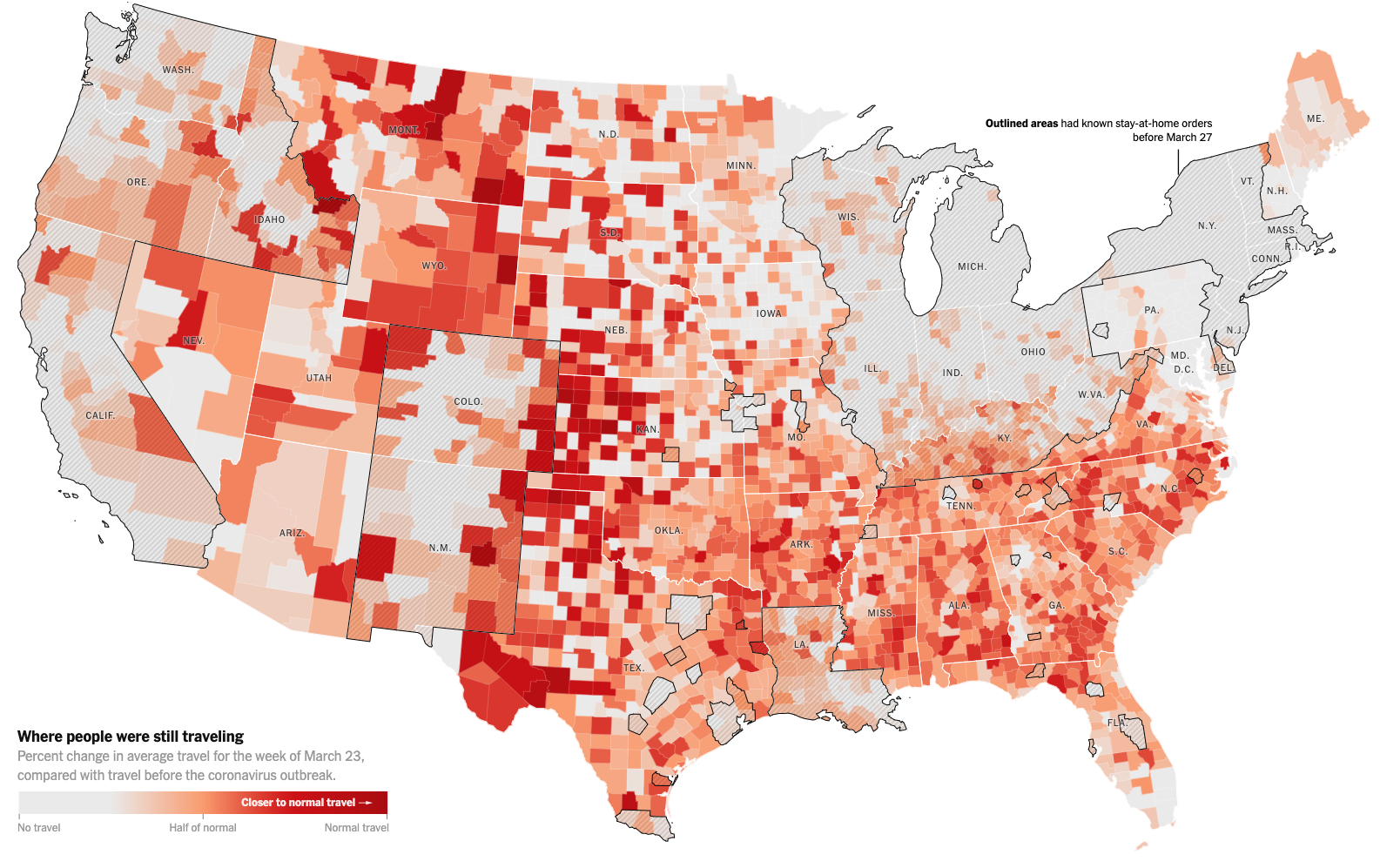
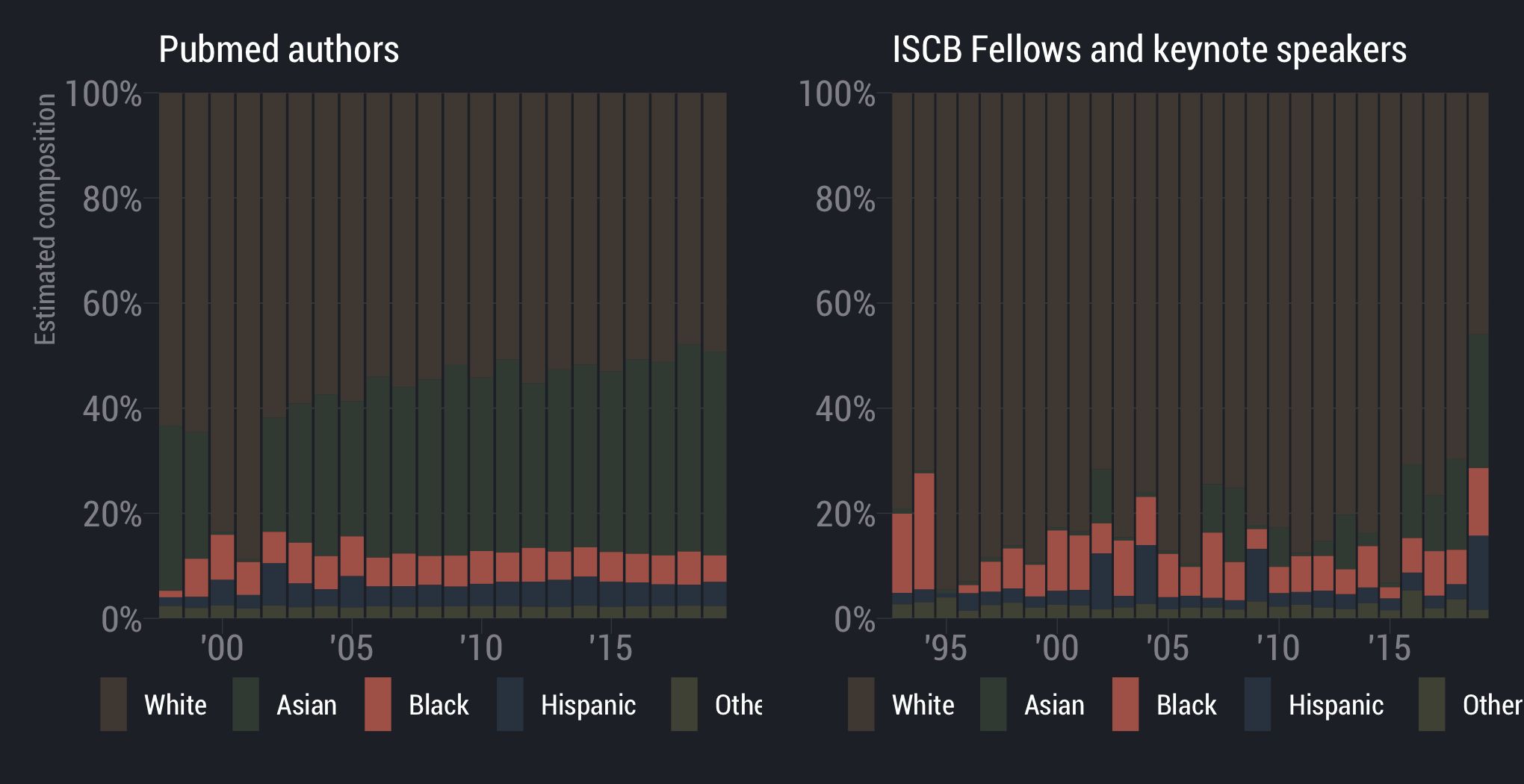
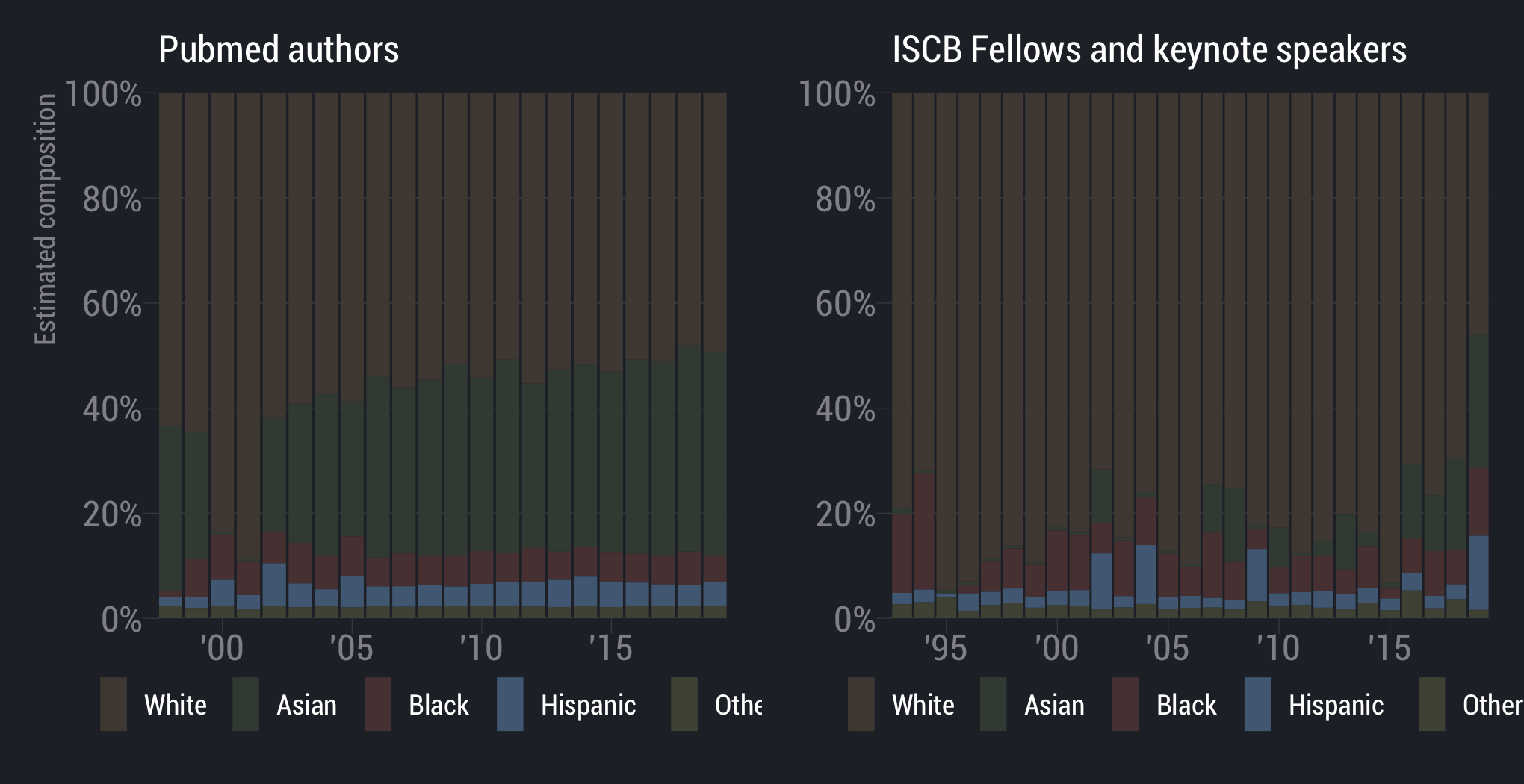
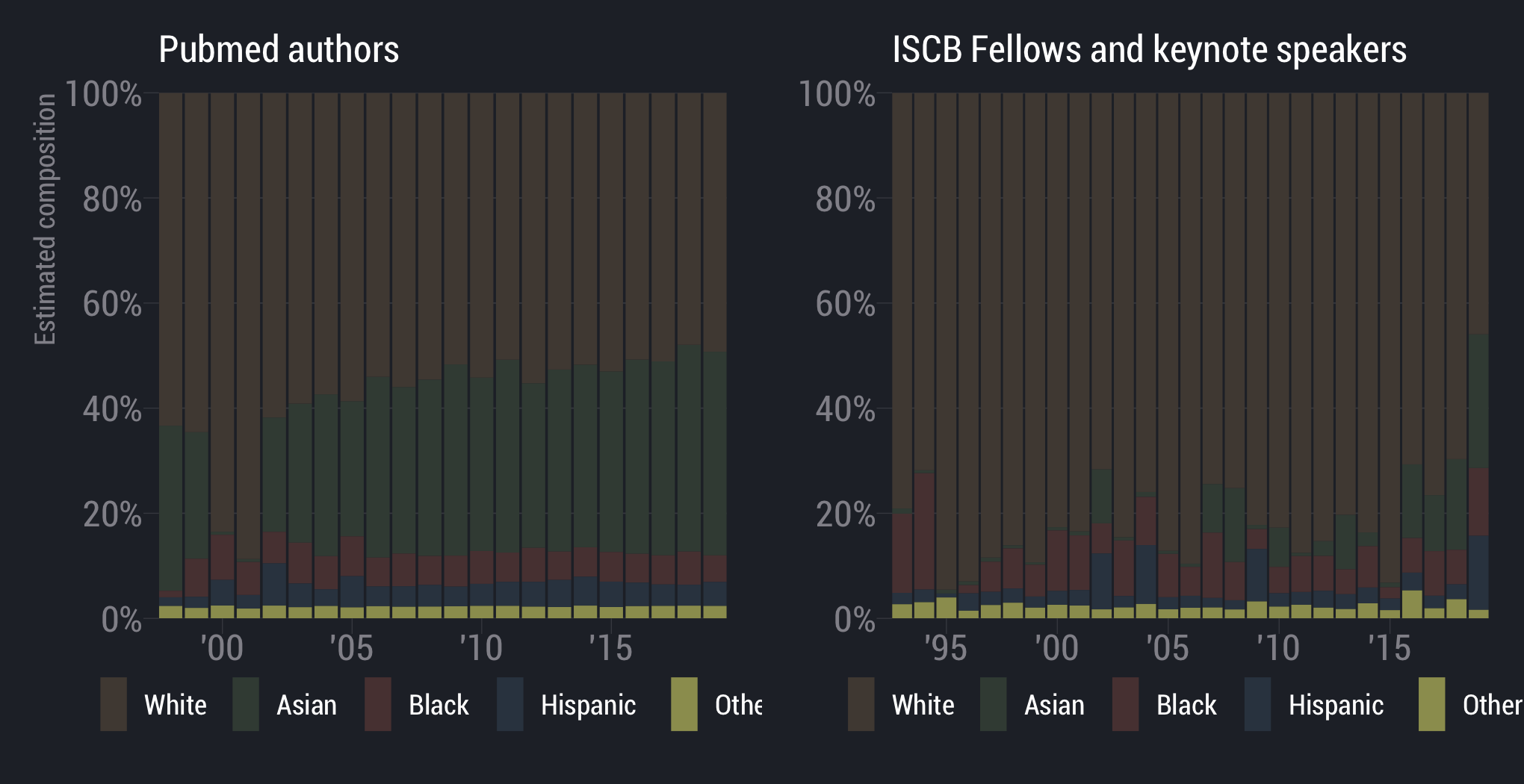
White scientists are overrepresented
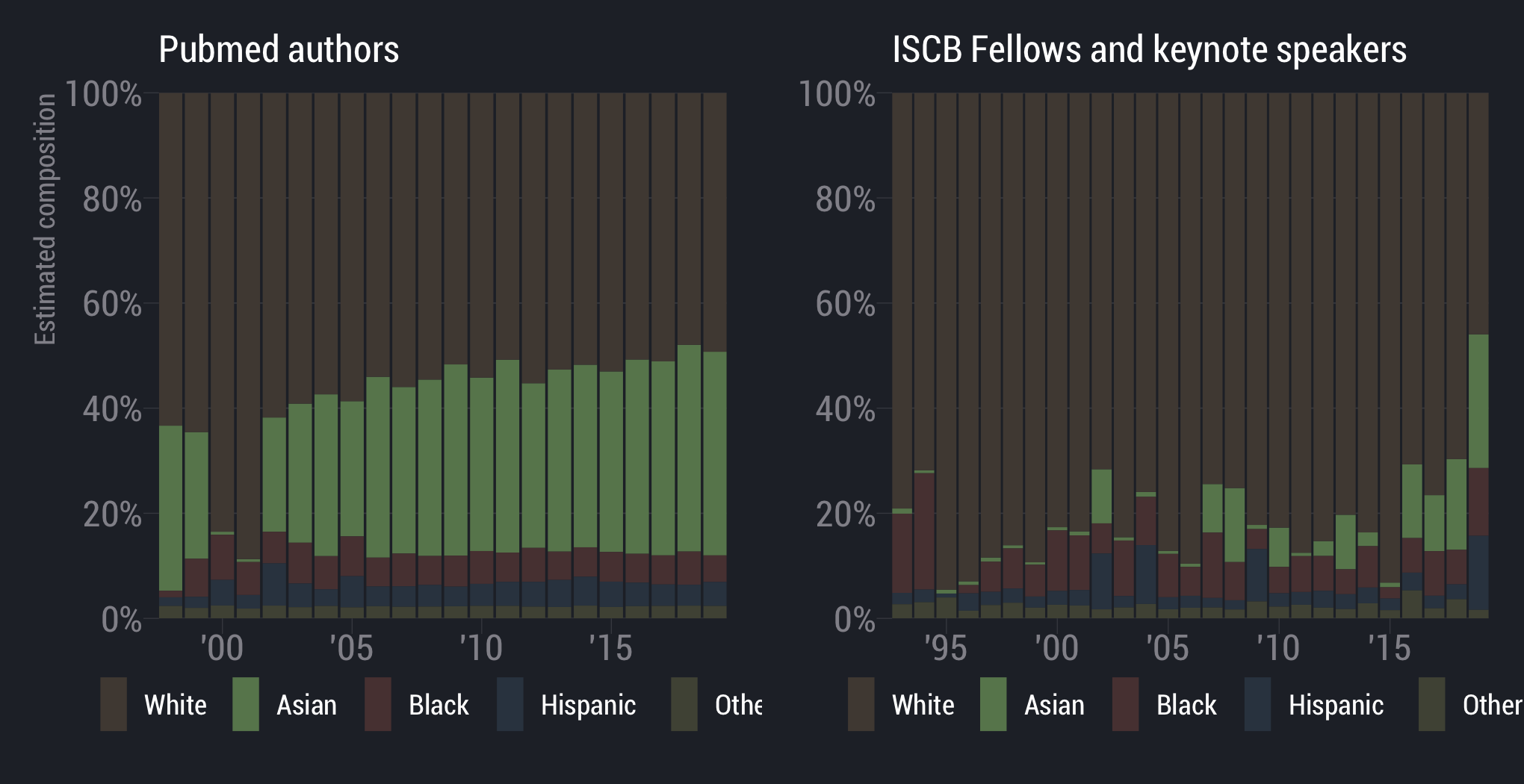
scientists are underrepresented
Asian

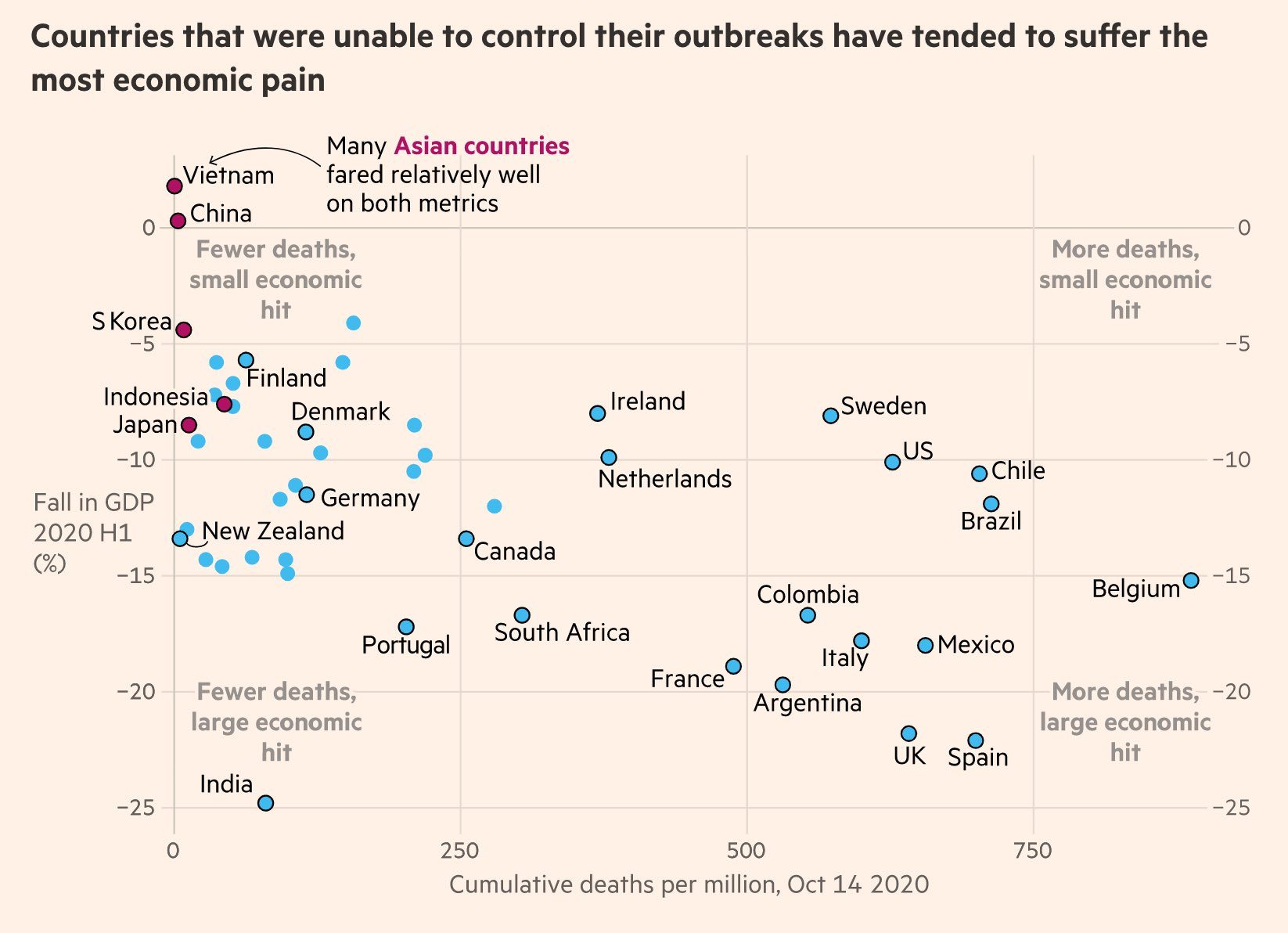
annotation
theme_set(theme_minimal())
theme_update(
legend.title = element_blank(),
panel.grid.minor = element_blank())
)
at the beginning of your script
theme_set()ggplot(df, aes(x = x, y = y)) +
geom_point() +
theme_bw() +
NULLggplot(df, aes(x = x, y = y)) +
geom_point() +
# theme_bw() +
NULLin magrittr pipe:
%>% {.}scale_y_continuous(
breaks = seq(0, 1, 0.2),
labels = scales::percent_format(accuracy = 0.1),
expand = expansions(mult = c(0, .1)))+ coord_fixed(ratio = 1)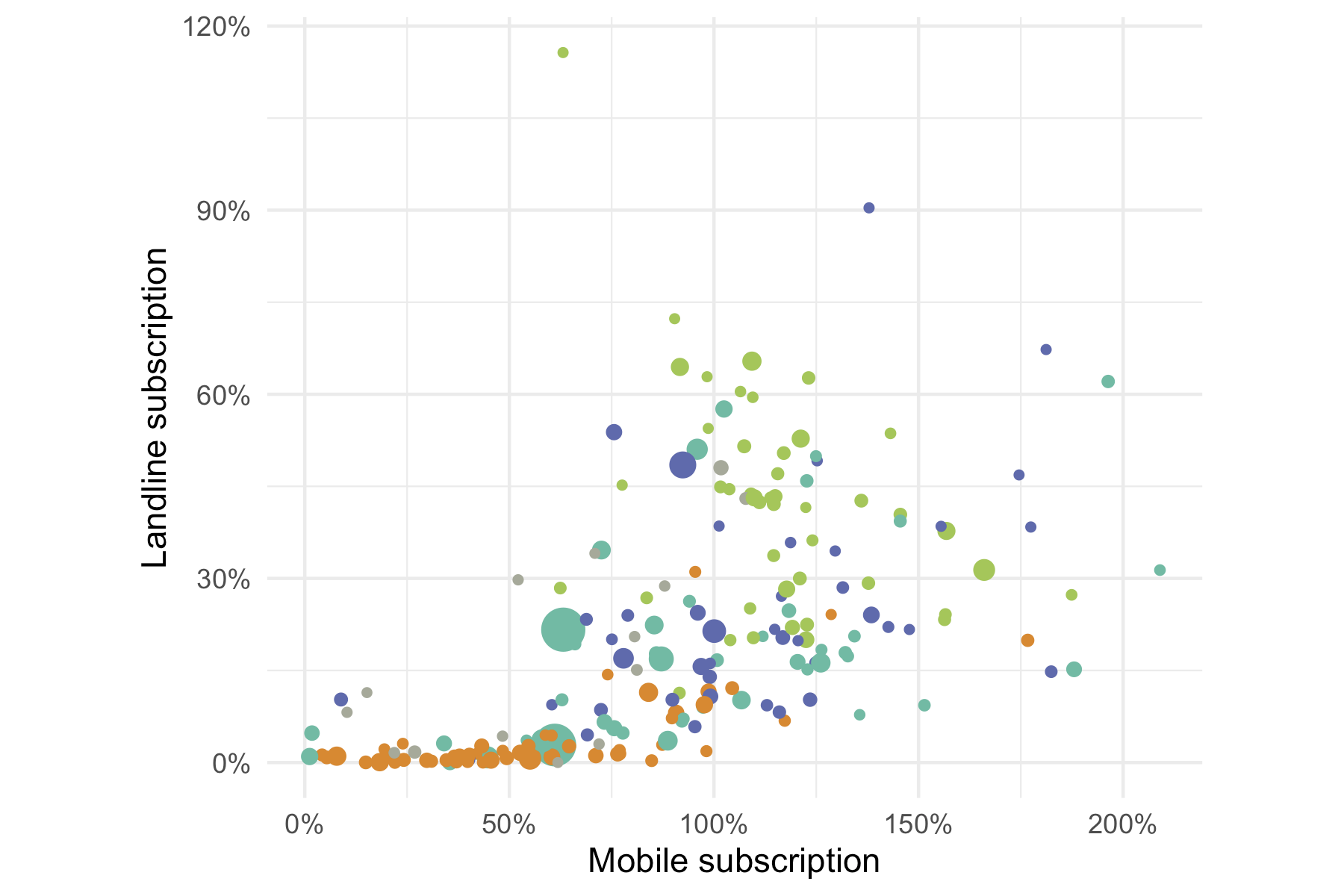
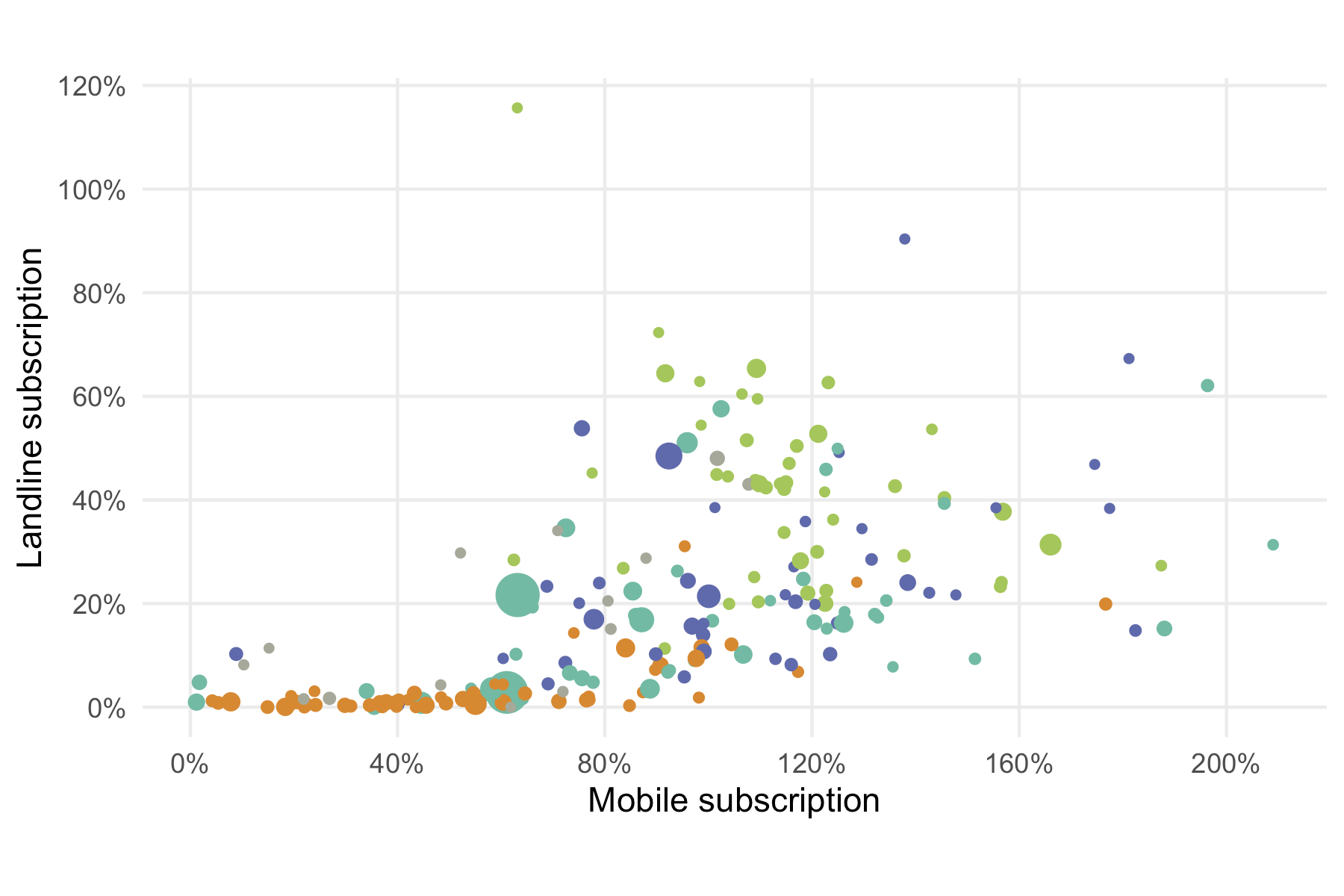
ggplot(mobile) +
geom_sf(aes(fill = mobile_subs)) +
coord_sf(crs = "+proj=eqearth +wktext")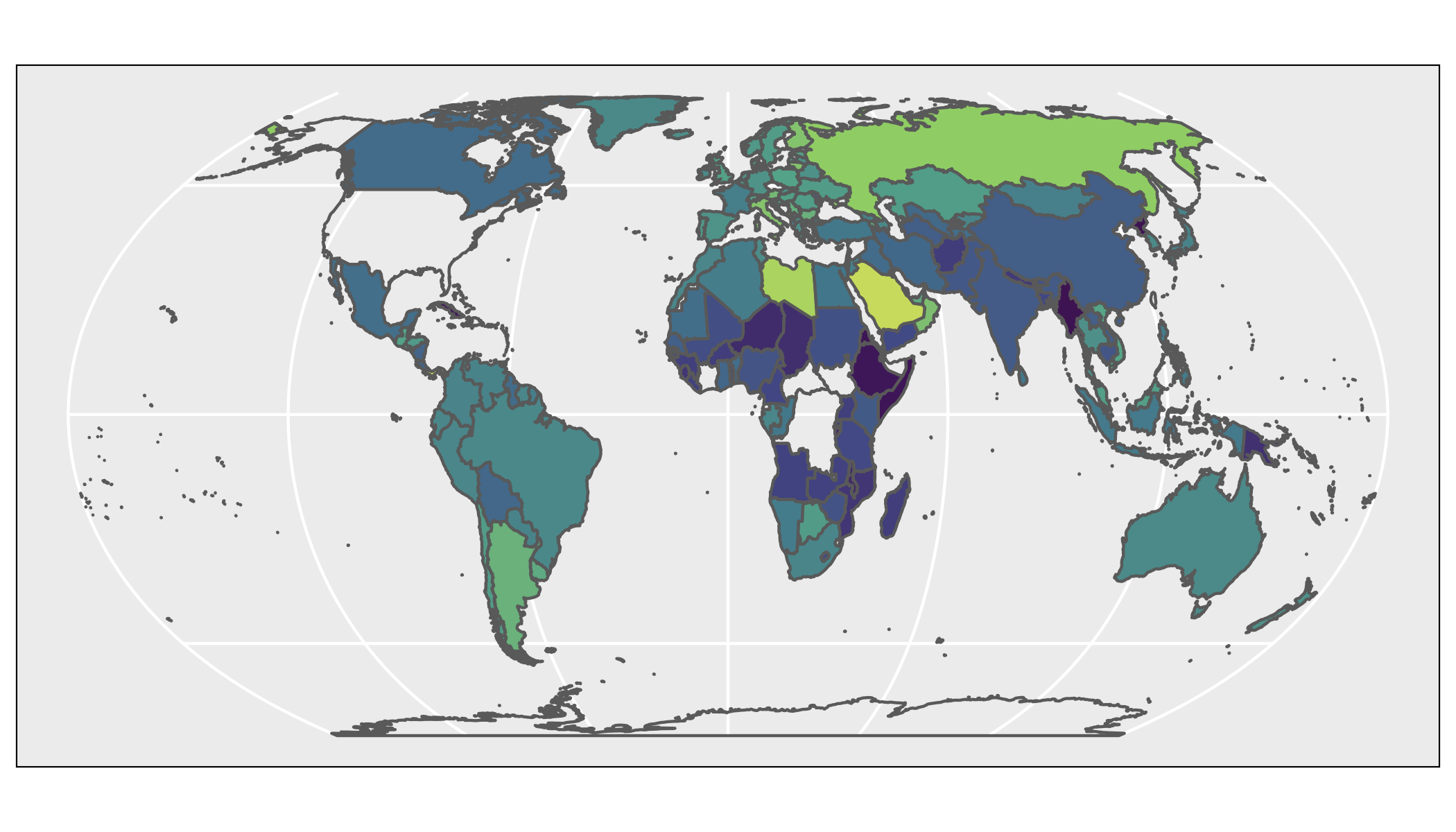
+ labs(y = NULL, fill = NULL)NULL, FALSE, NA and element_blank()
+ theme(axis.title.y = element_blank(),
legend.title = element_blank())# is equivalent to
+ labs(y = '', fill = '')+ scale_fill_continuous(guide = FALSE)+ guides(fill = FALSE)# is equivalent to
+ guides(fill = "none")+ scale_fill_continuous(guide = "none")# Setting the limits with the scale discards
# all data outside the range.
lims(x = c(20, NA), y = c(NA, 40))
# As of June 2019, this is allowed (and recommended)
coord_cartesian(xlim = c(20, NA), ylim = c(NA, 40))# Setting the limits with the scale discards
# all data outside the range.
lims(x = c(20, NA), y = c(NA, 40))
# As of June 2019, this is allowed (and recommended)
coord_cartesian(xlim = c(20, NA), ylim = c(NA, 40))geom_col()know these differences
geom_bar()geom_histogram()geom_point()geom_count()geom_jitter()jitters both in the x and y direction by defaults
Very quick on colors
- sequential, quantitative, divergent
- colorblind friendly palettes
+ scale_color_viridis_c()
filld+ colorblindr::scale_fill_OkabeIto()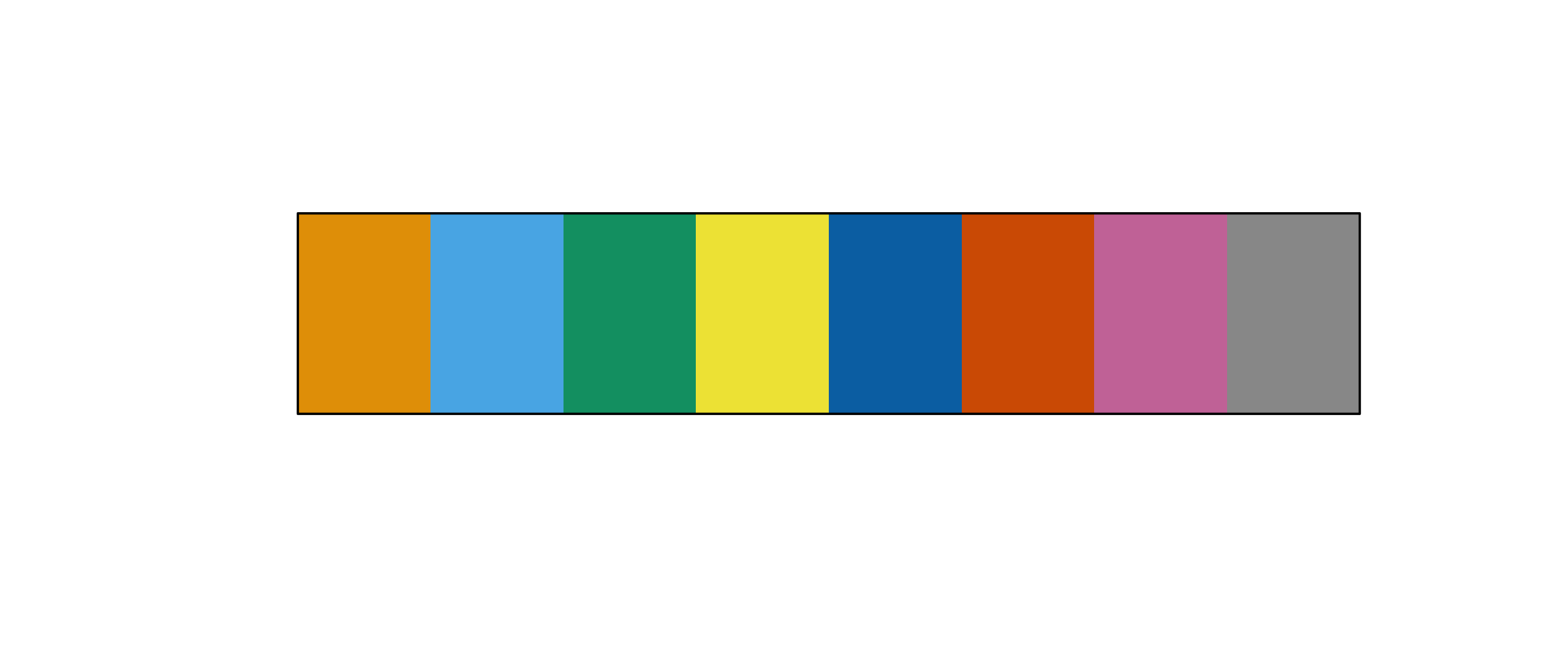
+ rcolorcarto::scale_color_carto_d()
Resources
your turn
Tools → Global Options → R Markdown:
uncheck Show output inline
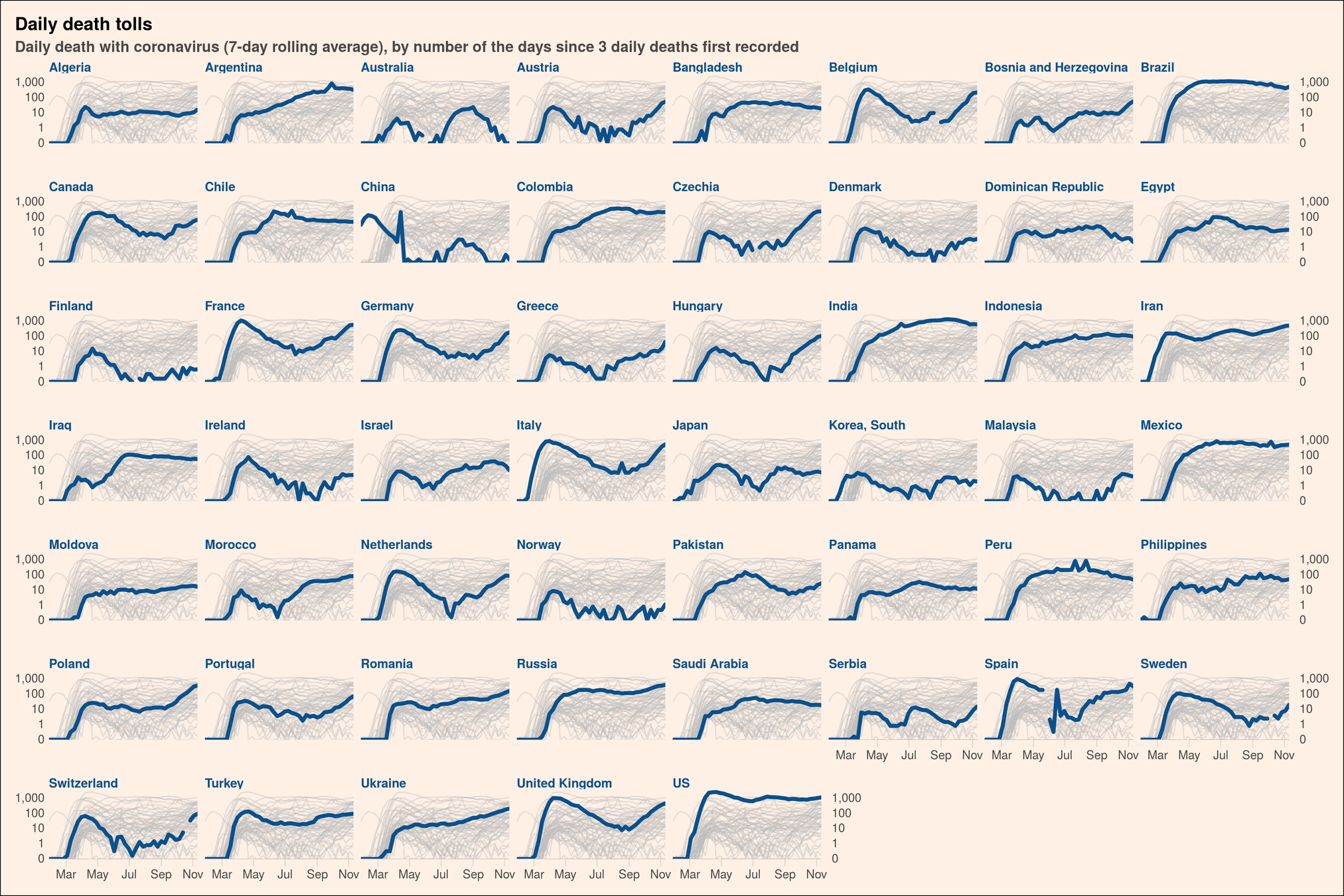
Great data stories
http://mkweb.bcgsc.ca/
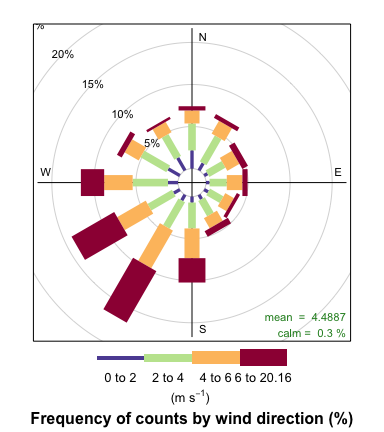
windRose(openair_data)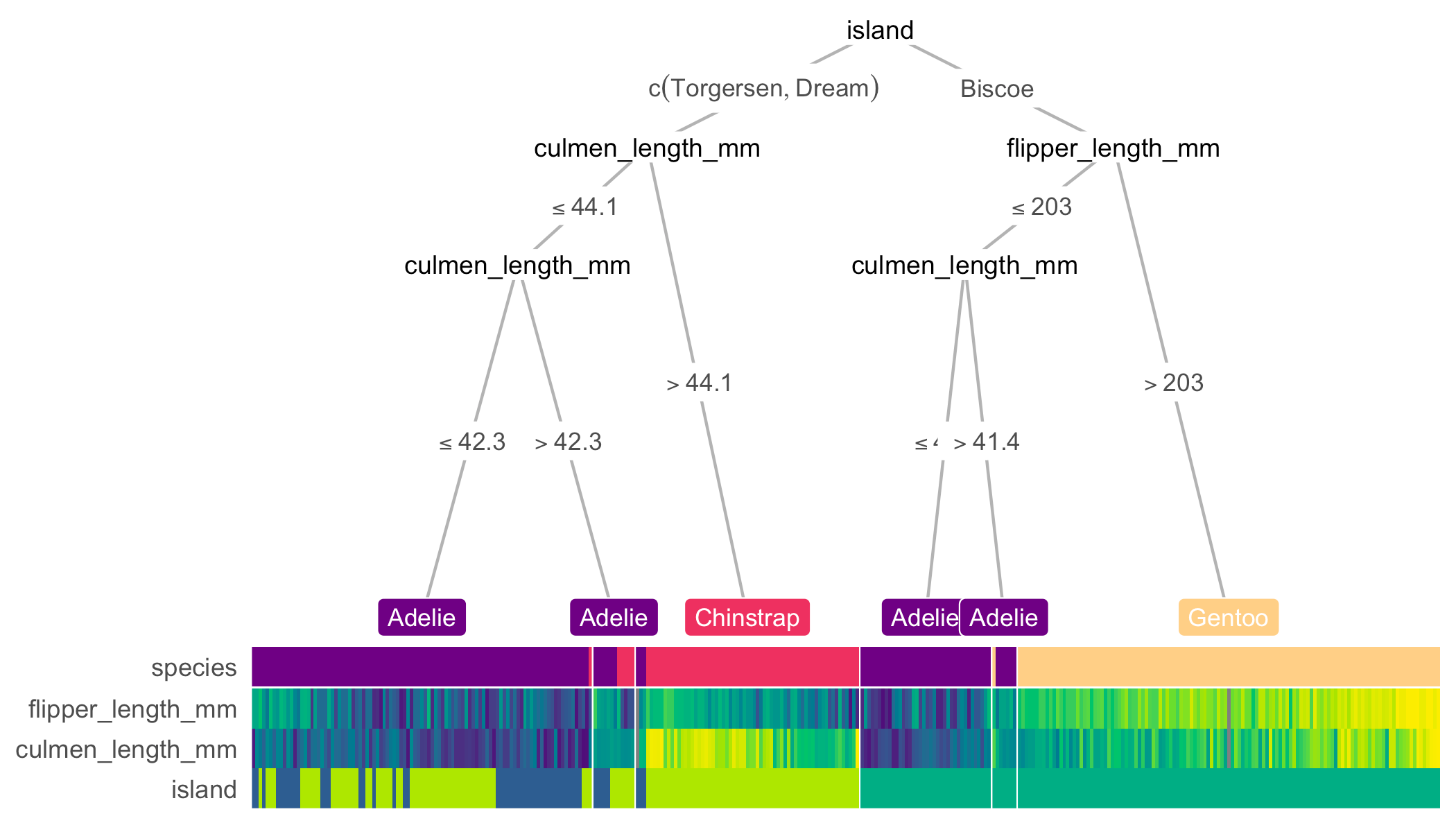
library(treeheatr)
heat_tree(penguins, target_lab = 'species')
How do you remove legend title?
What about y-axis title?