
Baysor: Cell segmentation in imaging-based spatial transcriptomics
Petukhov V, Xu RJ, Soldatov RA, Cadinu P, Khodosevich K, Moffit JR, Kharchenko P
Problem
How to map expression to space?
- MERFISH
- ISS
- osmFISH
- smFISH
- BaristaSeq
- Up to 10 cells
- Up to 10 transcripts
- Up to 10000 genes
Spatial protocols based on RNA-FISH or in situ sequencint
- DARTFISH
- ex-FISH
- StarMAP
- seq-FISH
- FISSEQ
Protocols
Data
6
7
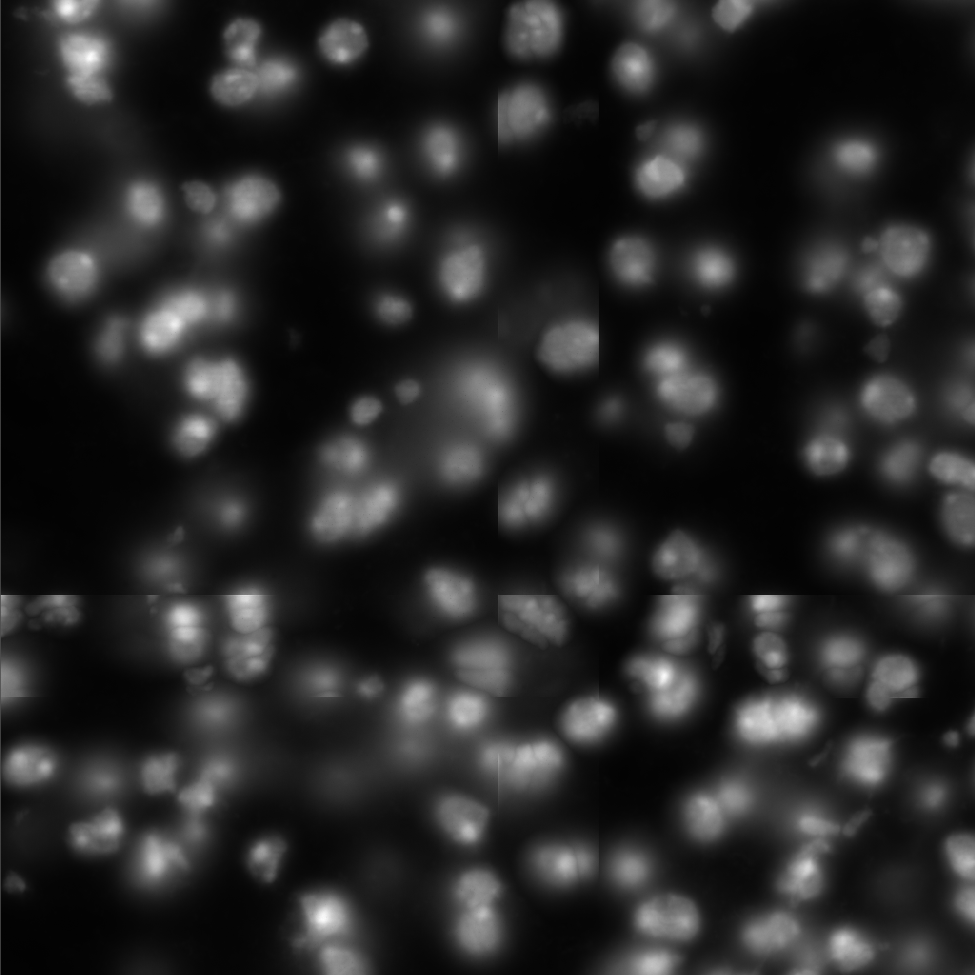
Segmentation problems

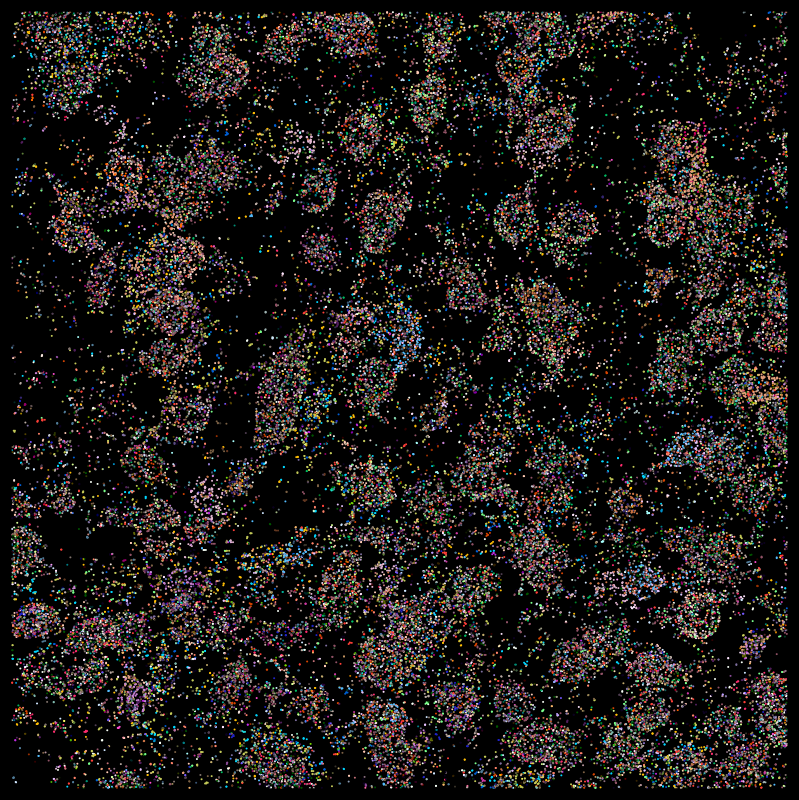
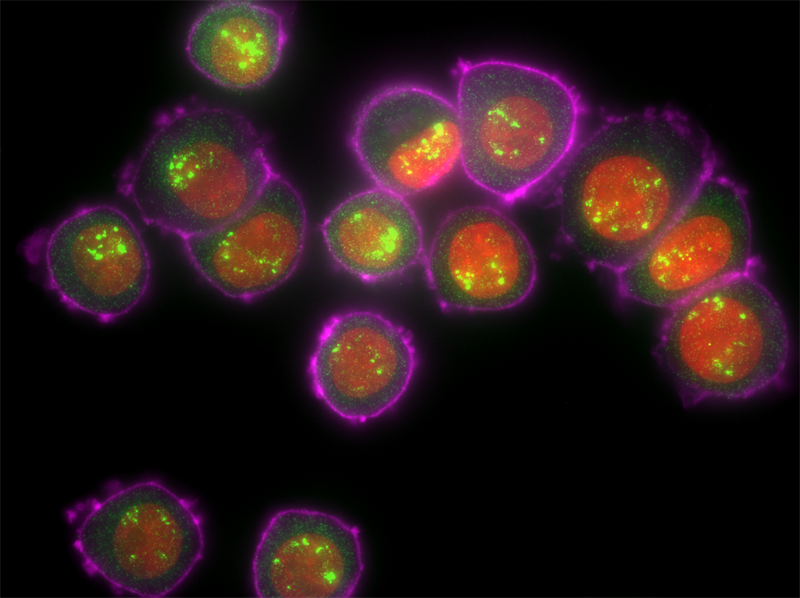
Molecules
DAPI
*Moffitt, J. R. et al. Molecular, spatial and functional single-cell profiling of the hypothalamic preoptic region. Science 362 (2018)
Segmentation problems
MERFISH
[1]
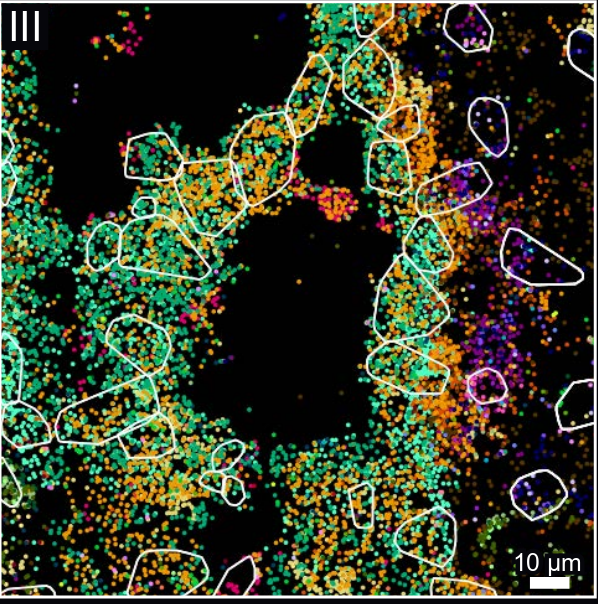
osm-FISH
[2]
1. Moffitt, J. R. et al. Molecular, spatial and functional single-cell profiling of the hypothalamic preoptic region. Science 362 (2018)
2. Simone Codeluppi, Lars E. Borm, Amit Zeisel, Gioele La Manno, Josina A. van Lunteren, Camilla I. Svensson & Sten Linnarsson. Nature Methods 15, 932–935 (2018)
Old results
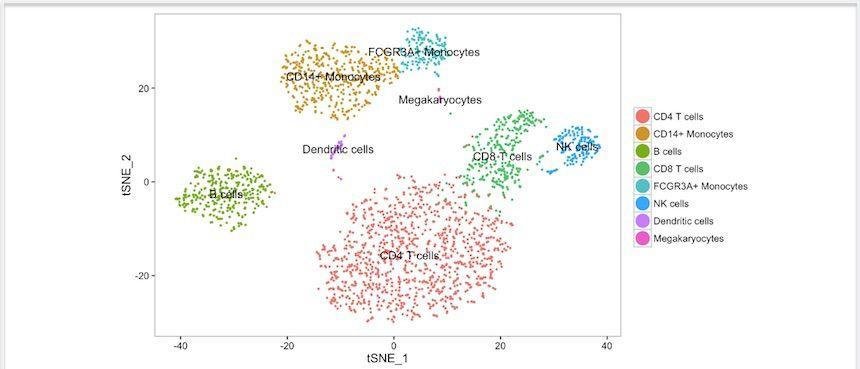
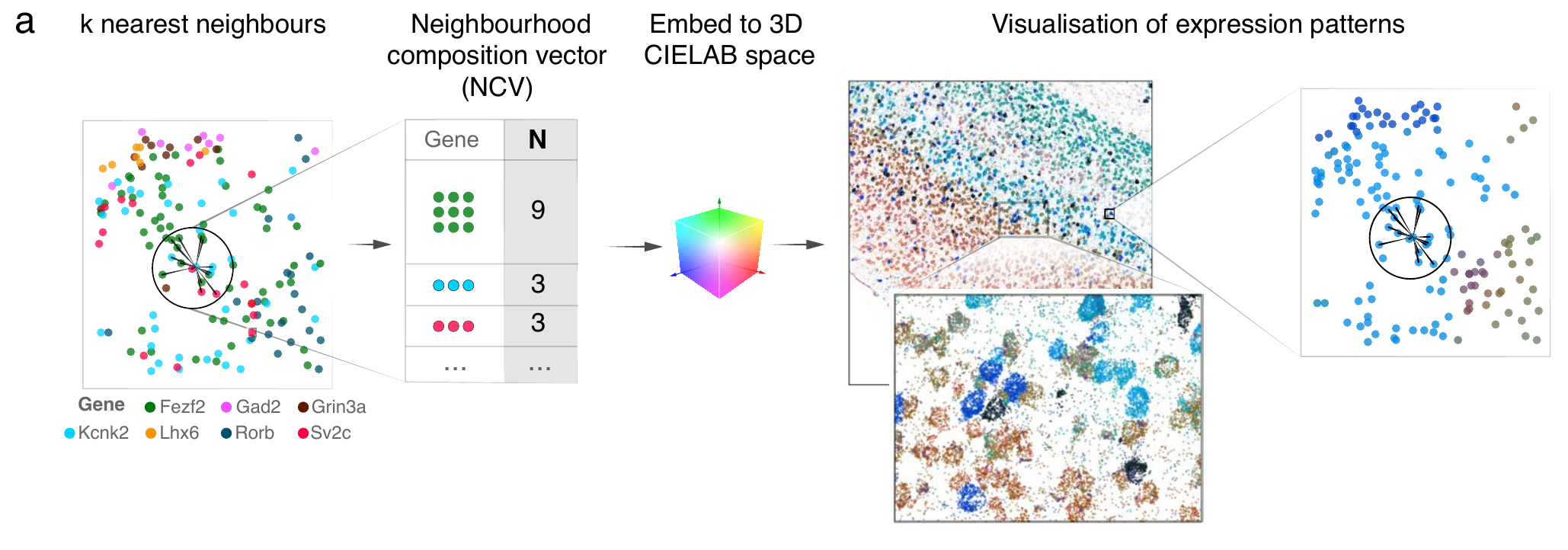
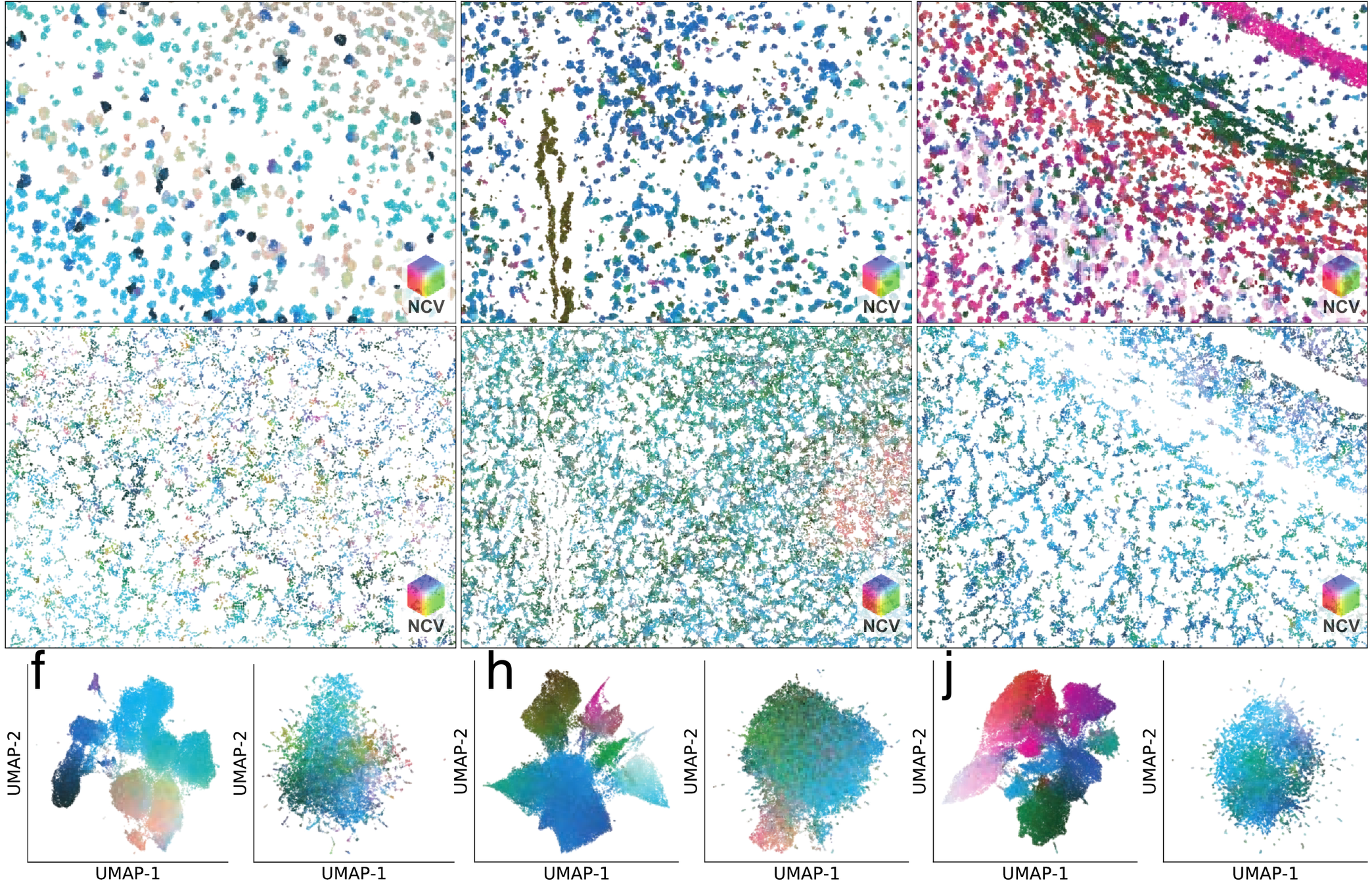
Neighborhood composition
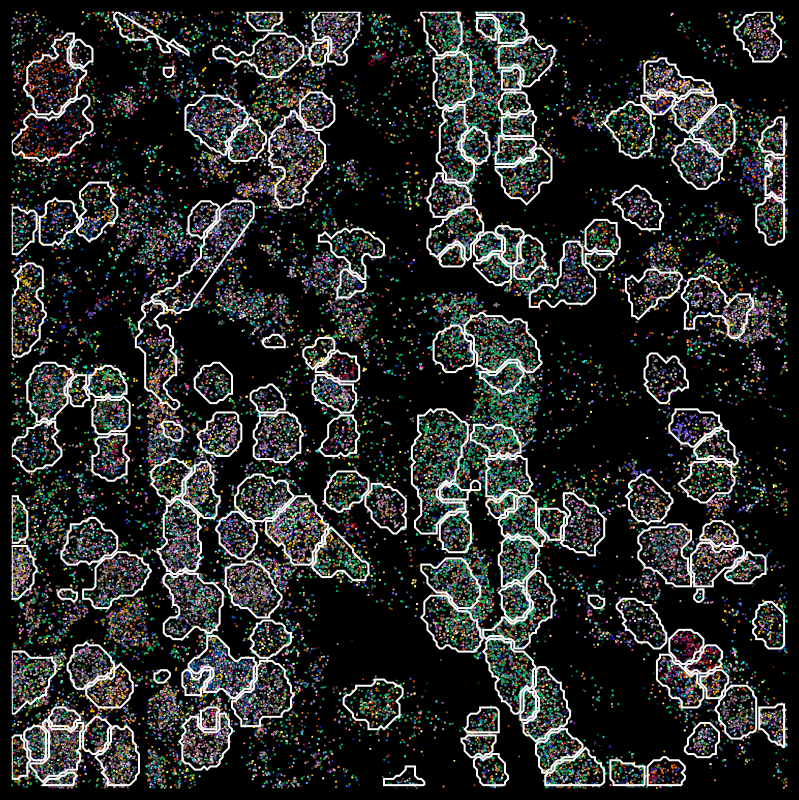
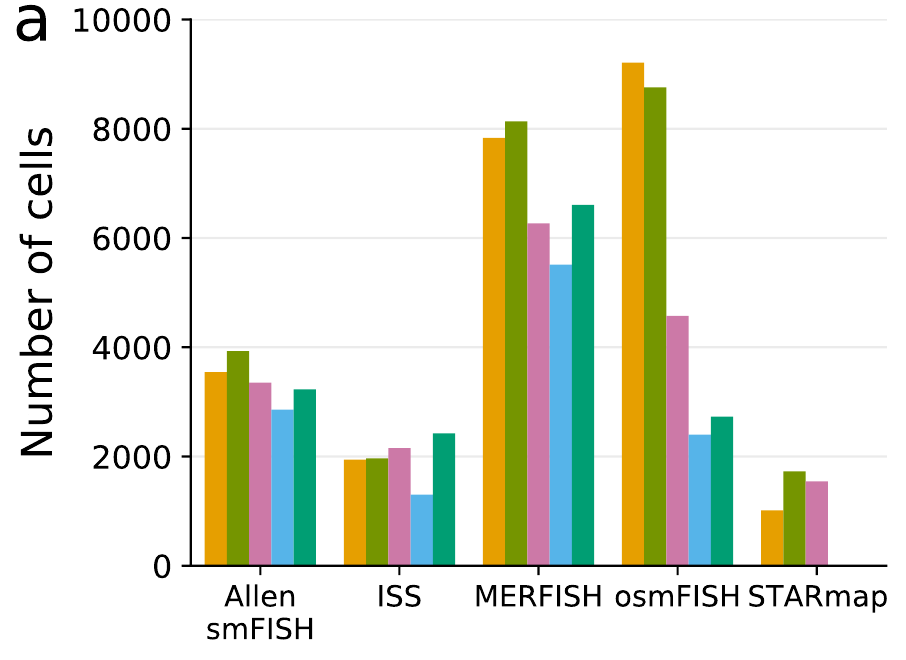
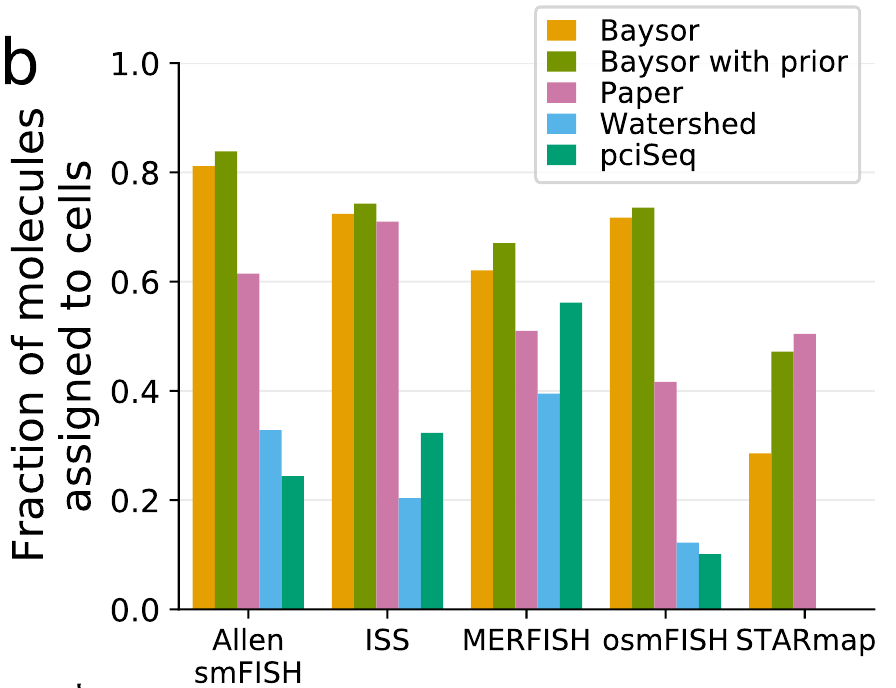
Segmentation across protocols
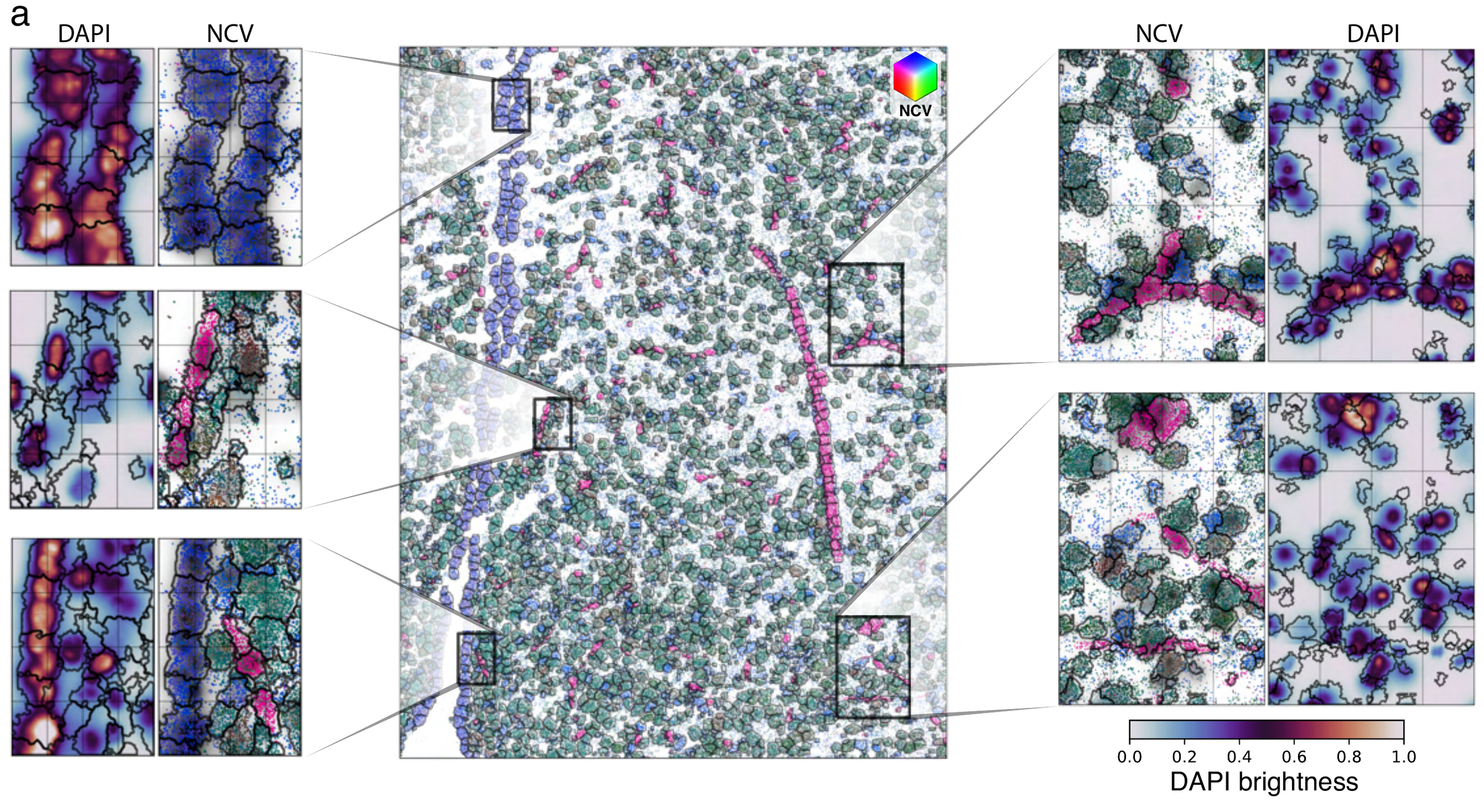
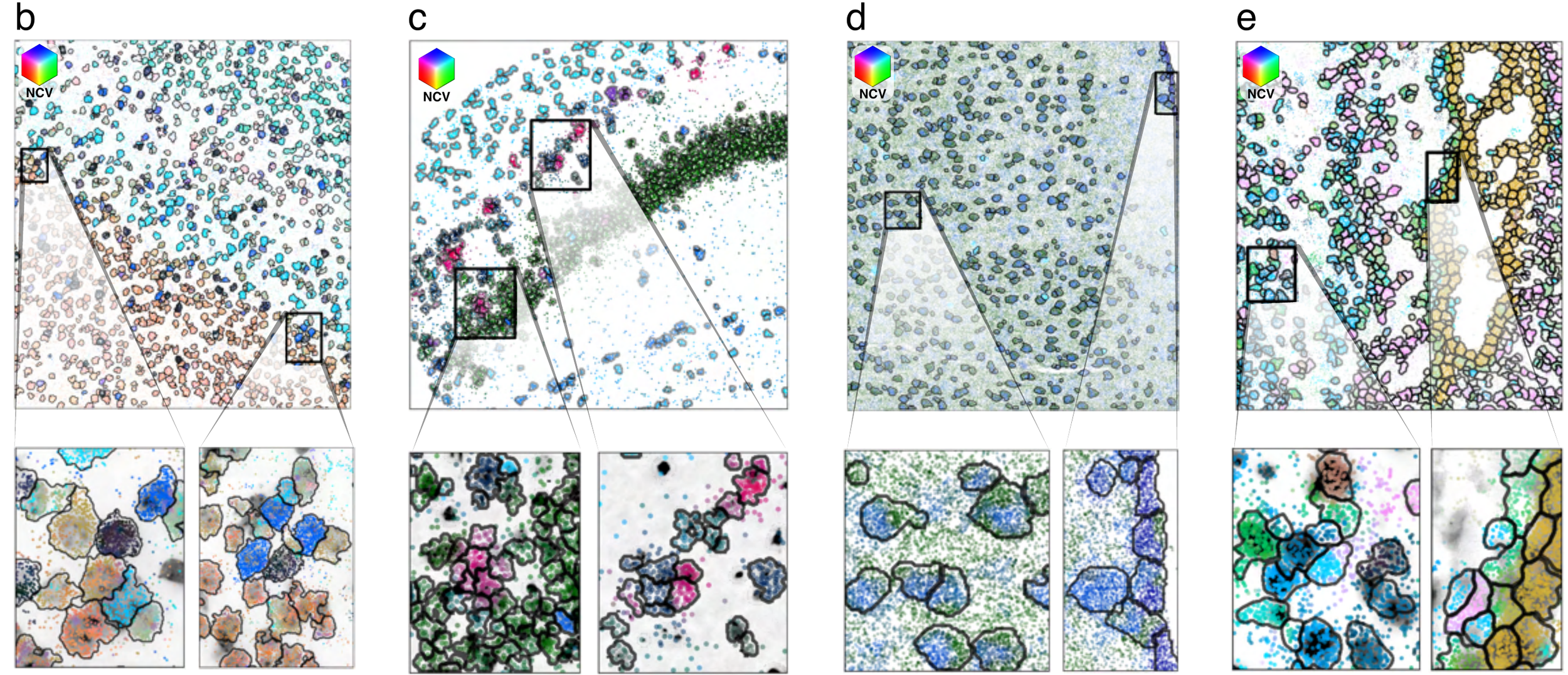
Segmentation across protocols
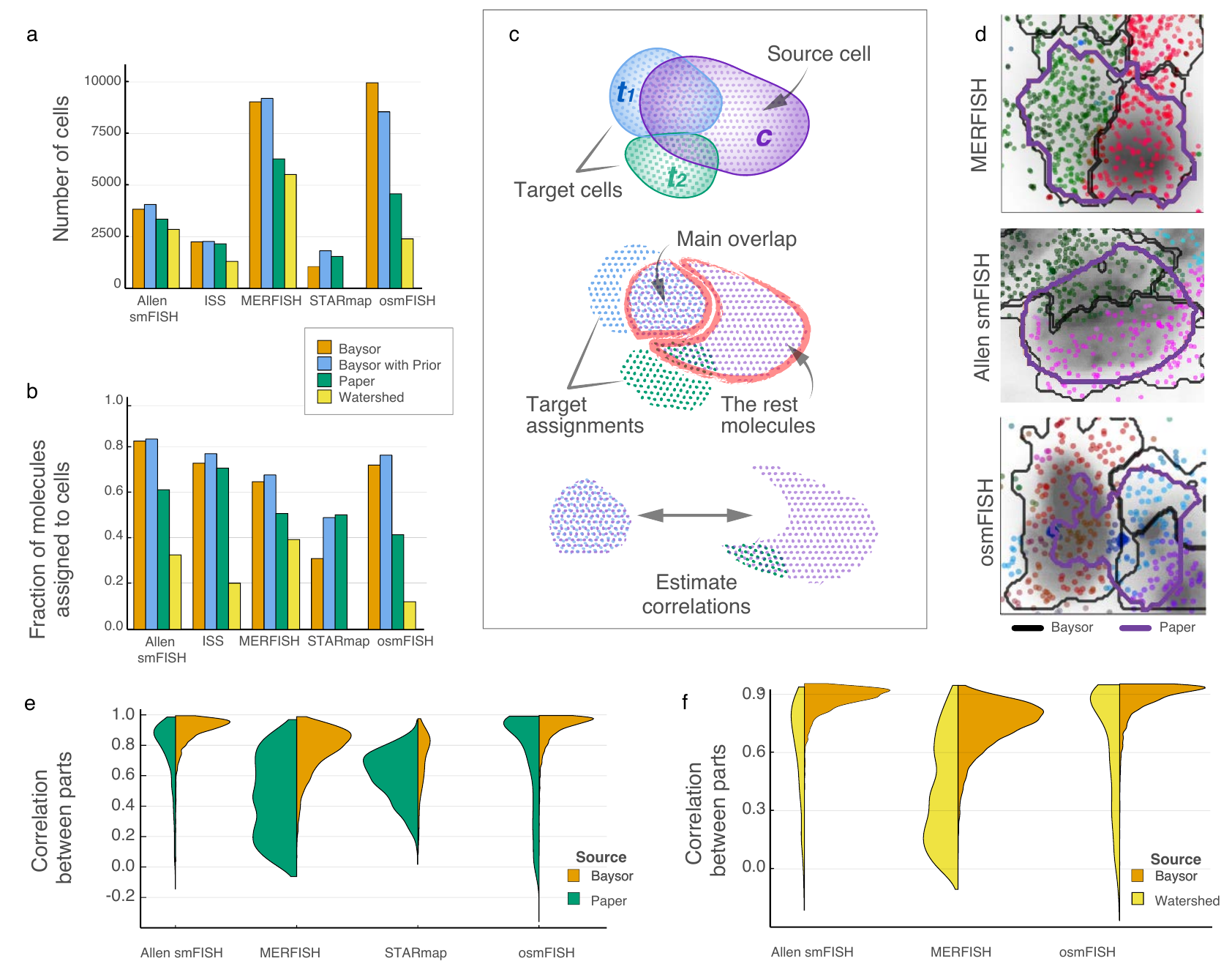
Benchmarking
New results
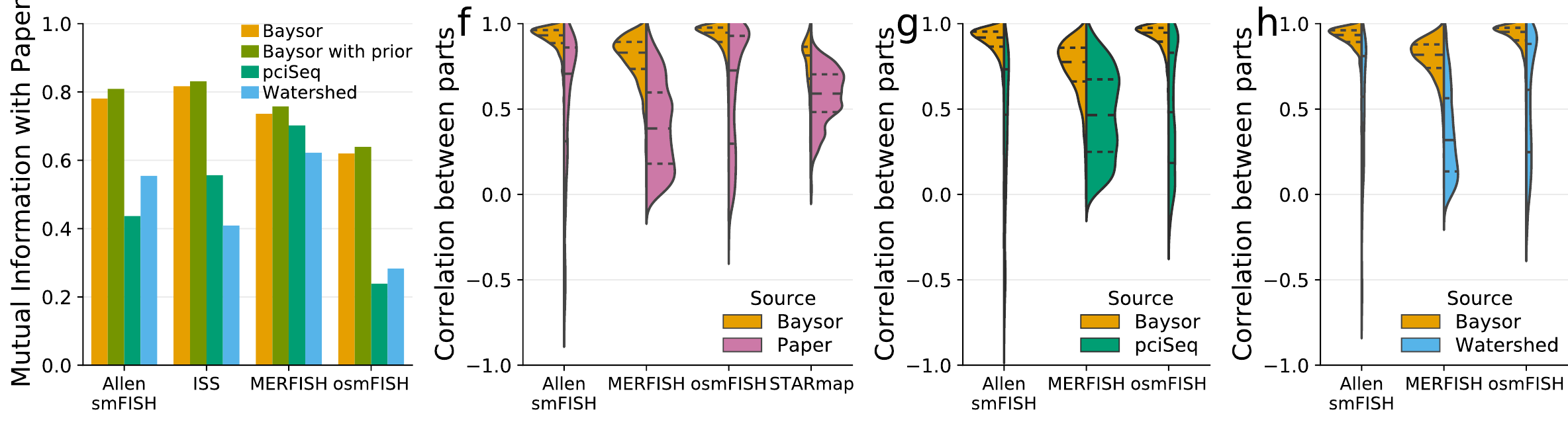
More benchmarks: pciSeq
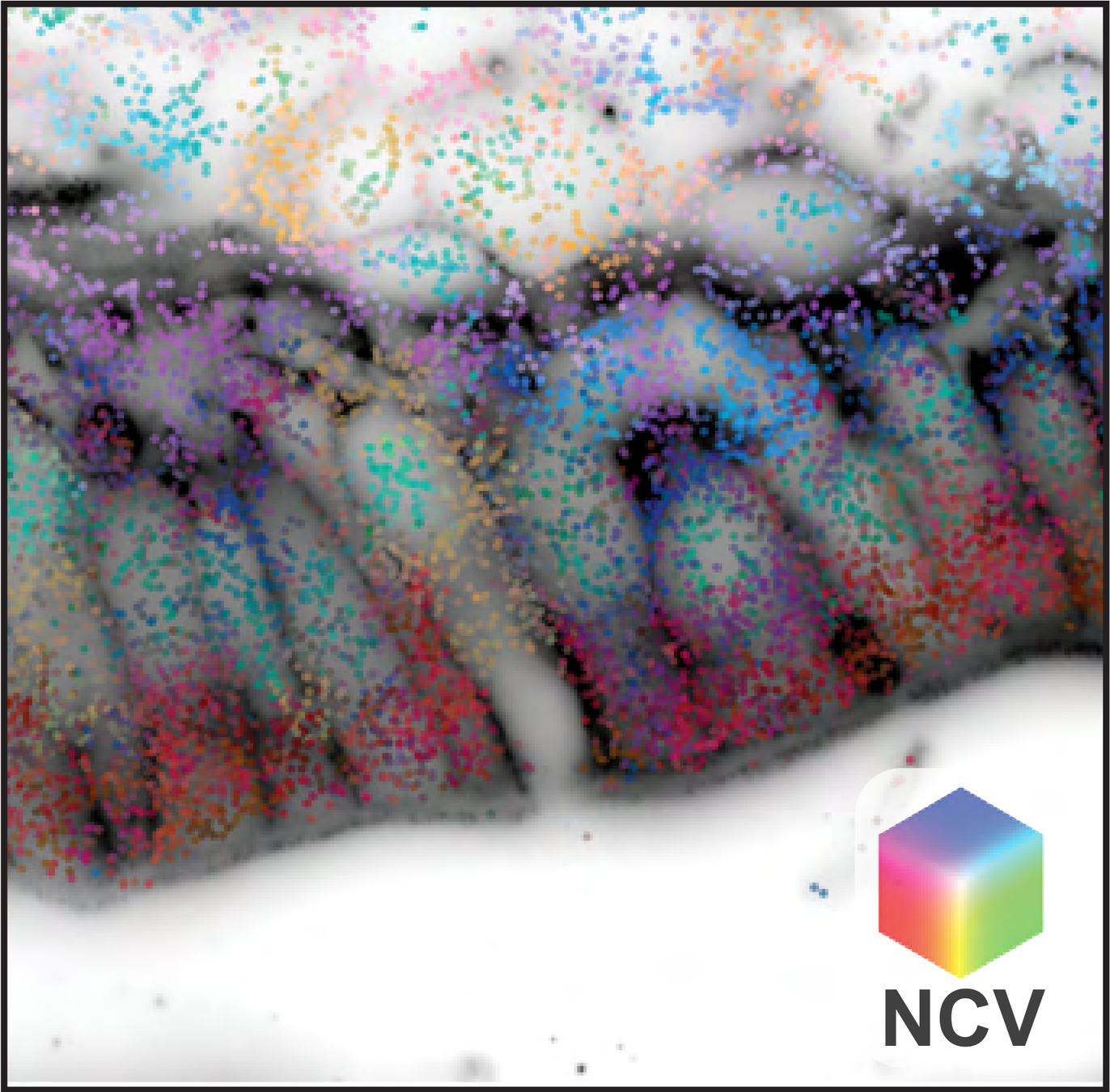
MERFISH gut data
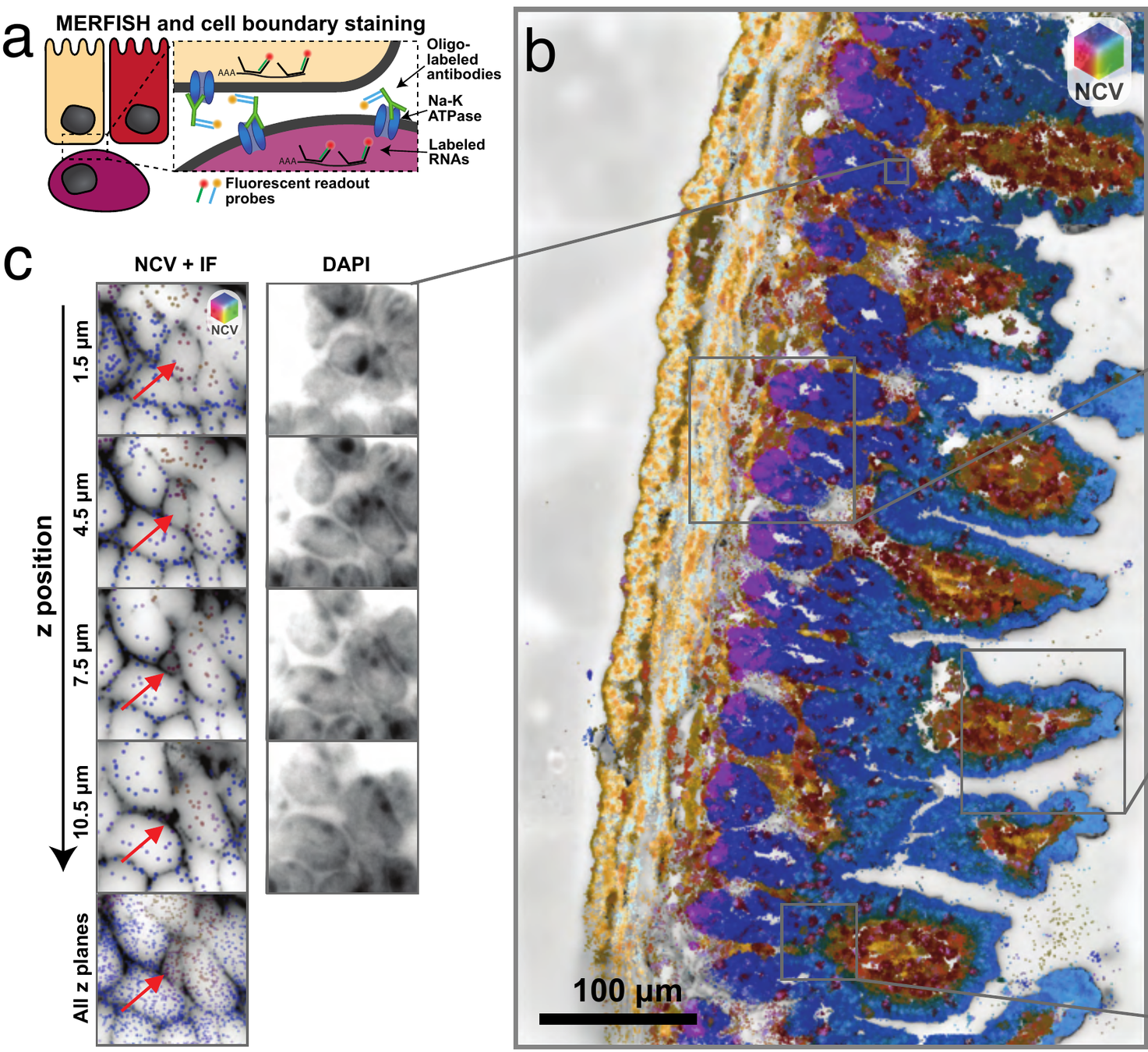
Segment nuclei against cytoplasm
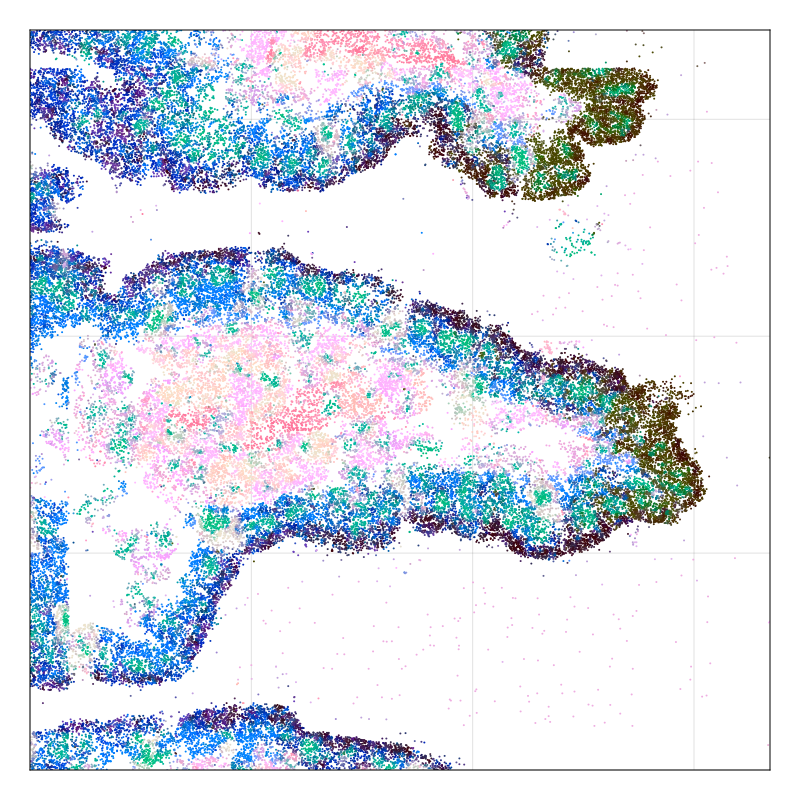
NCV
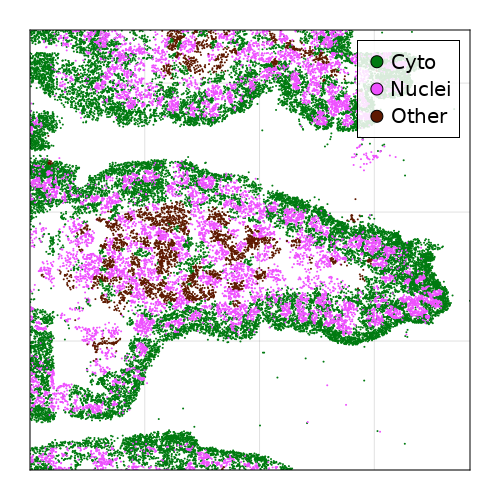
Compartments
Segment nuclei against cytoplasm
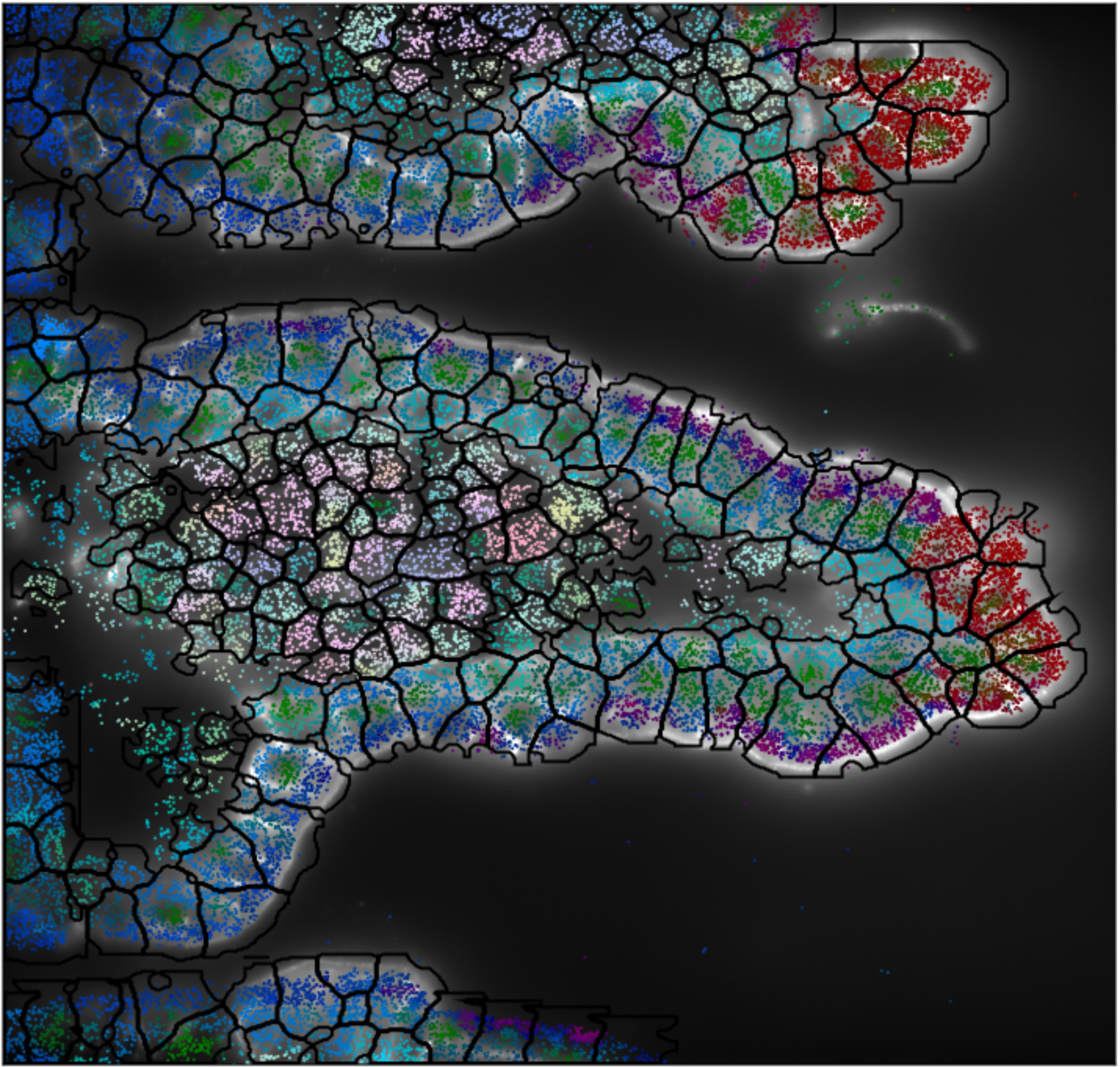
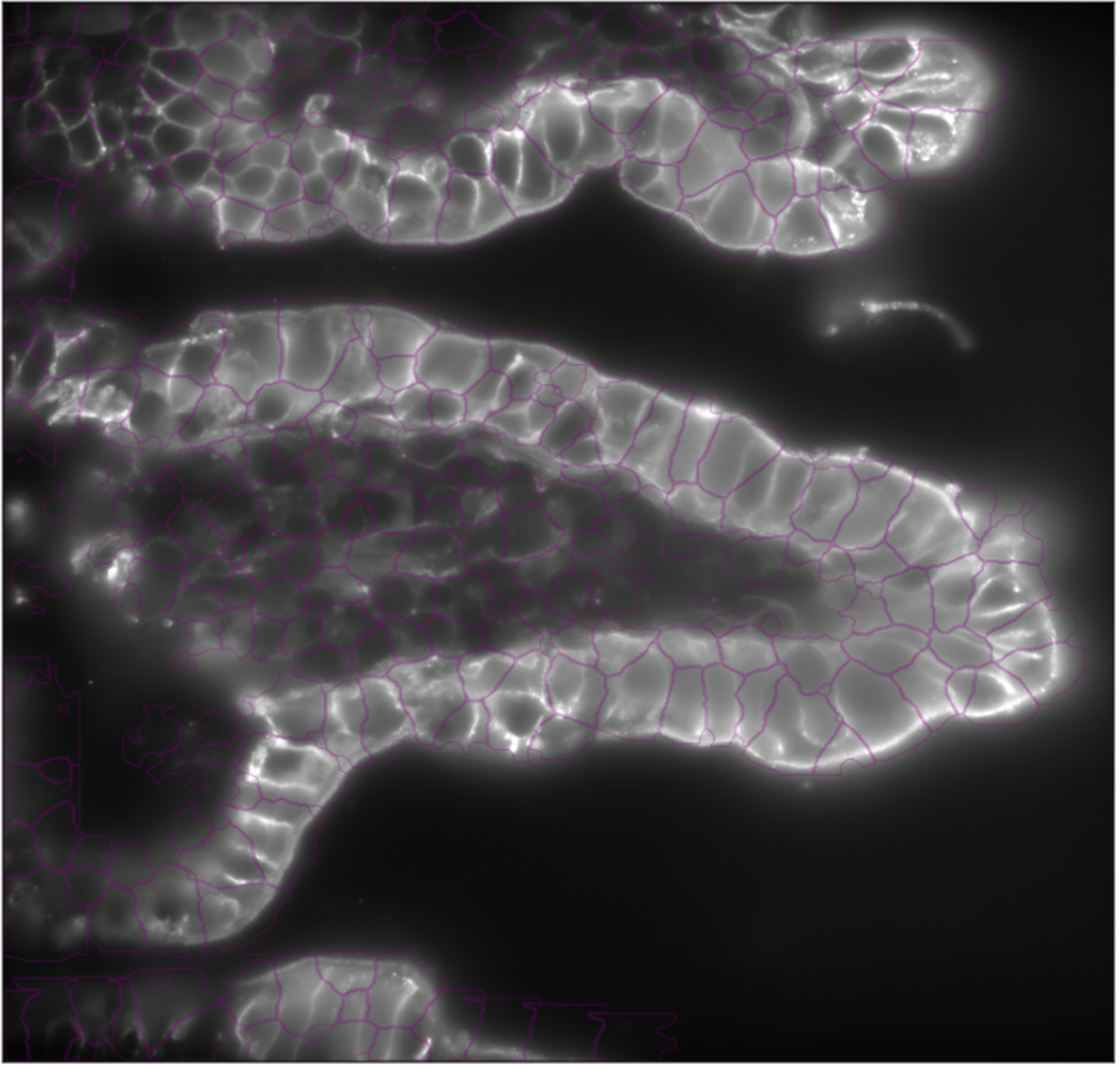
3D segmentation
Other plots
Thank you!
