
Baysor: segmentation of spatial transcriptomics data
Viktor Petukhov
Khodosevich lab, BRIC
How to map expression to space?
Spatial gene expression patterns

pciSeq
MERFISH
1. X. Qian, K.D. Harris, T. Hauling, D. Nicoloutsopoulos, A.M. Manchado, N. Skene, J. Hjerling-Leffler, M. Nilsson, bioRxiv 2018, 431957276097
2. Moffitt, J. R. et al. Molecular, spatial and functional single-cell profiling of the hypothalamic preoptic region. Science 362 (2018)
[1]
[2]
- MERFISH
- ISS
- osmFISH
- smFISH
- BaristaSeq
- Up to 10 cells
- Up to 10 transcripts
- Up to 10000 genes
Spatial protocols based on RNA-FISH or in situ sequencint
- DARTFISH
- ex-FISH
- StarMAP
- seq-FISH
- FISSEQ
Protocols
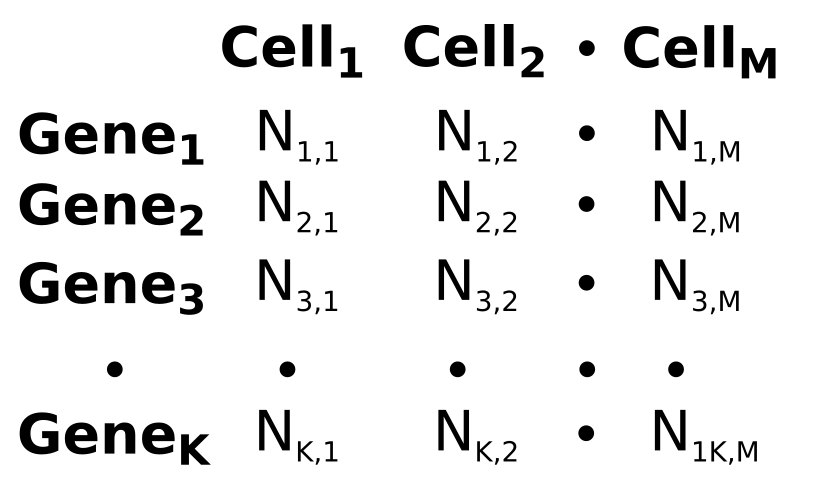
Data
6
7
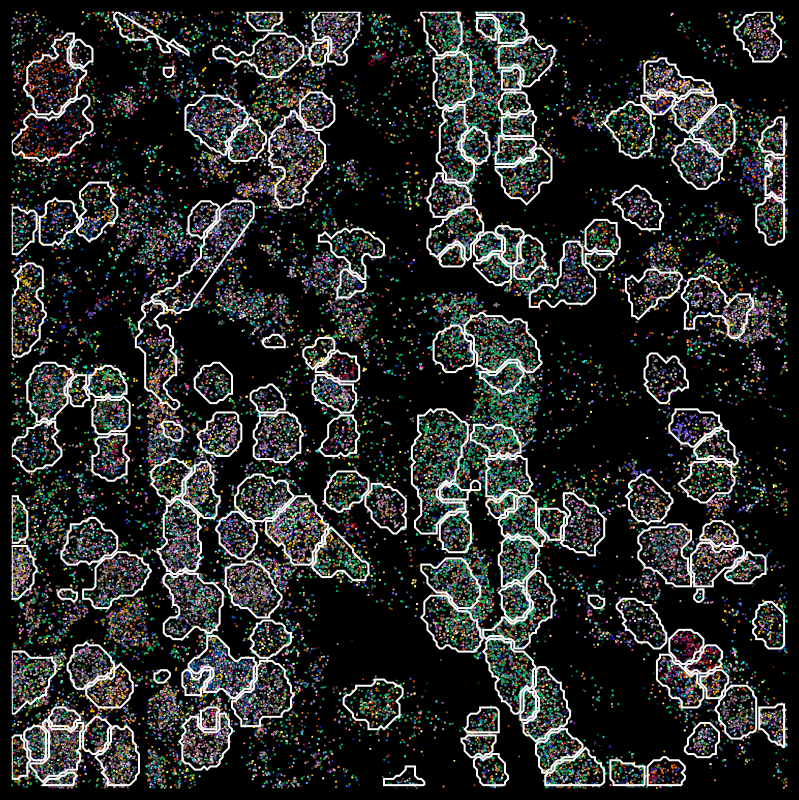
Segmentation problems
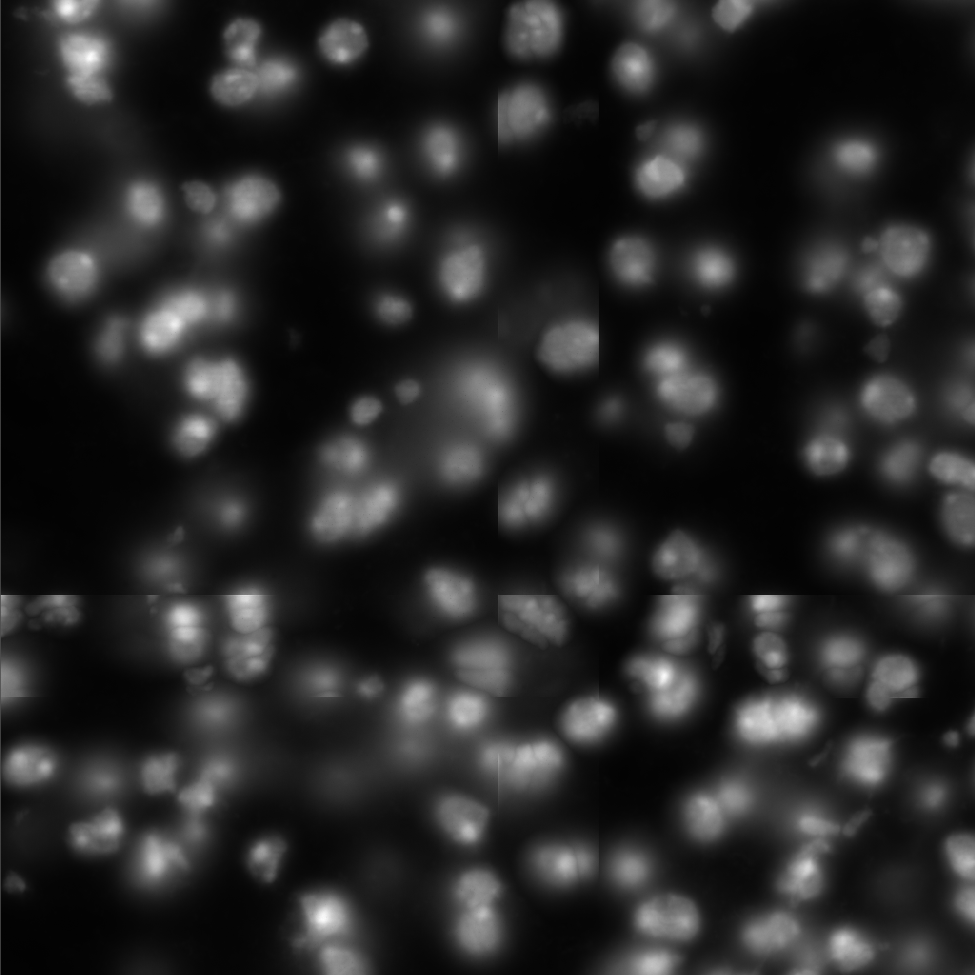

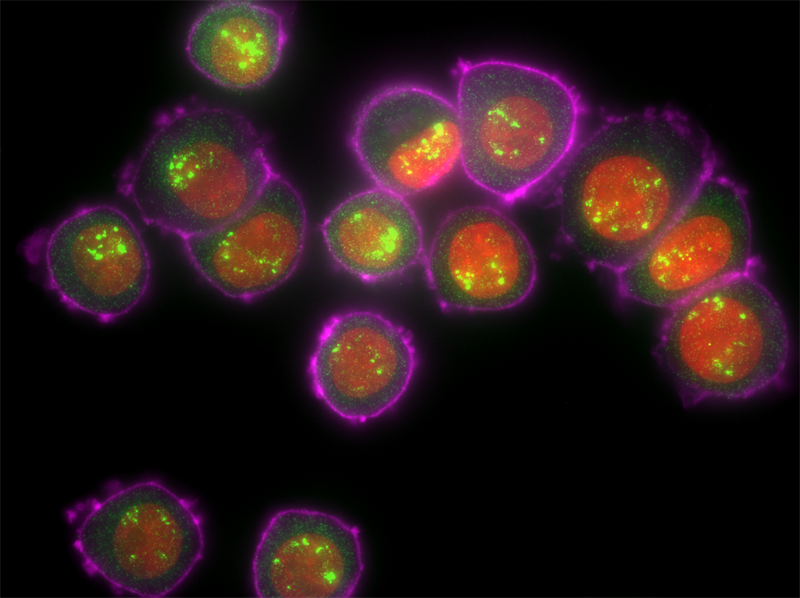
Molecules
DAPI
*Moffitt, J. R. et al. Molecular, spatial and functional single-cell profiling of the hypothalamic preoptic region. Science 362 (2018)
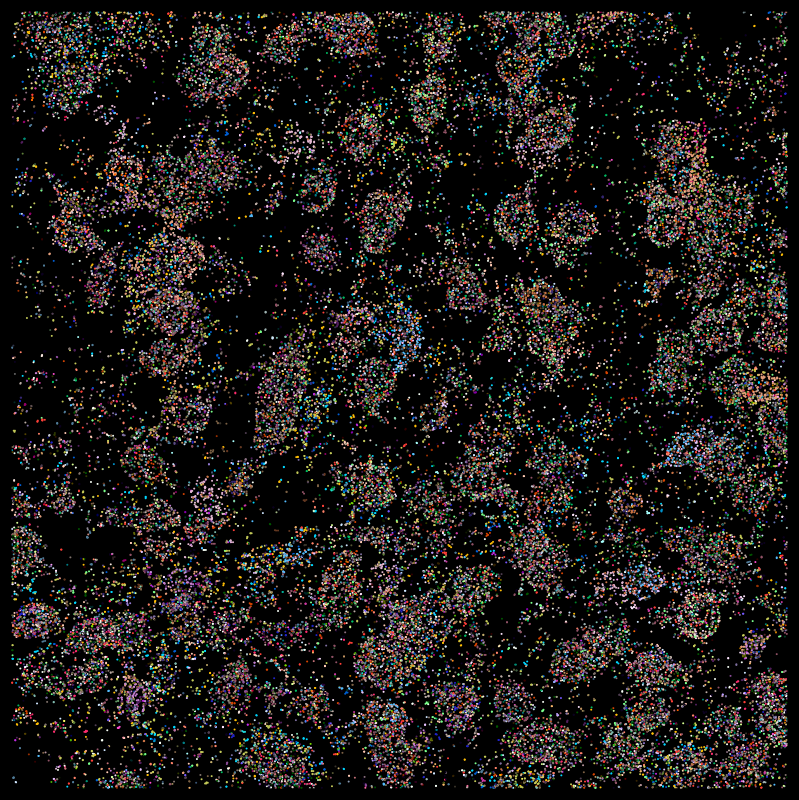
Segmentation problems


*Moffitt, J. R. et al. Molecular, spatial and functional single-cell profiling of the hypothalamic preoptic region. Science 362 (2018)
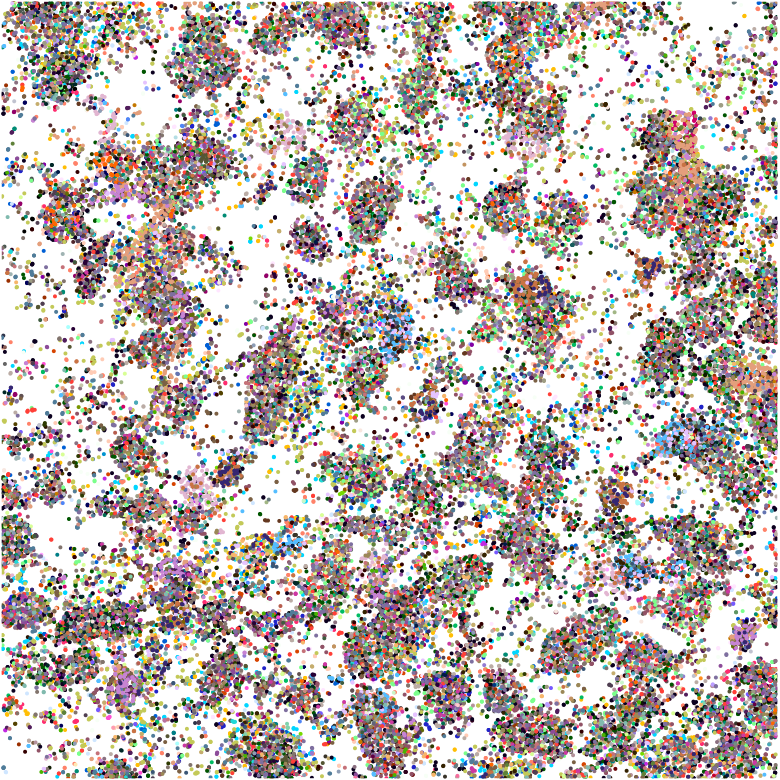
Segmentation problems
MERFISH
[1]
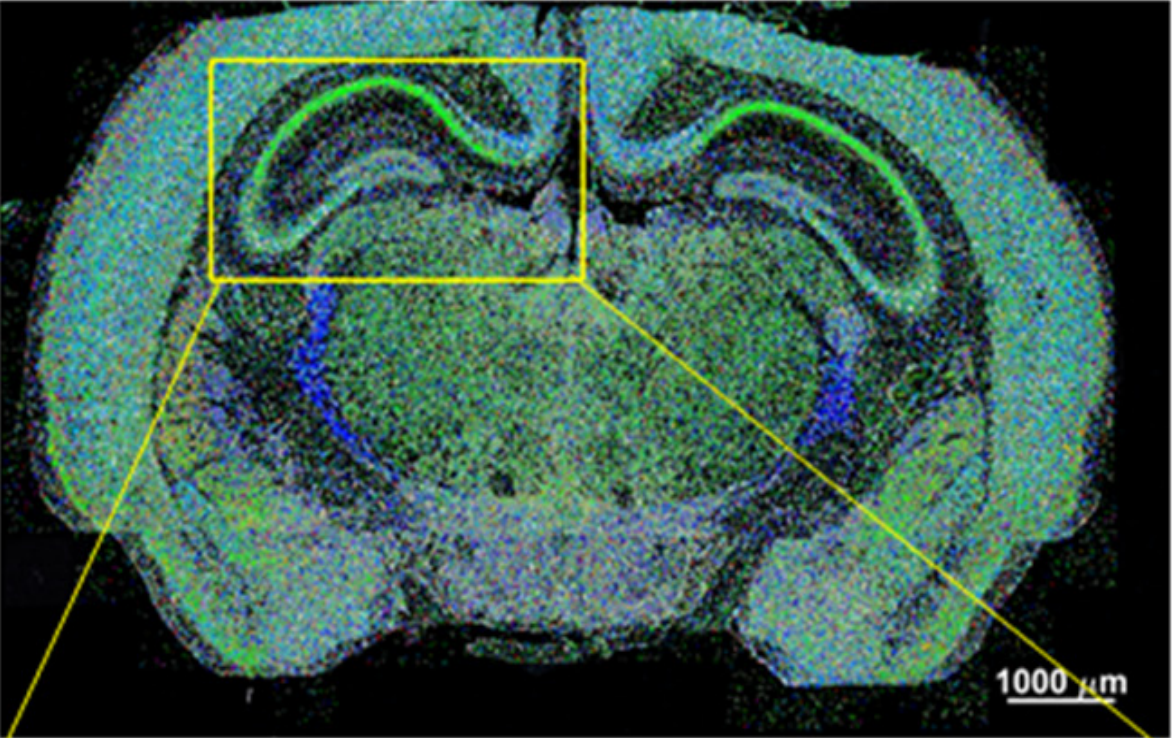
osm-FISH
[2]
1. Moffitt, J. R. et al. Molecular, spatial and functional single-cell profiling of the hypothalamic preoptic region. Science 362 (2018)
2. Simone Codeluppi, Lars E. Borm, Amit Zeisel, Gioele La Manno, Josina A. van Lunteren, Camilla I. Svensson & Sten Linnarsson. Nature Methods 15, 932–935 (2018)
Baysor: segmentation of spatial transcriptomics data

| X | Y | Gene |
|---|---|---|
| ... | ... | ... |
Expected
cell size
Transcript data
DAPI
Poly-A
staining
__
Optional
Baysor: Dirichlet Mixture Models over Markov Random Fields
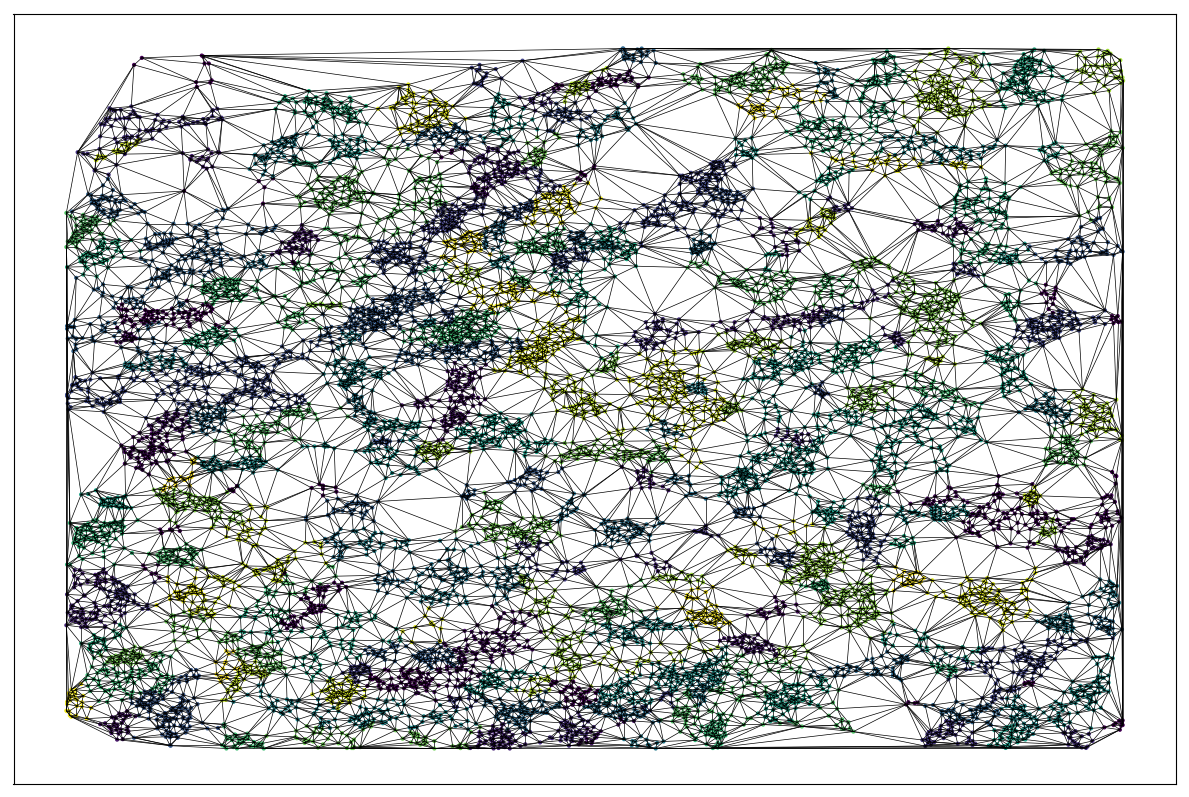
Random field via triangulation
Cell as a distribution

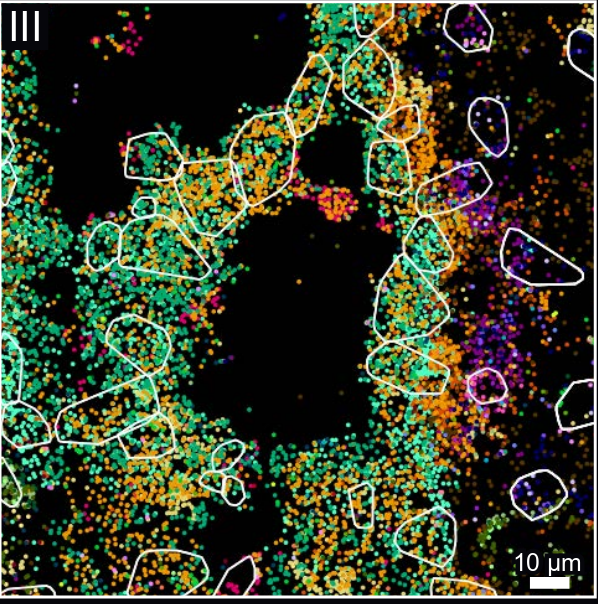
Baysor: Alorithm demonstration
Segmentation results
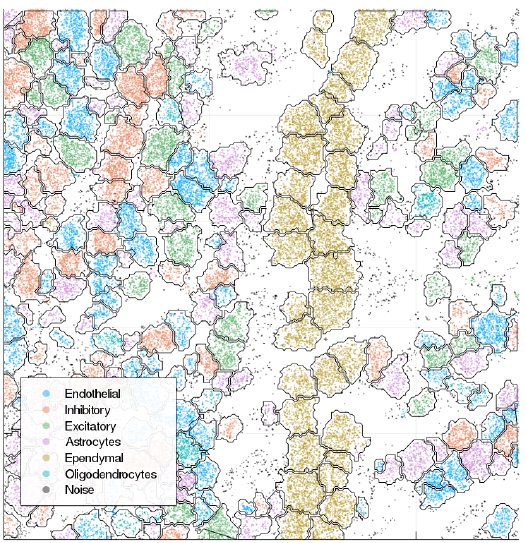
Local gene composition
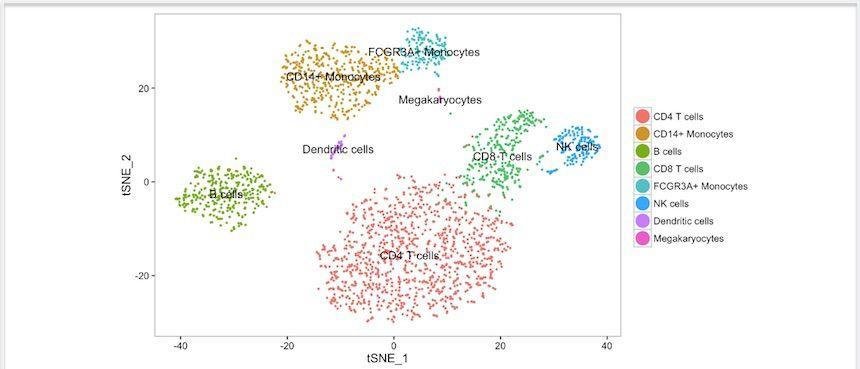
Cell type
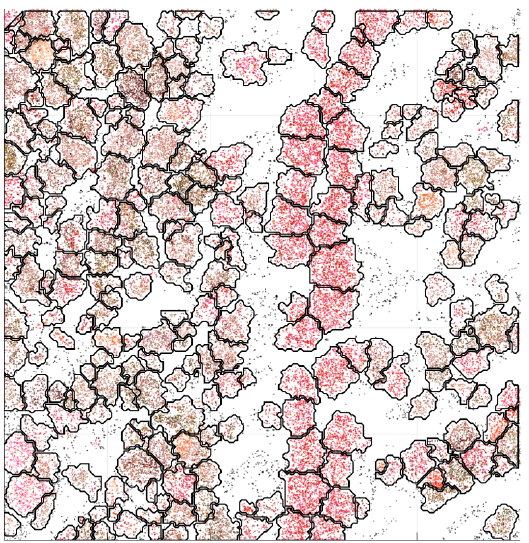
Segmentation results
| Protocol | Baysor | Staining |
|---|---|---|
| osm-FISH | 10059 | 3139 |
| Allen sm-FISH | 4435 | 2525 |
| MERFISH (subset) |
9279 | 6119 |
Number of segmented cells
%of assigned molecules
| Protocol | Baysor | Staining |
|---|---|---|
| osm-FISH | 87.4 | 44.1 |
| Allen sm-FISH | 79.6 | 61.6 |
| MERFISH (subset) |
75.5 | 47.4 |
Come to the poster for more details!

Acknowledgments


- Sten Linnarson lab
- Mats Nilsson lab
- Long Cai lab
- Jeffrey Moffitt lab
- SpaceTx consortium
Konstantin Khodosevich lab
Peter Kharchenko