
Hierarchical Automated Annotation of cell types
Viktor Petukhov , Peter Kharchenko
1,2
2,3
- University of Copenhagen, BRIC
- Harvard Medical School, DBMI
-
Harvard Stem Cell Institute
Problem
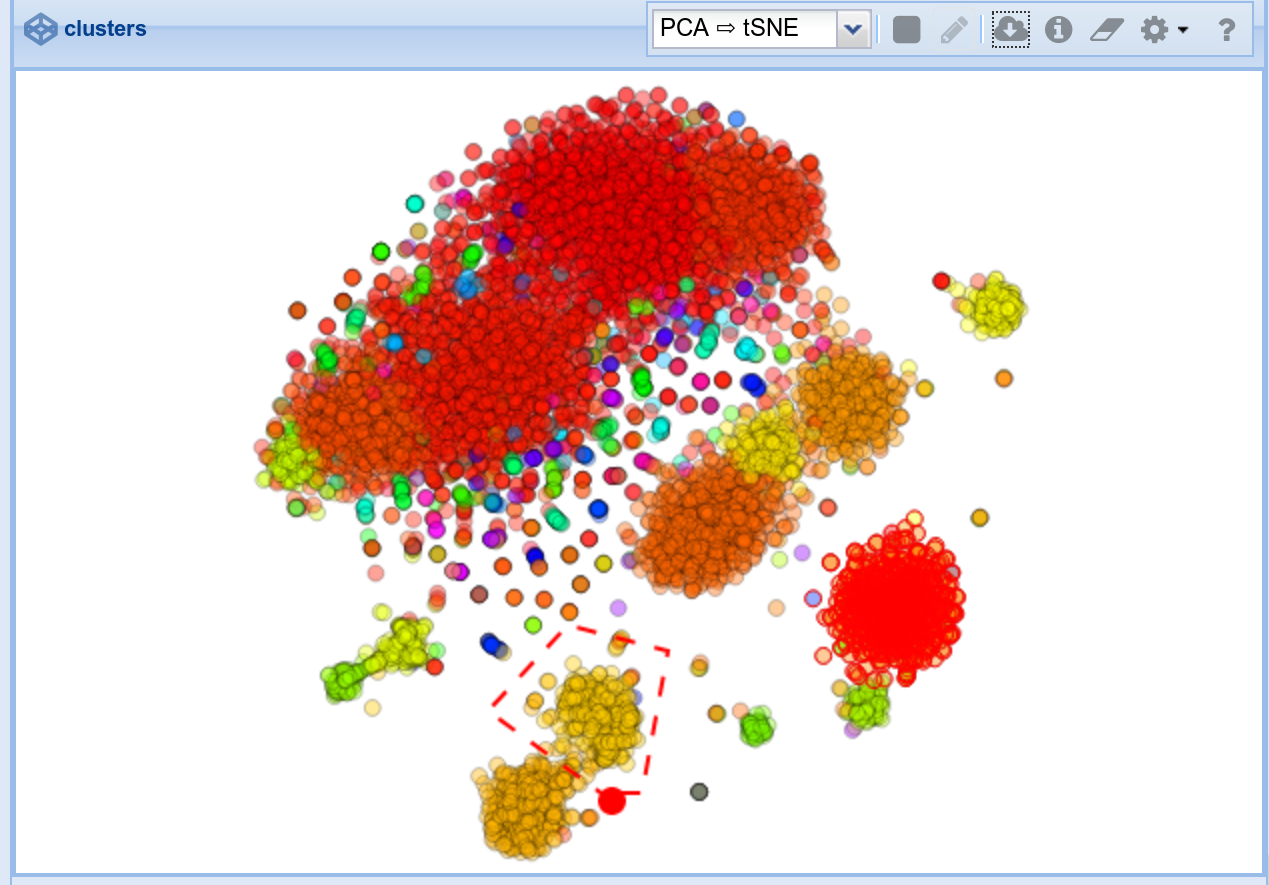
Manual annotation is painful!
Existing solutions
Based on
annotation
transfer
Based on
marker genes
Annotation transfer

Annotated cells
(e.g. published data)
Not-annotated
cells (e.g. your data)
Problems:
- Transfer between datasets suffers from batch-effect
- Isn't aware about cell type relations
- If it doesn't work, there is nothing you can do
Marker-based assignment

*Probabilistic cell-type assignment of single-cell RNA-seq for tumor microenvironment profiling, Nature Methods 2019
Benefits
- Intuitive
- Verifiable
- Transferable across datasets
Drawbacks
- Suffers from drop-outs
- Requires detailed annotation
- "Binarizes" expression matrix
Workflow
Inspired by Garnett*
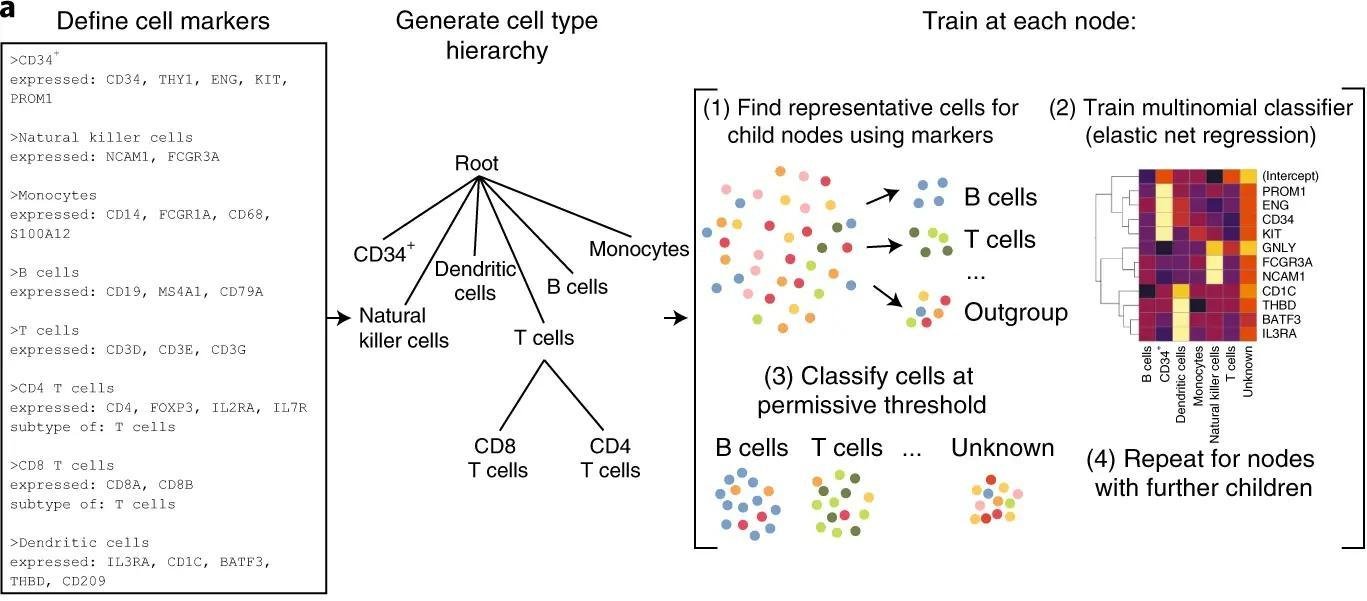
Modifications of the original idea
Cell Type scoring
- Estimate TF-IDF over expression matrix
- Normalize by features
- Estimate positive scores as sum over positive markers
- Negative score is a multiplier for positive score:
Modifications of the original idea
Label propagation
Graph diffusion (using Conos routines):
- ~100 times faster then Garnett GLMs
- Allow to work with alignment graph
- More pronounced work with hierarchies
- Workflow, optimized for easy marker selection
- Works on graphs, so can use joint graph from multiple samples
Modifications of the original idea
Other improvements
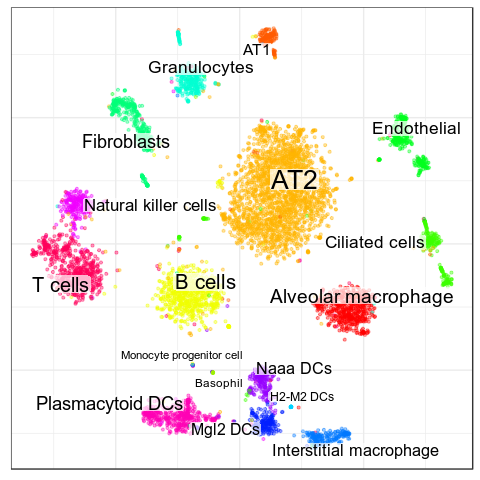
Benchmarks: MCA Lung
> AT2
expressed: Bex4
> AT1
expressed: Cryab
>Ciliated cells
expressed: Aldh1a1, Cyp2f2
> Interstitial macrophage
expressed: Apoe, Pf4
not expressed: Trbc2
> Alveolar macrophage
expressed: Ear1, Ear2
>T cells
expressed: Cd8b1, Trbc2
>Natural killer cells
expressed: Klra8, Nkg7
not expressed: Trbc2
> Naaa DCs
expressed: Naaa
> Mgl2 DCs
expressed: Mgl2> Plasmacytoid DCs
expressed: Plac8
> H2-M2 DCs
expressed: Epsti1, H2-M2
>Granulocytes
expressed: Il1b, Il1r2
>Endothelial
expressed: Pecam1, Flt1, Chd5, Kdr
>Fibroblasts
expressed: Dcn, Acta2, Inmt
>B cells
expressed: Cd19, Ms4a1, Cd79a
>Monocyte progenitor cell
expressed: Ctsg, Mpo
>Basophil
expressed: Ccl3, Ccl4
Cell Types
Garnett
Accuracy: 32.3%
Unclassified: 62.7%
Average TPR: 40.1%
Average Precision: 73.2%
Our code
Accuracy: 96.6%
Average TPR: 93.9%
Average Precision: 90.3%
Benchmarks: MCA Lung
Paper
CellAnnotatoR
Garnett
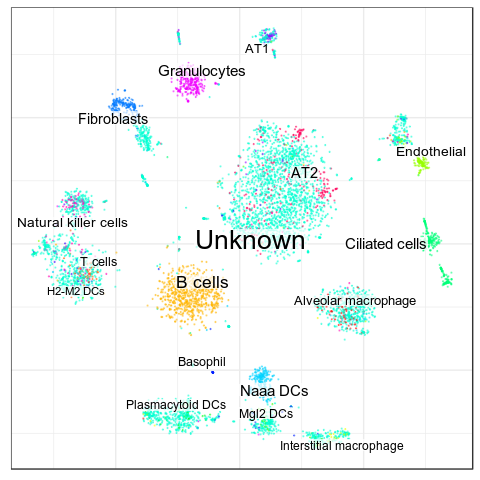
Benchmarks: MCA Lung, CellAssign
We couldn't get good results with CellAssign
(and we're not alone in this: Issue #35 "About results reproducibility")
CellAssign also doesn't use info about negative markers and cell type hierarchies
Accuracy: 7.0%
Average TPR: 10.2%
Average Precision: 6.2%
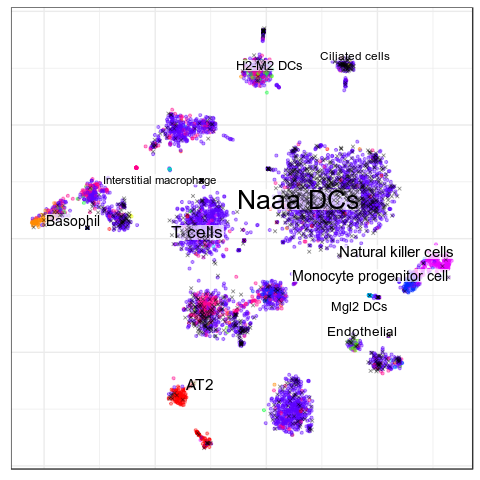
Benchmarks: MERFISH
Cell Types
>Inhibitory
expressed: Gad1
not expressed: Slc17a7
>Excitatory
expressed: Slc17a6, Slc17a7, Sema3c
not expressed: Gad1
>OD Mature
expressed: Ttyh2, Mbp, Opalin
not expressed: Pdgfra
>OD Immature
expressed: Pdgfra, Mki67
>Astrocyte
expressed: Aqp4
>Microglia
expressed: Selplg
>Ependymal
expressed: Cd24a
not expressed: Gad1
>Endothelial
expressed: Fn1
>Pericytes
expressed: Myh11
>Endothelial 1
expressed: Igf1r
subtype of: Endothelial
>Endothelial 2
expressed: Bmp7, Lepr
subtype of: Endothelial
>Endothelial 3
expressed: Ace2
subtype of: Endothelial
>OD Immature 1
expressed: Traf4
subtype of: OD Immature
>OD Immature 2
expressed: Mki67
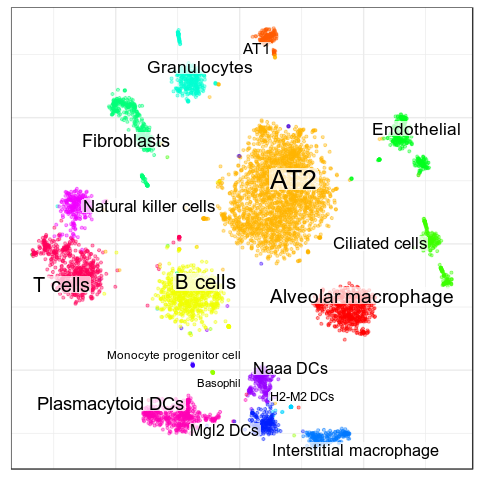
subtype of: OD Immature> AT2
expressed: Bex4
> AT1
expressed: Cryab
>Ciliated cells
expressed: Aldh1a1, Cyp2f2
> Interstitial macrophage
expressed: Apoe, Pf4
not expressed: Trbc2
> Alveolar macrophage
expressed: Ear1, Ear2
>T cells
expressed: Cd8b1, Trbc2
>Natural killer cells
expressed: Klra8, Nkg7
not expressed: Trbc2
> Naaa DCs
expressed: Naaa
> Mgl2 DCs
expressed: Mgl2Garnett
Accuracy: 23.3%
Unclassified: 75.1%
Average TPR: 8.4%
Average Precision: 26.2%
CellAnnotatoR
Accuracy: 90.0%
Average TPR: 84.6%
Average Precision: 83.7%
Benchmarks: MERFISH
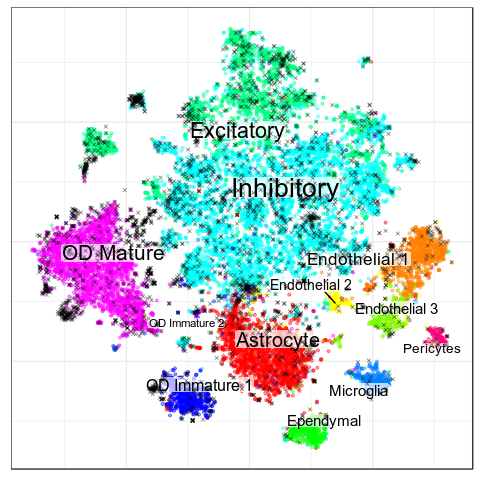
Paper
CellAnnotatoR
Garnett
Black crosses are ambiguous
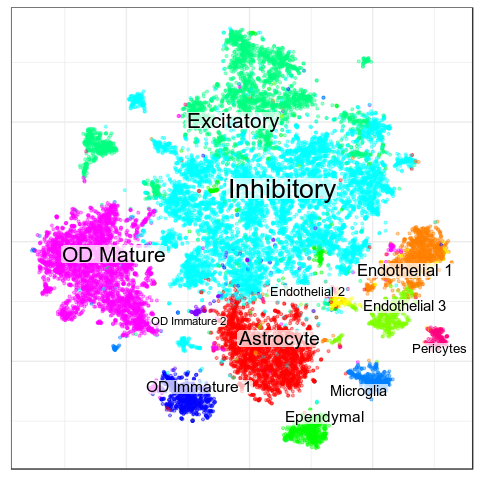
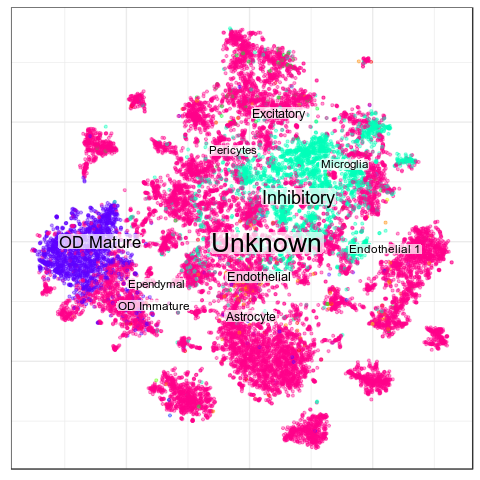
Benchmarks: Human Cortex
(our data)
>Astrocytes
expressed: SLC1A3, GJB6, FGFR3
not expressed: RBFOX3, SYP
> Microglia
expressed: CX3CR1, GPR34, P2RY12, MRC1
not expressed: RBFOX3, SYP
>Oligodendrocytes
expressed: MOG, ERMN
not expressed: RBFOX3, SYP
>Oligodendrocyte Precursors
expressed: CSPG4, PDGFRA, VCAN
not expressed: RBFOX3, SYP
>Vascular
expressed: DCN, PTGDS, ATP1A2, ITIH5, FLT1
not expressed: RBFOX3, SYP
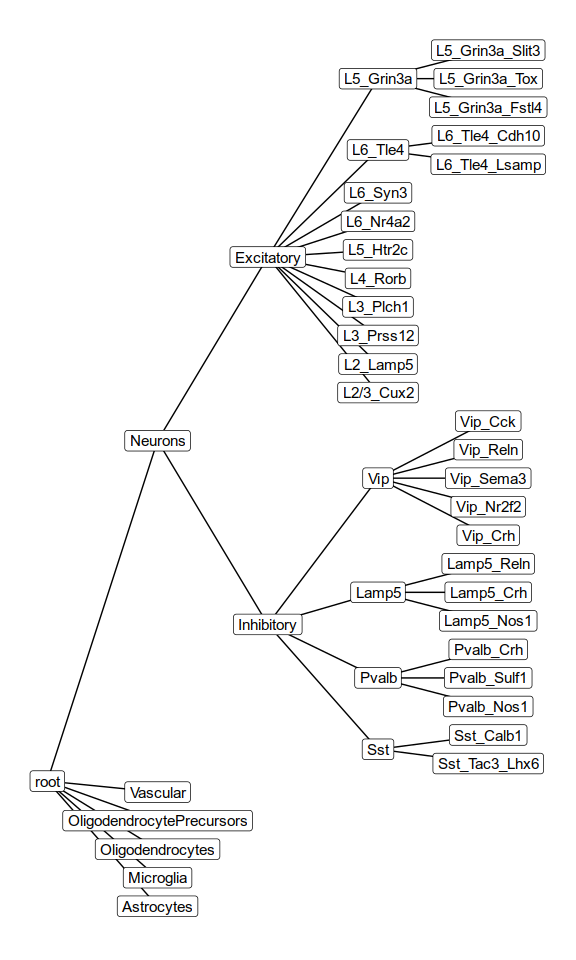
>Neurons
expressed: SYT1, SYP, SNAP25, RBFOX3
not expressed: MOG, ERMN, SLC1A3, CX3CR1, GPR34
# Neurons
>Inhibitory
expressed: GAD1, GAD2, SOX6, PVALB, SST, VIP, LHX6, NDNF, CALB2, SULF1
not expressed: SLC17A7, SATB2
subtype of: Neurons
>Excitatory
expressed: SLC17A7, SATB2, RORB, CUX2, TLE4, NR4A2, SEMA3C
not expressed: GAD1, GAD2, SOX6, PVALB
subtype of: Neurons
# Inhibitory
>Pvalb
expressed: PVALB, NOS1, SULF1, LHX6, KCNS3, CRH, PLEKHH2
not expressed: LAMP5, ID2, SST, FAM89A, RELN, SEMA6A, TAC3, DDR2, VIP
subtype of: Inhibitory
>Lamp5
expressed: ID2, LAMP5, SV2C, PDGFD, CCK, RELN
not expressed: VIP, CALB2, SST, FAM89A, DDR2, NR2F2
subtype of: Inhibitory
>Sst
expressed: SST, NOS1, SEMA6A, FAM89A, LHX6
not expressed: VIP, CALB2, CRH, CHAT, CCK, LAMP5, ID2, SV2C, PDGFD, PVALB, KCNS3
subtype of: Inhibitory
>Vip
expressed: VIP, TAC3, CALB2, NR2F2, LAMA3, COL5A2, SEMA3C, FAM19A1
not expressed: ID2, NOS1, LAMP5, PDGFD
subtype of: Inhibitory
## PVALB
>Pvalb_Nos1
expressed: NOS1
not expressed: CRH
subtype of: Pvalb
>Pvalb_Sulf1
expressed: SULF1
not expressed: NOS1, CRH
subtype of: Pvalb
>Pvalb_Crh
expressed: CRH, PLEKHH2
not expressed: NOS1, RGS5
subtype of: Pvalb
## LAMP5
>Lamp5_Nos1
expressed: NOS1, SFRP1
not expressed: LAMA3
subtype of: Lamp5
>Lamp5_Crh
expressed: CRH, SFRP1
subtype of: Lamp5
>Lamp5_Reln
expressed: RELN, LAMA3
not expressed: ID2
subtype of: Lamp5
## SST
>Sst_Tac3_Lhx6
expressed: TAC3, LHX6
not expressed: CALB1
subtype of: Sst
>Sst_Calb1
expressed: CALB1
not expressed: TAC3
subtype of: Sst
## VIP
>Vip_Crh
expressed: CRH, TAC3, IGFBP5
not expressed: SEMA3C, SEMA6A, NR2F2
subtype of: Vip
>Vip_Nr2f2
expressed: CRH, NR2F2, IGFBP5
not expressed: SEMA3C, SEMA6A, TAC3, RELN
subtype of: Vip
>Vip_Sema3
expressed: SEMA3C, SEMA6A, COL5A2
not expressed: CRH, RELN
subtype of: Vip
>Vip_Reln
expressed: RELN, DDR2
not expressed: TAC3, SEMA3C, IGFBP5
subtype of: Vip
>Vip_Cck
expressed: CCK, FAM19A1, NR2F2
not expressed: RELN, TAC3, IGFBP5, SEMA3C
subtype of: Vip
# Excitatory
>L2/3_Cux2
expressed: LAMP5, CUX2, COL5A2
not expressed: PDGFD, FAT4, PARD3, PRSS12, GABRG1, COBLL1, PXDN
subtype of: Excitatory
>L2_Lamp5
expressed: LAMP5, CUX2, PDGFD, PARD3
not expressed: RORB, GABRG1, COL5A2, PXDN
subtype of: Excitatory
>L3_Prss12
expressed: PRSS12, RORB, COBLL1, CUX2
not expressed: LAMP5, GABRG1, GRIN3A, CMTM8, PXDN, OPRK1, PDGFD, FAT4, PDZD2
subtype of: Excitatory
>L3_Plch1
expressed: PRSS12, RORB, COBLL1, PLCH1
not expressed: LAMP5, GABRG1, GRIN3A, CMTM8, PXDN, OPRK1
subtype of: Excitatory
>L4_Rorb
expressed: RORB, GABRG1, CUX2
not expressed: PRSS12, CMTM8, PXDN, OPRK1, LAMP5
subtype of: Excitatory
>L5_Grin3a
expressed: GRIN3A, TLL1, CMTM8, RORB, TOX
not expressed: HTR2C, CUX2, PXDN, OPRK1, GABRG1
subtype of: Excitatory
>L5_Htr2c
expressed: HTR2C, PARD3, NXPH2, TLE4
not expressed: CMTM8, PXDN, LGR6
subtype of: Excitatory
>L6_Nr4a2
expressed: NR4A2, POSTN, HTR2C
not expressed: PRSS12, KCNIP1, NXPH2, PXDN
subtype of: Excitatory
>L6_Syn3
expressed: PXDN, OPRK1
not expressed: CUX2, RORB, HTR2C, CMTM8
subtype of: Excitatory
>L6_Tle4
expressed: TLE4, LGR6
not expressed: CUX2, RORB, HTR2C
subtype of: Excitatory
## L5_Grin3a
> L5_Grin3a_Fstl4
expressed: FSTL4, PRKG1
not expressed: FAM19A1, NTM, RGS6, SLIT3
subtype of: L5_Grin3a
> L5_Grin3a_Tox
expressed: TOX, DCC
not expressed: FAM19A1, NTM, ROBO2, RGS6, SLIT3
subtype of: L5_Grin3a
> L5_Grin3a_Slit3
expressed: FAM19A1, NTM, ROBO2, RGS6, SLIT3
subtype of: L5_Grin3a
## L6_Tle4
> L6_Tle4_Lsamp
expressed: LSAMP, RYR2
not expressed: CDH10, CNTN4
subtype of: L6_Tle4
> L6_Tle4_Cdh10
expressed: CDH10, CNTN4
not expressed: LSAMP, RYR2
subtype of: L6_Tle4Cell Types
Cell type hierarchy
Benchmarks: Human Cortex
(our data)
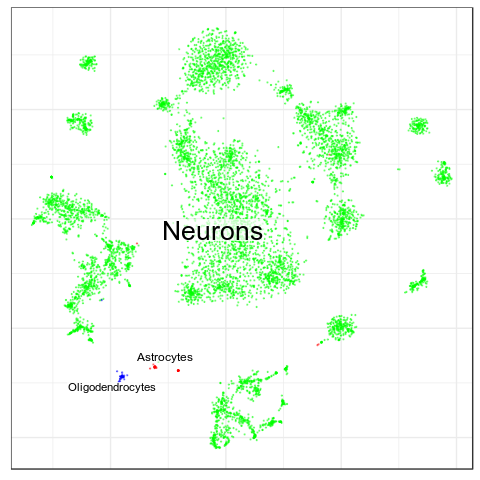
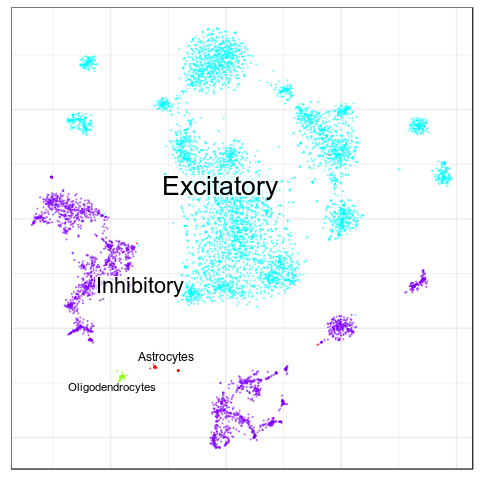
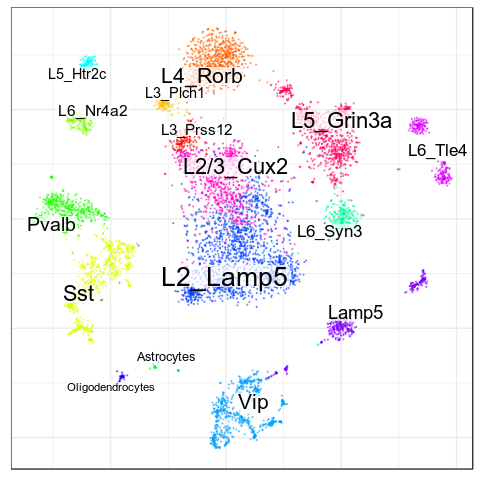
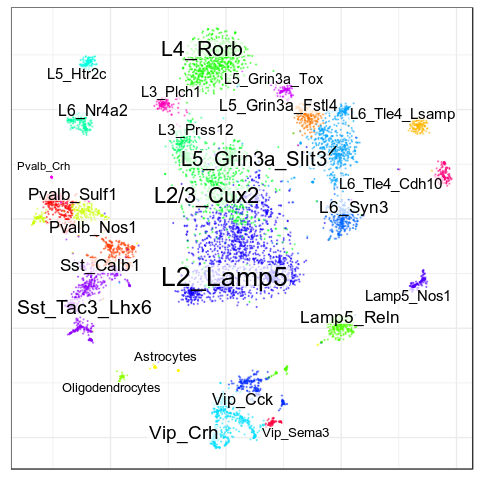
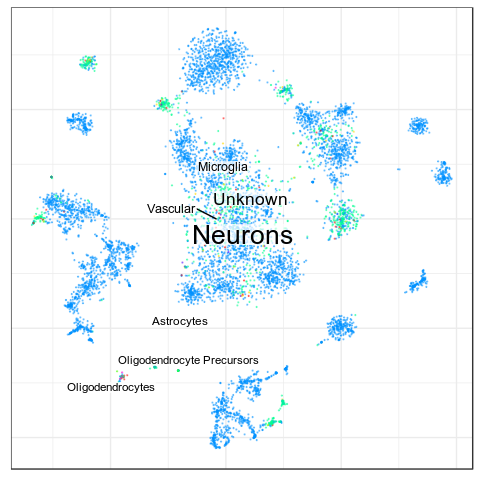
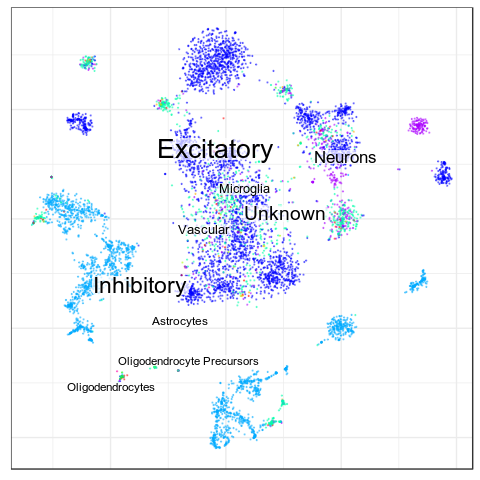
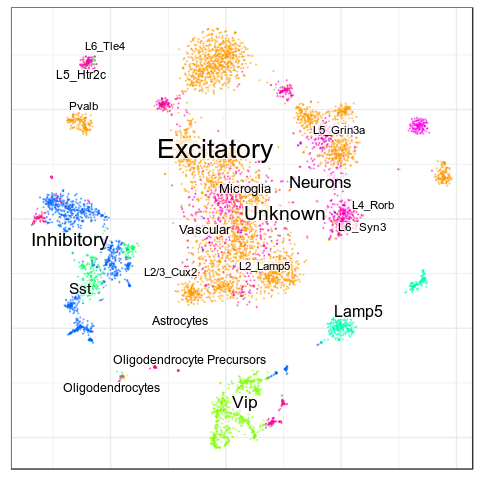
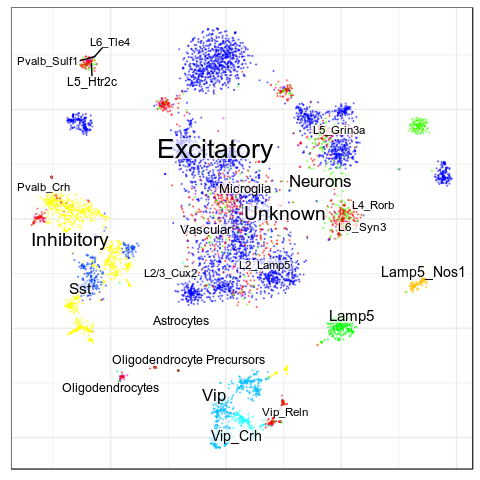
CellAnnotatoR
Garnett
"Recognized" cells:
85.0%
78.4%
16.6%
4.0%
No "ground truth" here, but we validated our annotation with the corresponding markers.
"Recognized cells" mean fraction of cells, which has at least some label from the corresponding level
Possible improvements
- Multiple types ("tags") per cell
- Automated selection of marker genes
- Good uncertainty estimates
- More robust way of working with gene ids