{treeheatr} and {pmlbr}
visualizing decision trees
on benchmark datasets
Trang Le
University of Pennsylvania
RLadies Miami 2020-11-19
@trang1618
Google doc: https://tiny.cc/treeheatr
Take a bad chart and make it better: a data visualization workshop
part 1
Your decision tree may be cool, but what if I tell you
you can make it hot?
🌳
🔥
http://tiny.cc/treeheatr
decision tree + heatmap
http://tiny.cc/treeheatr
rpart.plot
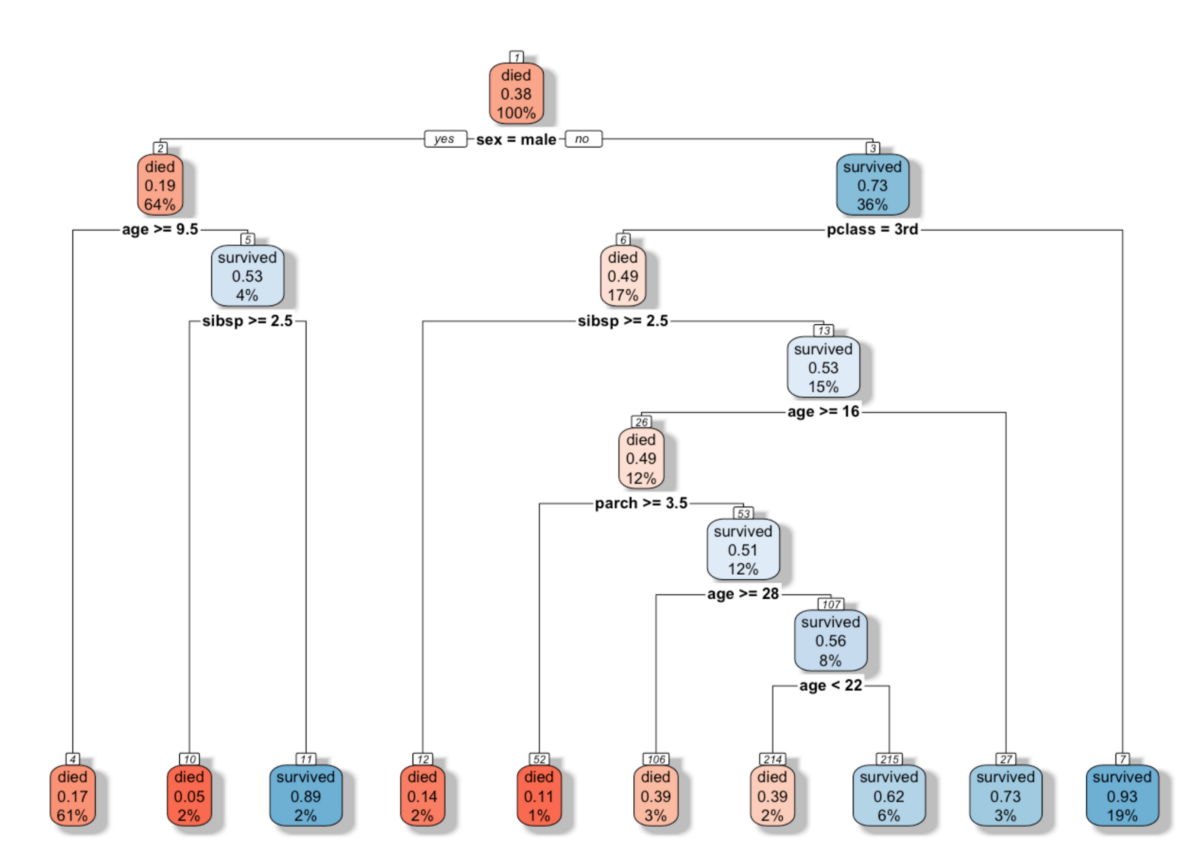
Terry Therneau, Beth Atkinson, Brian Ripley
visNetwork::visTree()
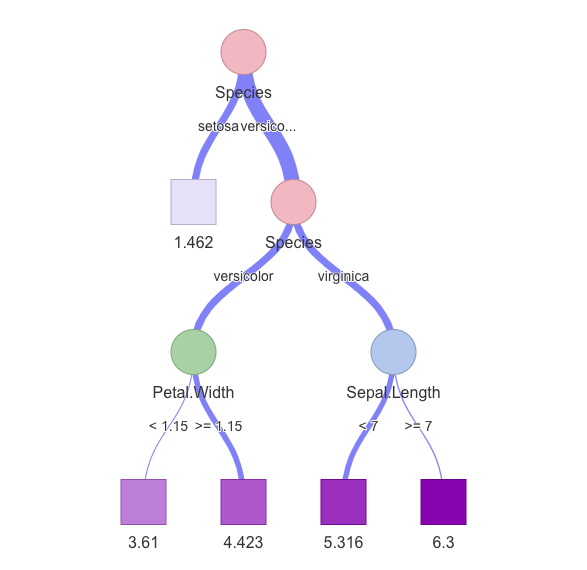
Almende B.V., Benoit Thieurmel , Titouan Robert
partykit::plot.party()
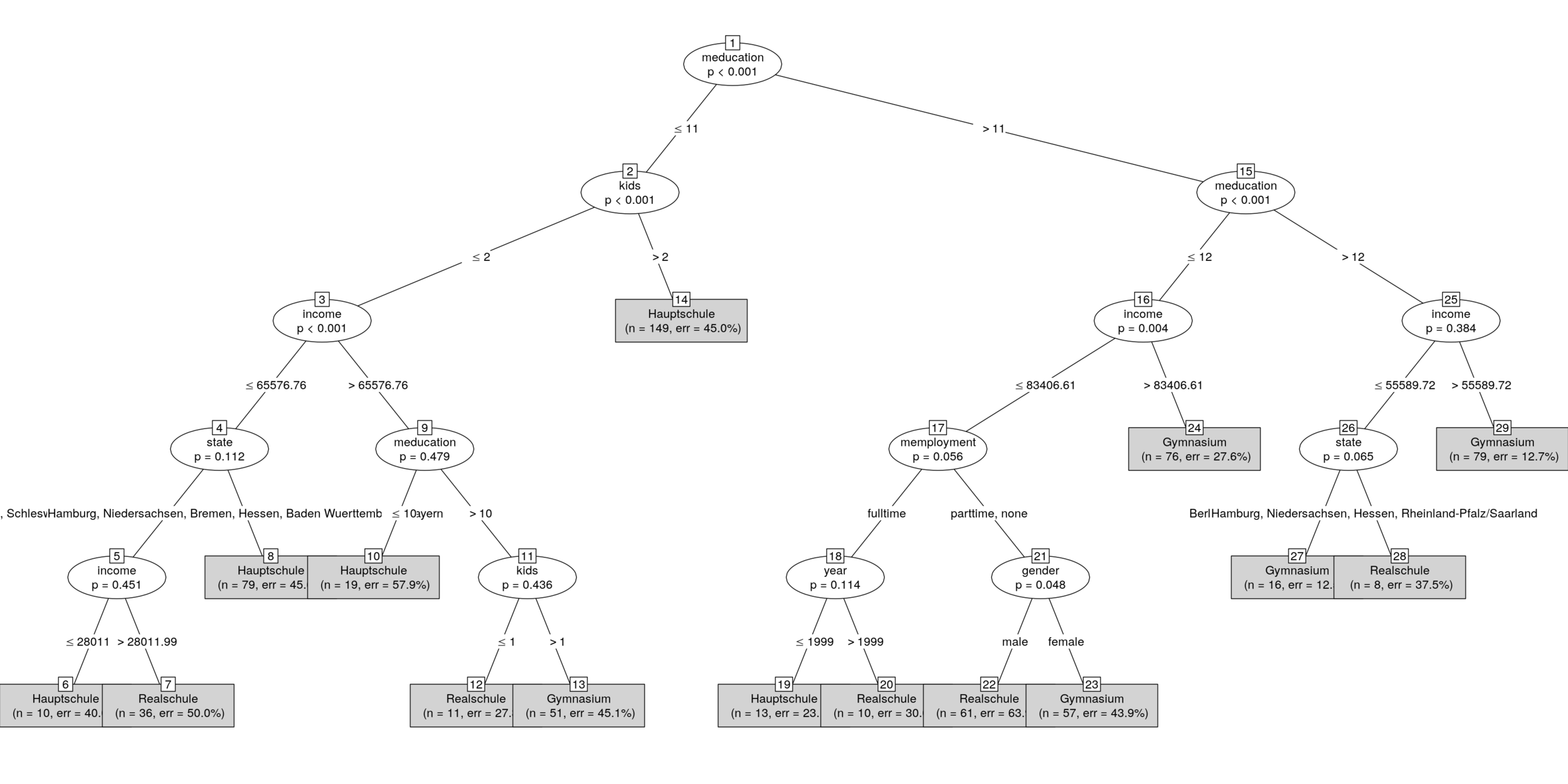
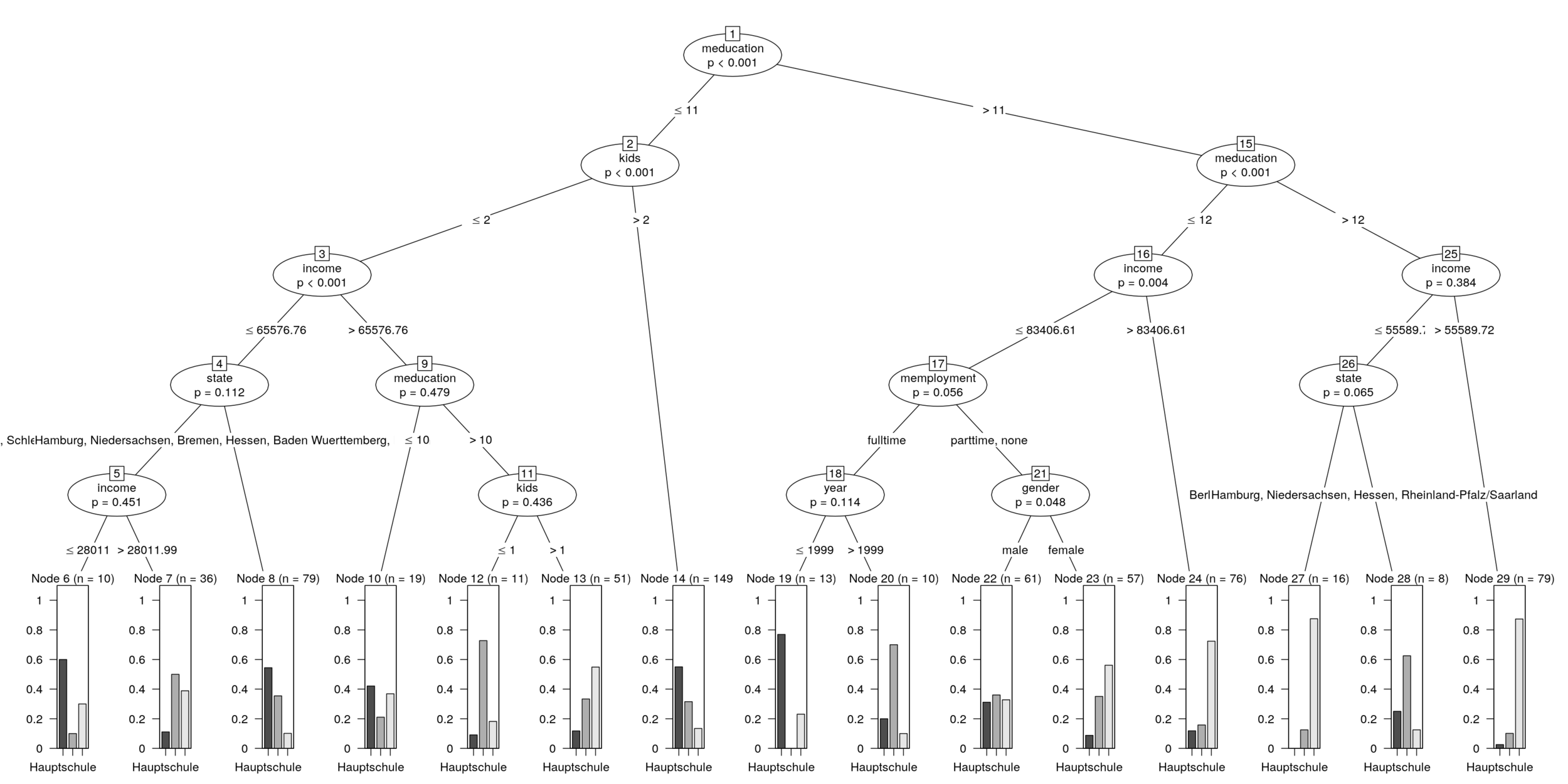
Torsten Hothorn, Heidi Seibold, Achim Zeileis
ggparty
Martin Borkovec, Niyaz Madin, et al.

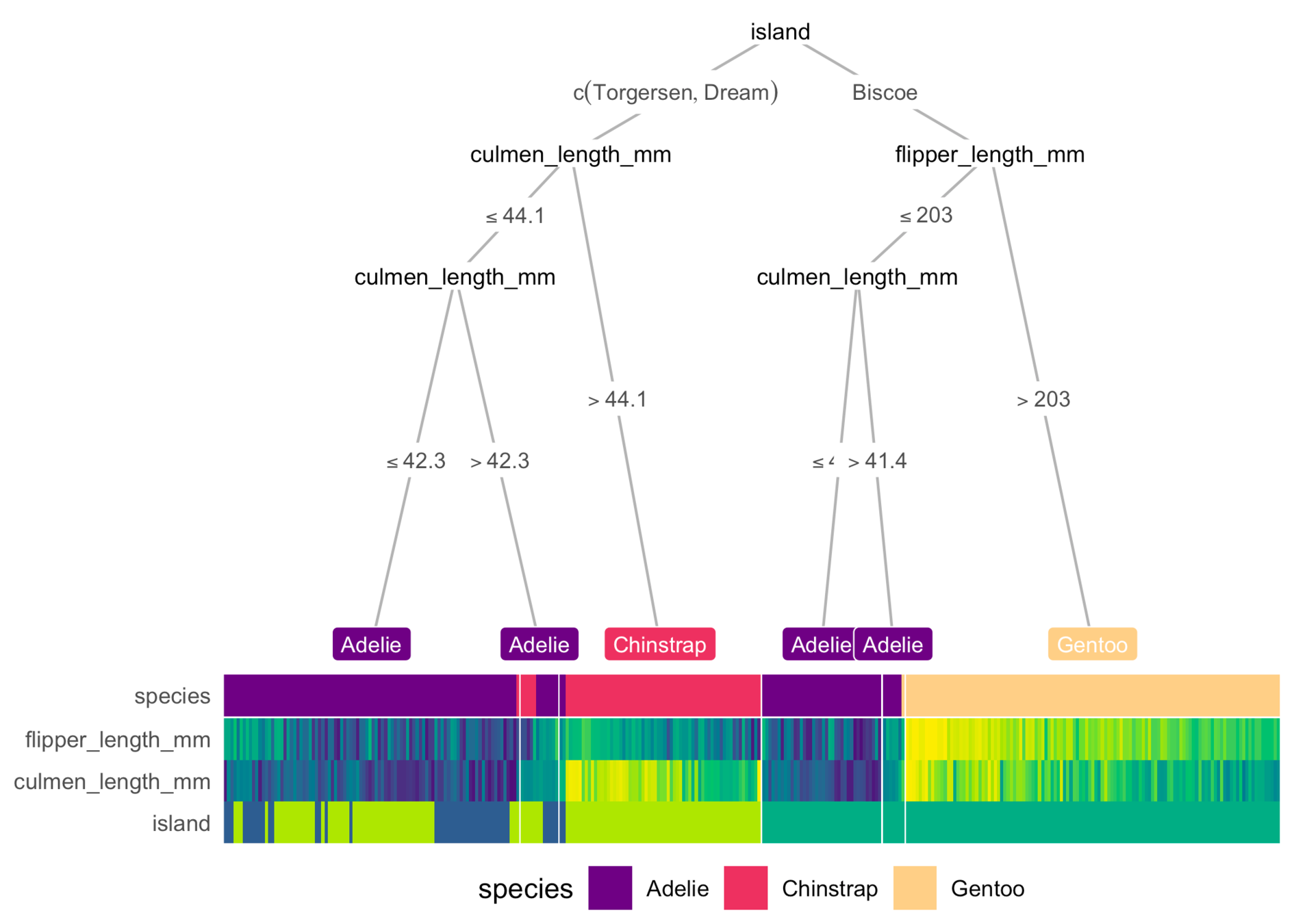
dtreeviz
(Python)
Terence Parr, Prince Grover
treeheatr
target
featuresdependent variable/outcome/phenotype
predictors/variables
treeheatr::heat_tree(x)can be of object
-
party (or constparty)
specifying the precomputed tree -
partynode
specifying the manual tree - data.frame (or tibble)
heat_tree(x, target_lab = 'Outcome')351 blood samples (January 10 - February 18)
3 features:
- lactic dehydrogenase (LDH)
- lymphocyte levels
- high-sensitivity C-reactive protein (hs_CRP)
COVID-19 patient data
http://tiny.cc/treeheatr
heat_tree(x = covid, target_lab = 'Outcome')data.frame
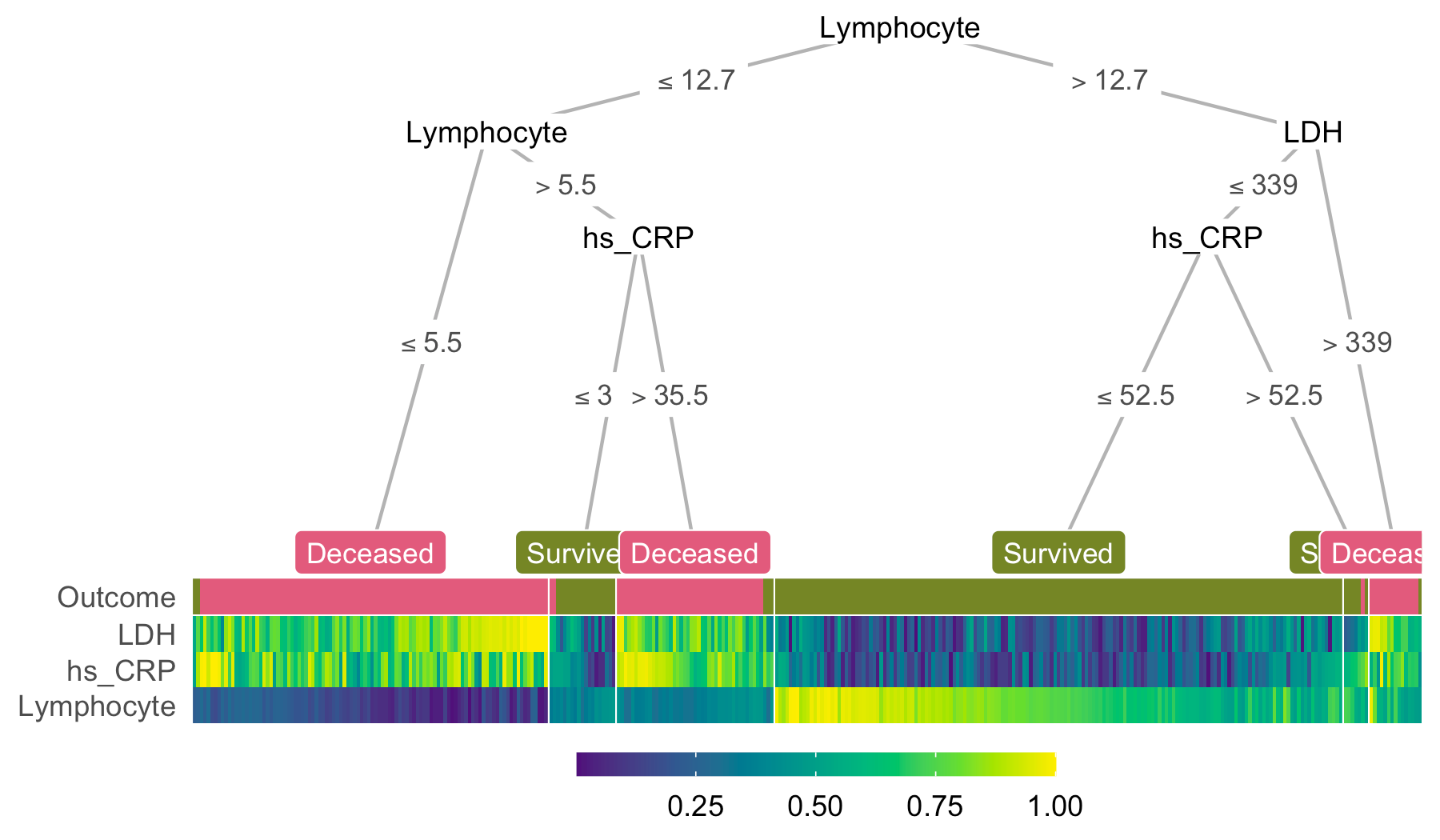
heat_tree(covid, target_lab = 'Outcome',
feats = NA)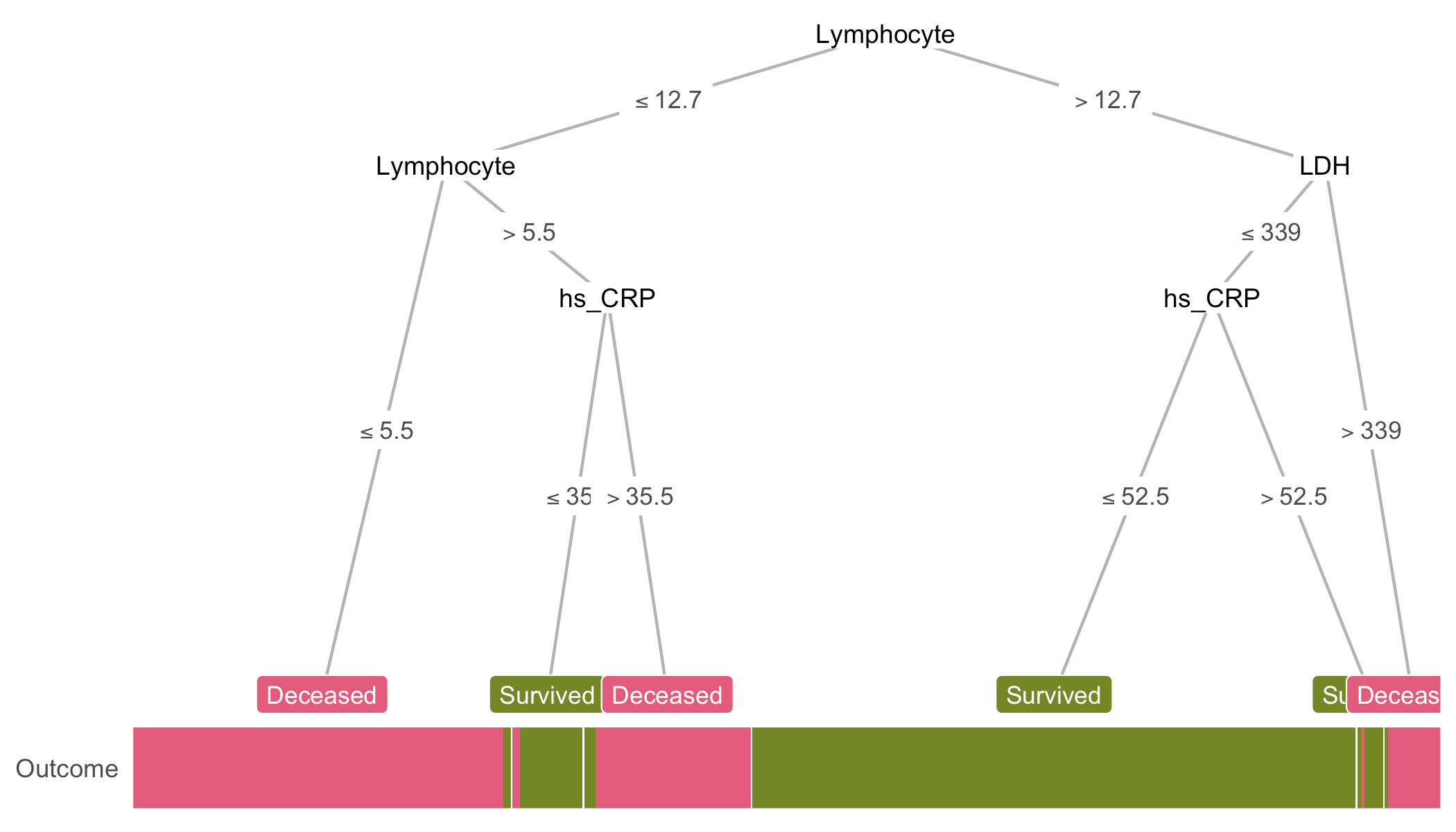
split_ldh <- partysplit(1L, breaks = 365)
split_crp <- partysplit(2L, breaks = 41.2)
split_lymp <- partysplit(3L, breaks = 14.7)
custom_tree <- partynode(1L, split = split_ldh , kids = list(
partynode(2L, split = split_crp, kids = list(
partynode(3L, info = 'Survival'),
partynode(4L, split = split_lymp, kids = list(
partynode(5L, info = 'Death'),
partynode(6L, info = 'Survival'))))),
partynode(7L, info = 'Death')))
heat_tree(x = custom_tree, data_test = covid, target_lab = 'Outcome')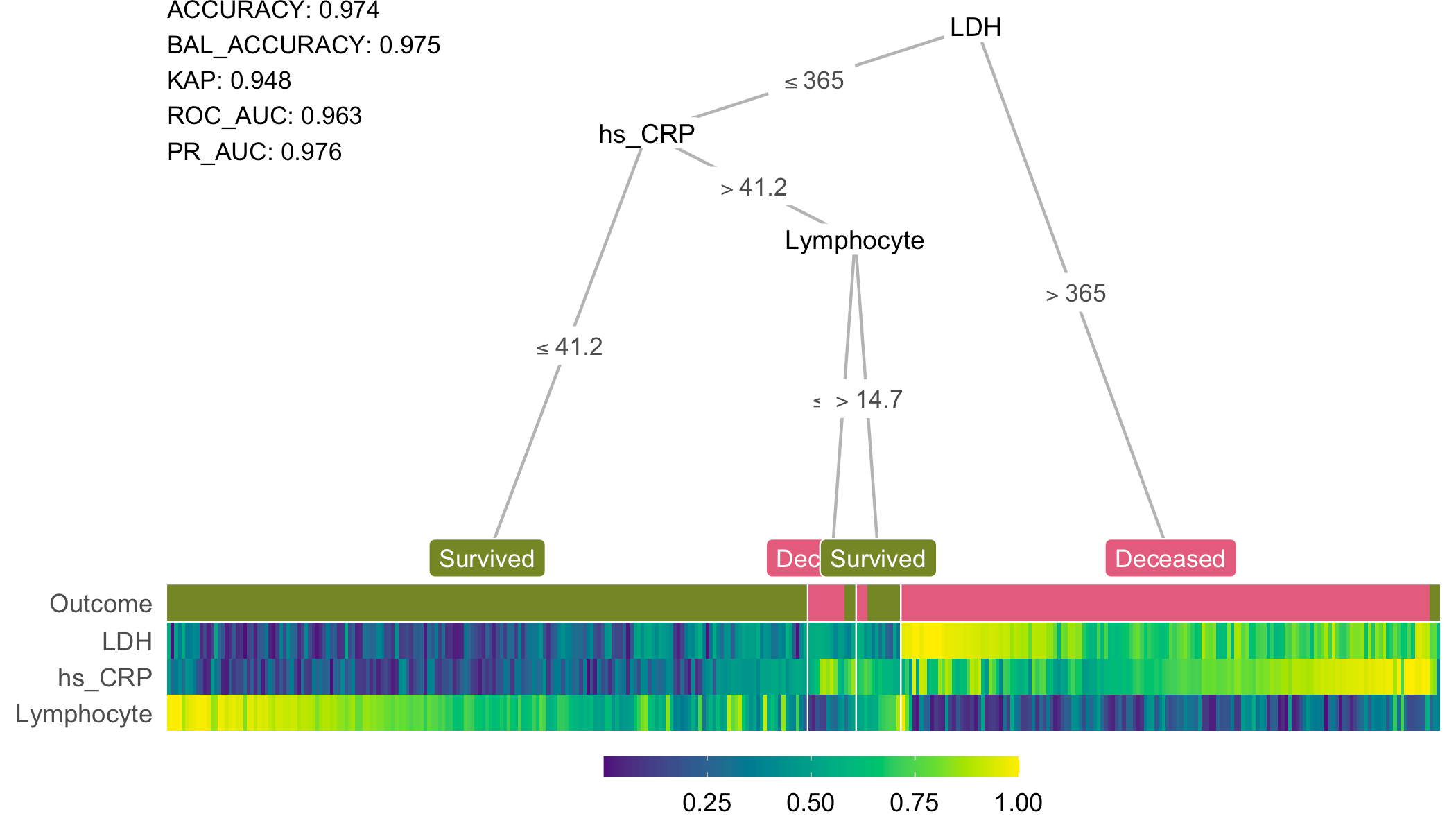
part 2
what is benchmarking?
a standard practice to illustrate the strengths and weaknesses of algorithms with regards to different problem characteristics
target
featuresdependent variable/outcome/phenotype
predictors/variables
pmlbr::fetch_data(x)character object
name of the dataset to fetch from PMLB
fetch_data('wine_quality_red')| fixed.acidity | volatile.acidity | ... | target |
|---|---|---|---|
| 7.4 | 0.700 | 5 | |
| 7.8 | 0.880 | 5 | |
| 7.8 | 0.760 | 5 | |
| 11.2 | 0.280 | 6 | |
| 7.4 | 0.700 | 5 | |
| 7.4 | 0.660 | 5 | |
| 7.9 | 0.600 | 5 |
fetch_data('wine_quality_red')PMLB for other purposes
teaching
first time open source contributors
Thanks!
@trang1618



Funding
NIH LM010098
NIH AI116794
R packages
ggplot2
partykit
ggparty
heatmaply
People
Jason Moore
Anonymous reviewers