Case-control analysis of single-cell RNA-seq studies
Petukhov V, Igolkina A, Rydbirk R, Mei S, Christoffersen L, Kharchenko P, Khodosevich K
viktor.petukhov@pm.me
Structure
Overview of the problem
State of the field
Cacoa: case-control analysis
Epilepsy data analysis
Slide complexity
Papers we analyzed
Epilepsy
Cacoa
Overview
State
Papers we analyzed
Epilepsy
Cacoa
Overview
State
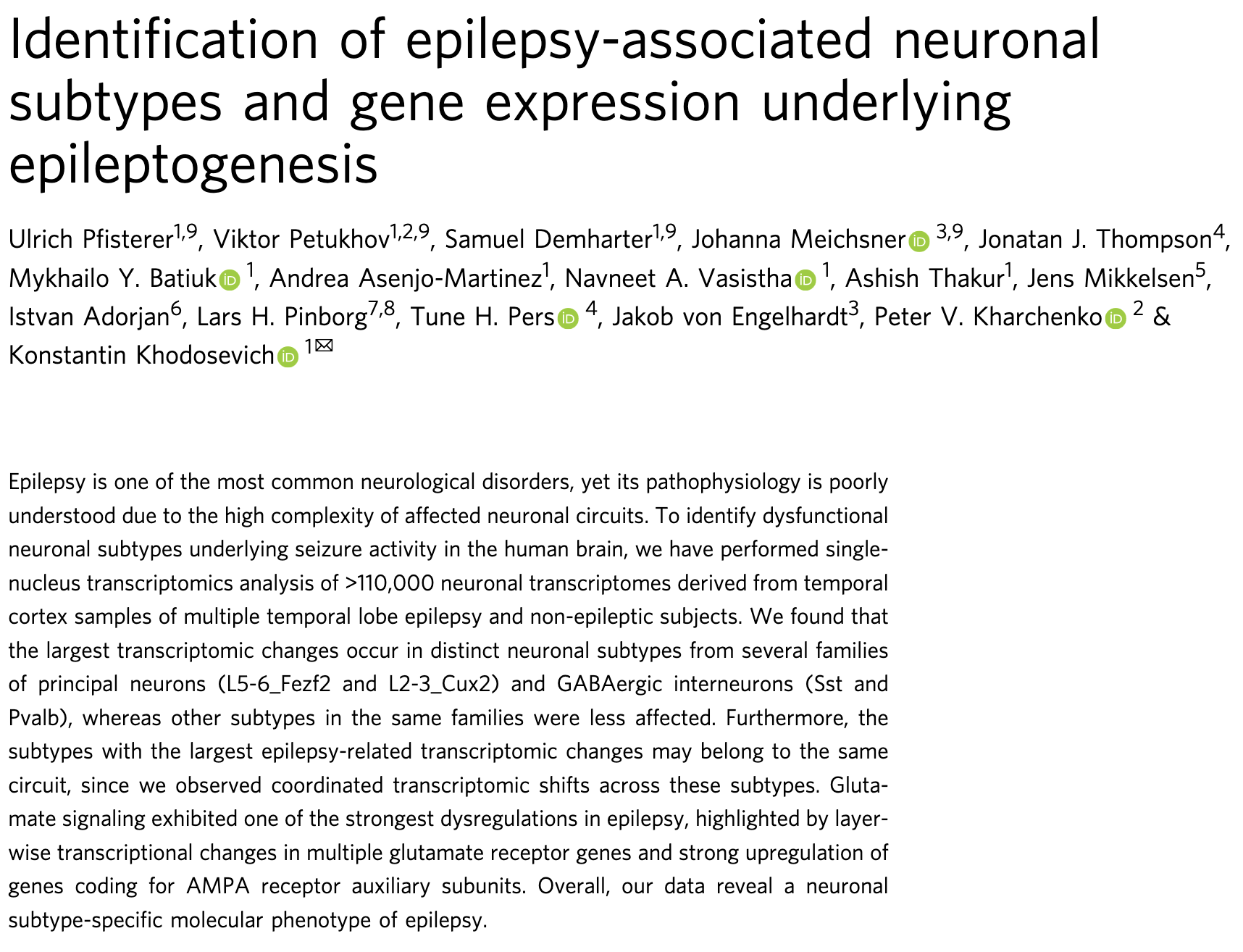
scRNA-seq to study Temporal Lobe Epilepsy
- Temporal Cortex
- 9 control vs 10 epilepsy samples
- 117k nuclei
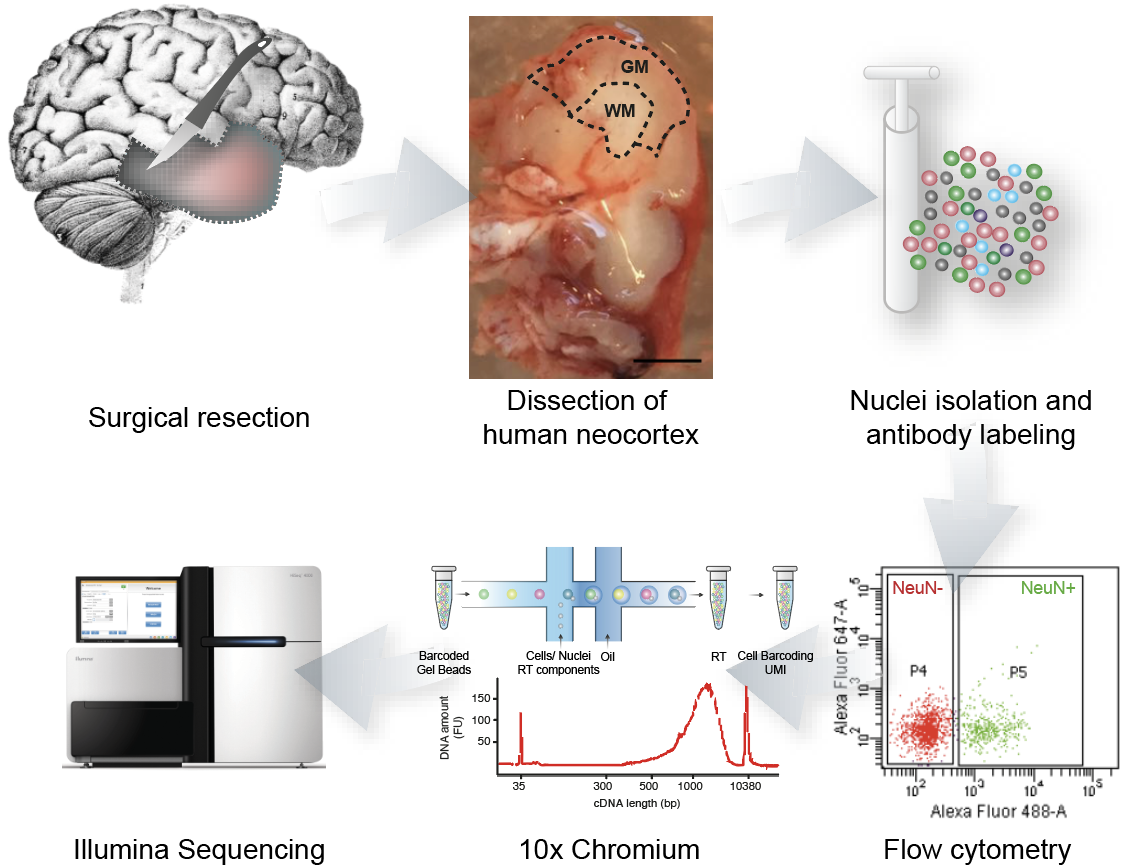
Our data: single-cell RNA-sequencing (2015)
DNA:
RNA molecules
Genes
}
Expression
levels
(3;
1;
5)

Genes
DNA:
(3;
1;
5)
}
Expression
levels







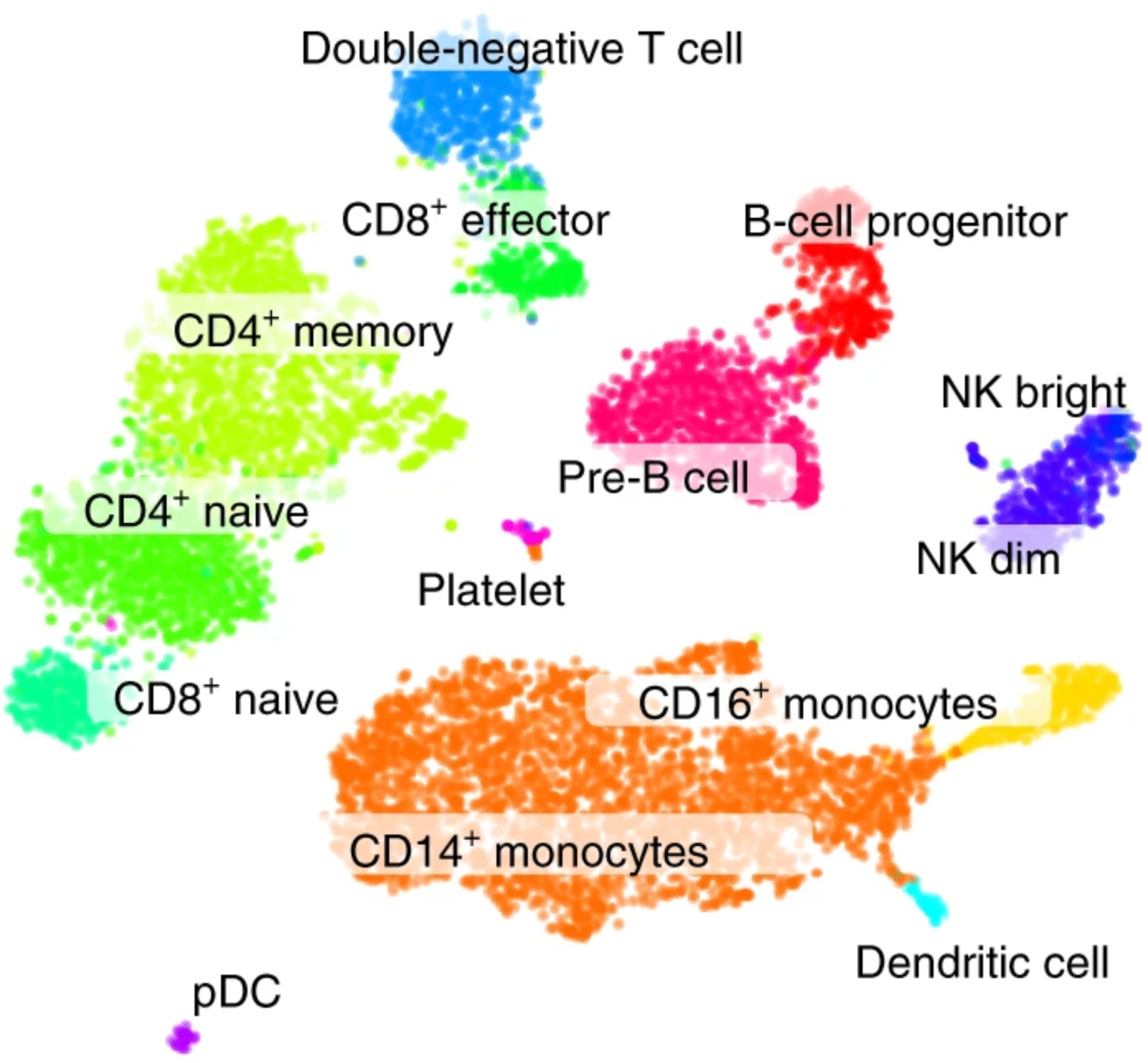
t-SNE 1
t-SNE 2
Our data: single-cell RNA-sequencing (2015)
~10k cells x 20k genes







Genes
DNA:
(3;
1;
5)
}
Expression
levels






































































































t-SNE 1
t-SNE 2
Our data: single-cell RNA-sequencing (2015)





Our data: multiple samples (2018)
Problem: multiple conditions (2020)
Control patients
Case patients










Problem: multiple conditions (2020)
Control patients
Case patients


















































Cells
























Cell
types
Problem: multiple conditions (2020)
Control patients
Case patients










Cell
types
Cells
































































Cells
Genes


Cells
Genes


Cells
Genes


Cells
Genes


Cells
Genes


Cells
Genes


Cells
Genes


Cells
Genes


How to compare?
Goal: develop a comprehensive set of methods for analysis of scRNA-seq case-control experiments
State of the art: no methods were published when we started, several competitors exist now
Problem: how to measure changes between conditions?
Case-control analysis of single-cell RNA-seq studies
Existing solutions
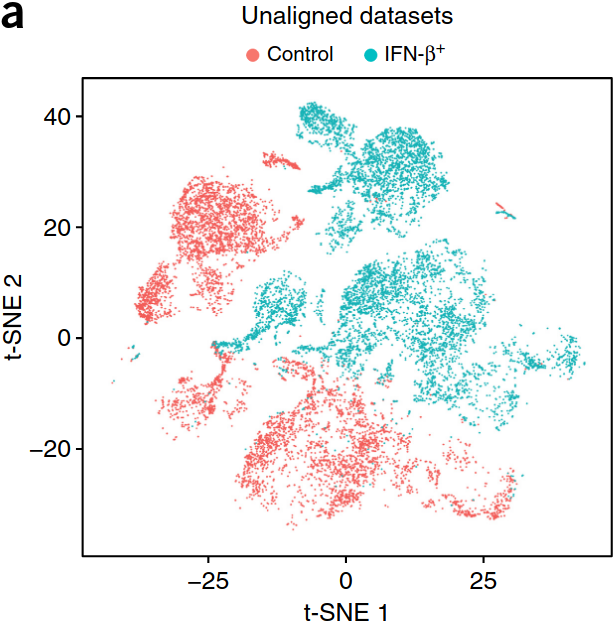
Epilepsy
Cacoa
Overview
State
Existing solutions
Align samples
scVI, Conos, ..., Seurat
See the review from the Theis lab
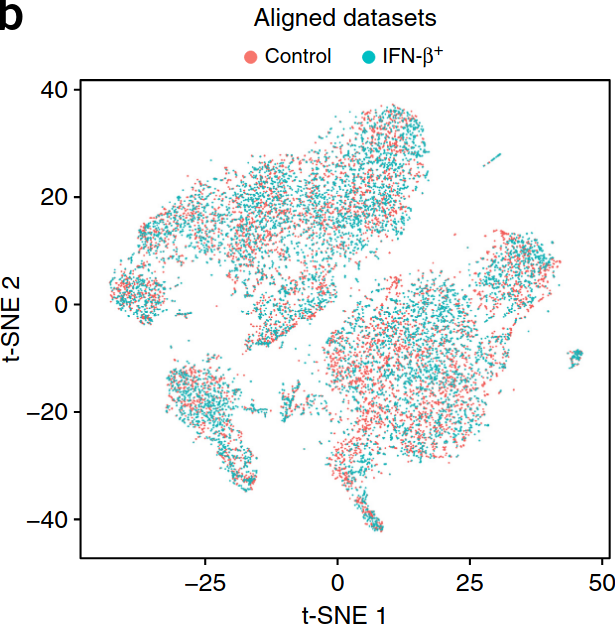
Existing solutions
Align samples
Perform joint annotation
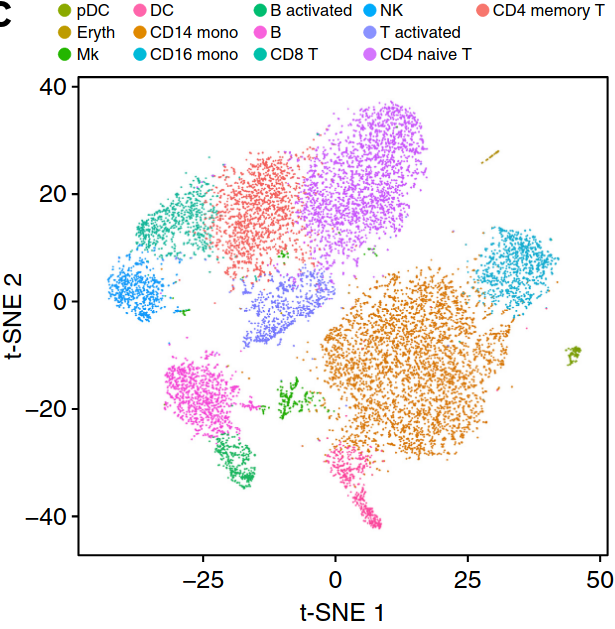
Existing solutions
Align samples
Perform joint annotation
Run differential expression
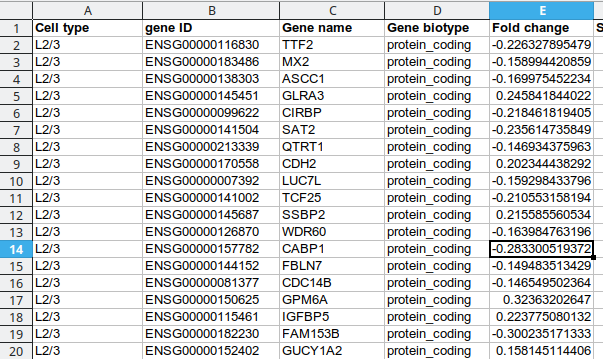
Existing solutions
Align samples
Perform joint annotation
Run differential expression
Run Gene Ontology analysis

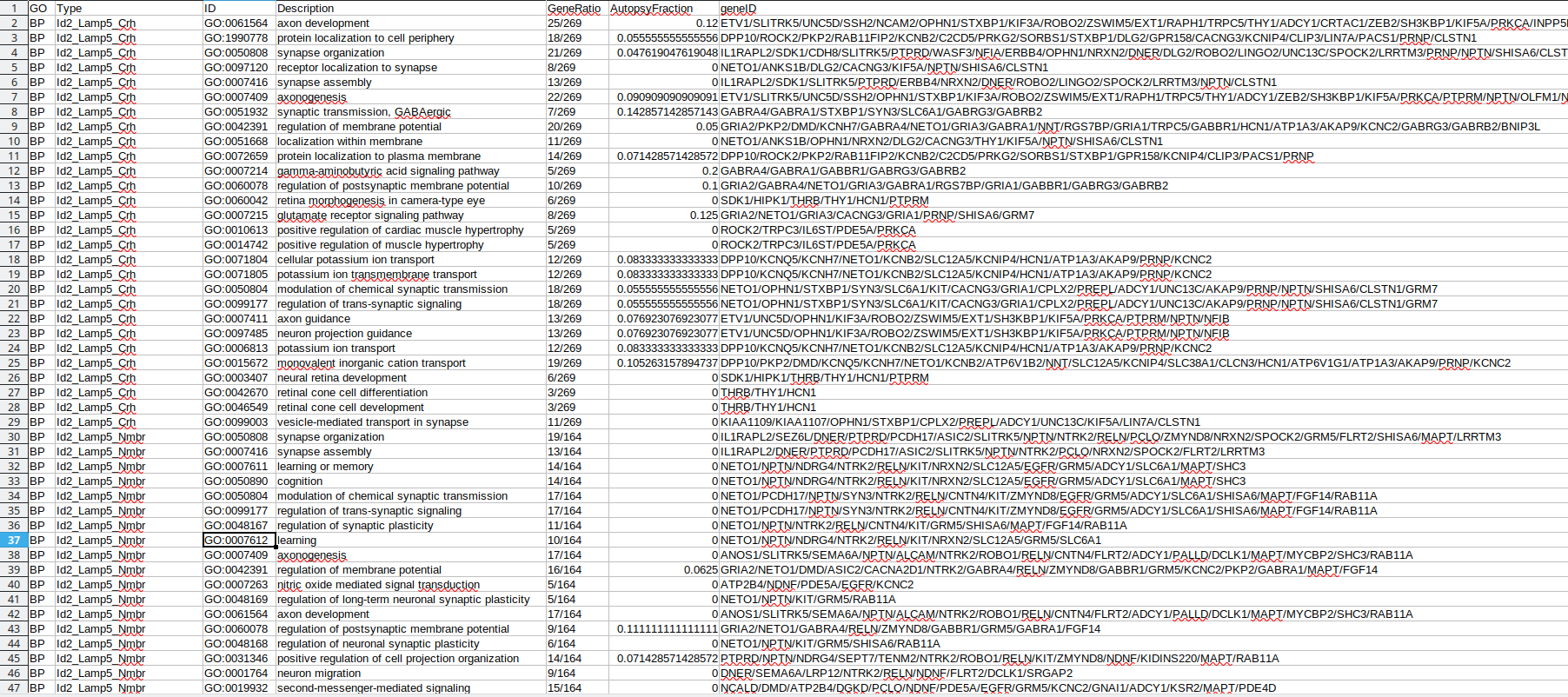
Existing solutions
Align samples
Perform joint annotation
Run differential expression
Run Gene Ontology analysis
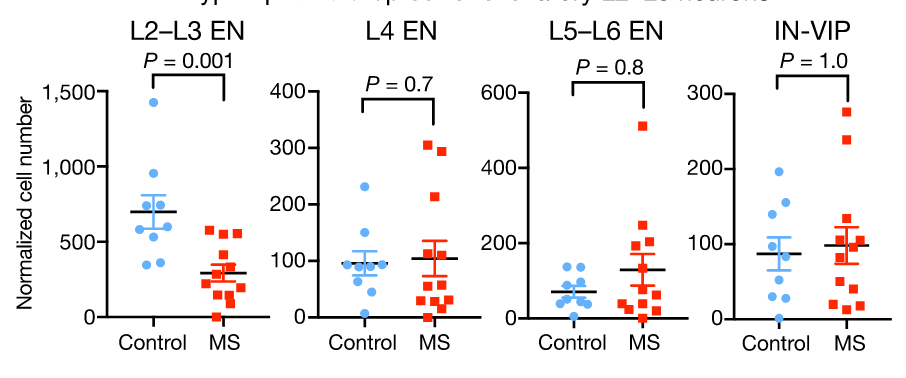
Compare cell type proportions
Can we do better?
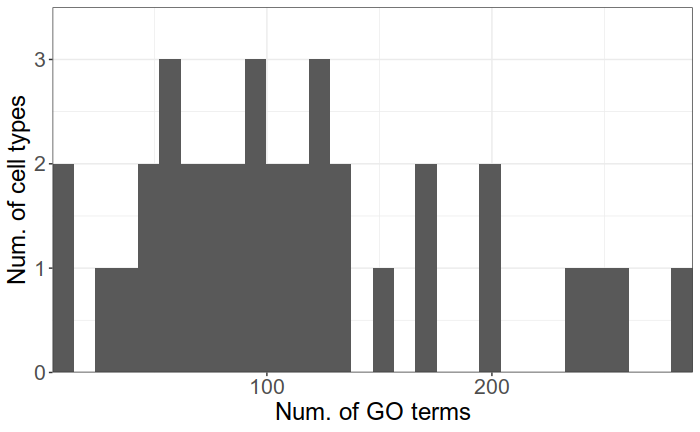
There are up to hundreds significant GO terms per type
There are up to 1000 significant DE genes per type
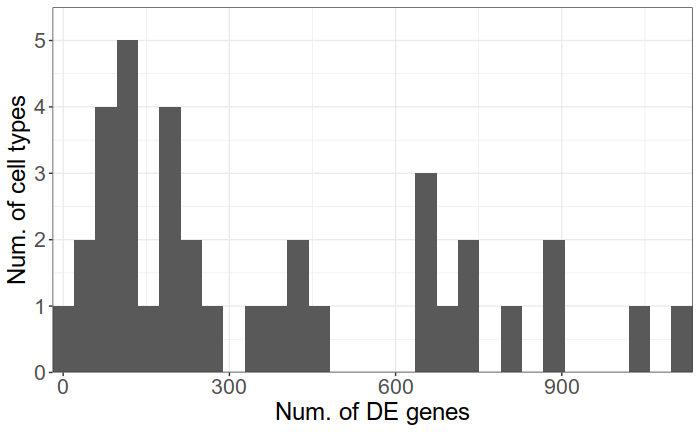
Case-control analysis of single-cell RNA-seq studies
Compositional analysis


Gene expression analysis

What can we possibly do?
Epilepsy
Cacoa
Overview
State
Case-control analysis of single-cell RNA-seq studies



Case-control analysis of single-cell RNA-seq studies
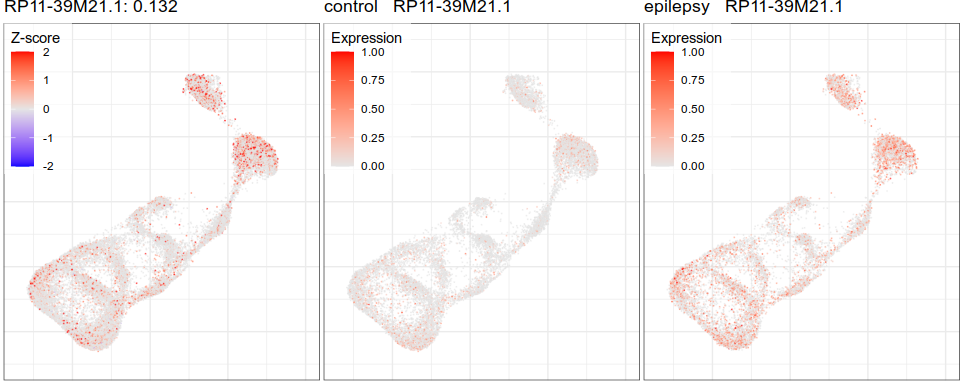
Gene expression analysis

Compositional analysis


Cluster-based
Cluster-free
Control
Multiple sclerosis
Composition analysis, cluster-based


Composition analysis, cluster-based
Problem: changes are not independent
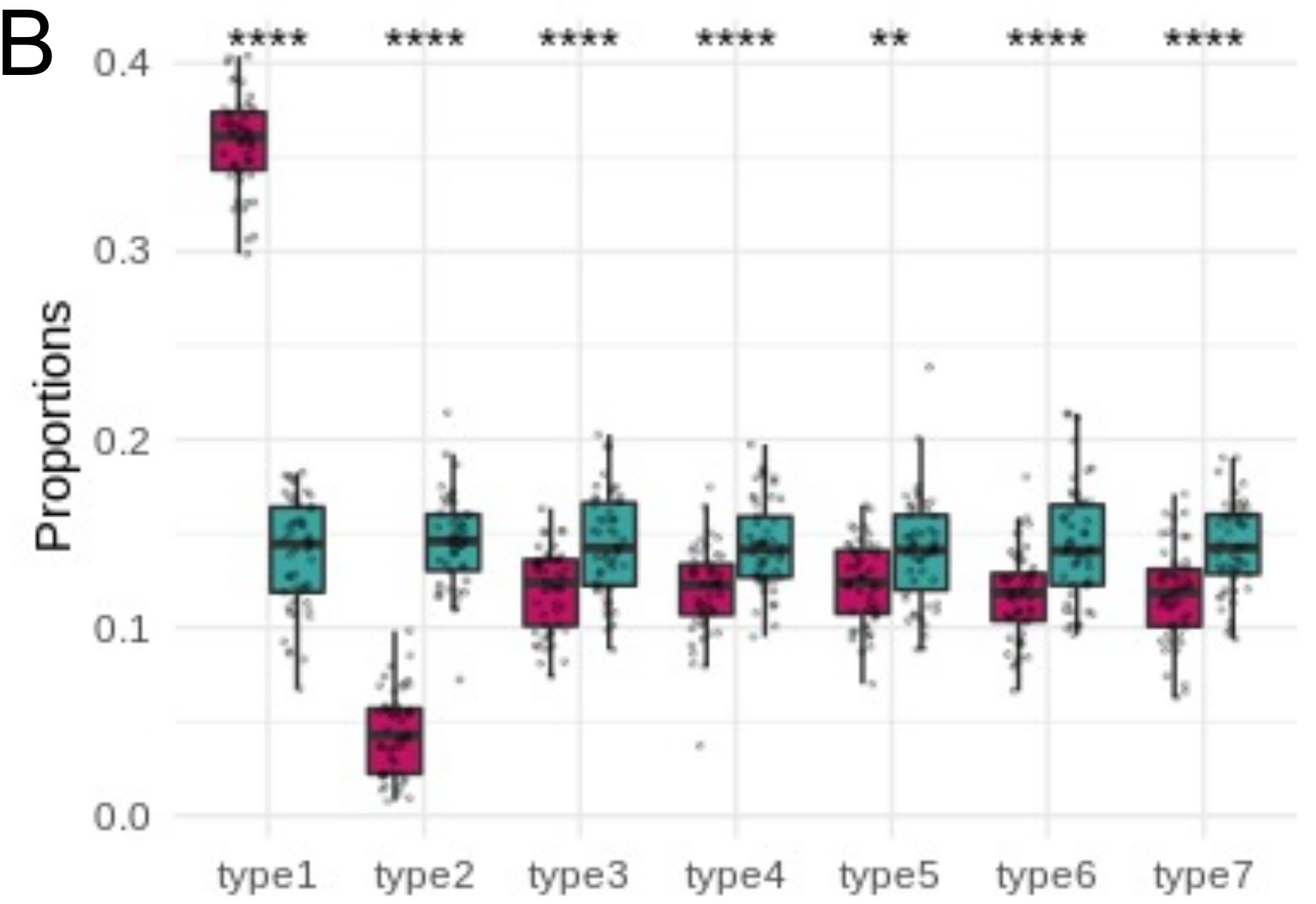
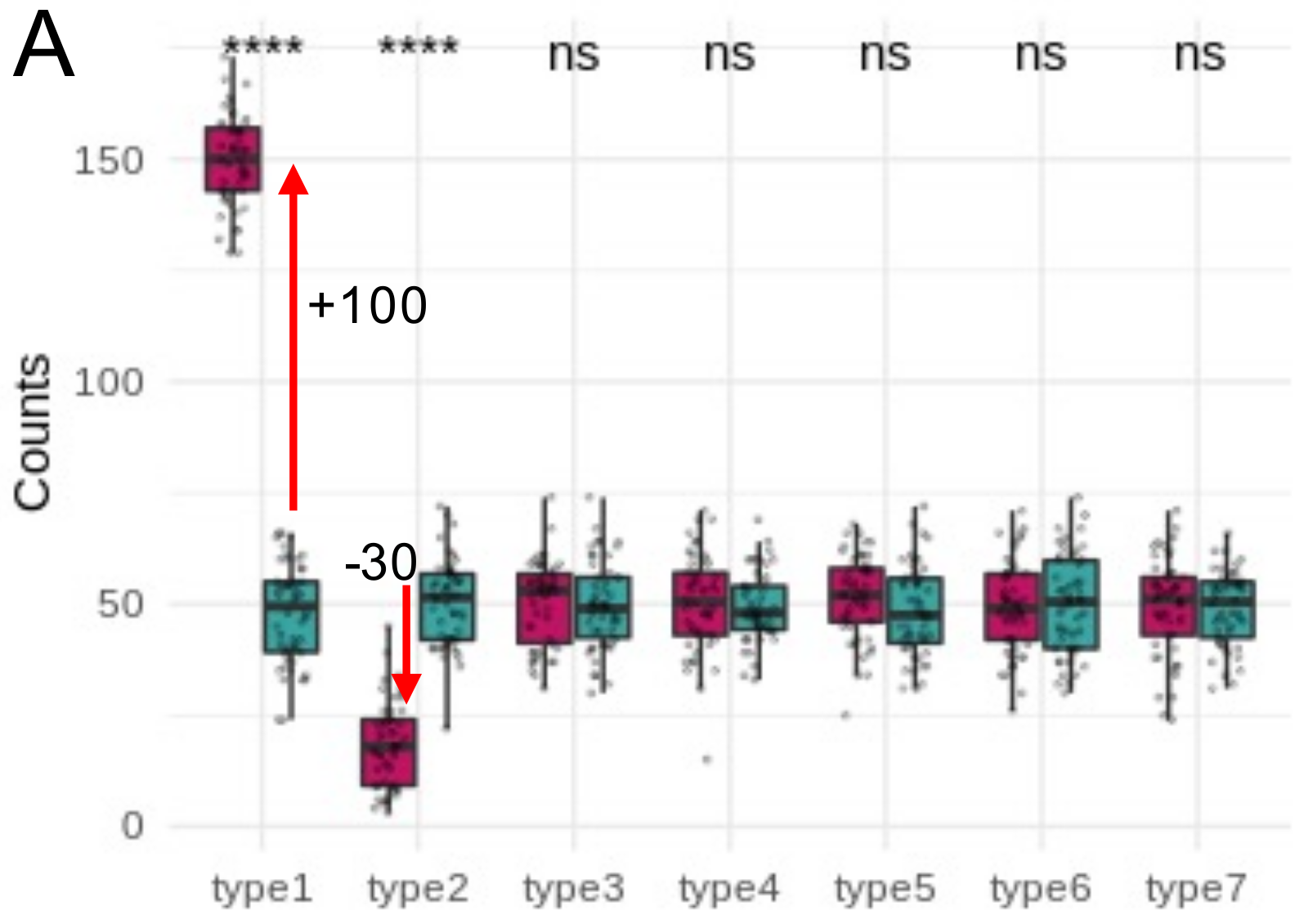
*Credit to Anna Igolkina
Composition analysis, cluster-based
Problem: changes are not independent



*Credit to Anna Igolkina
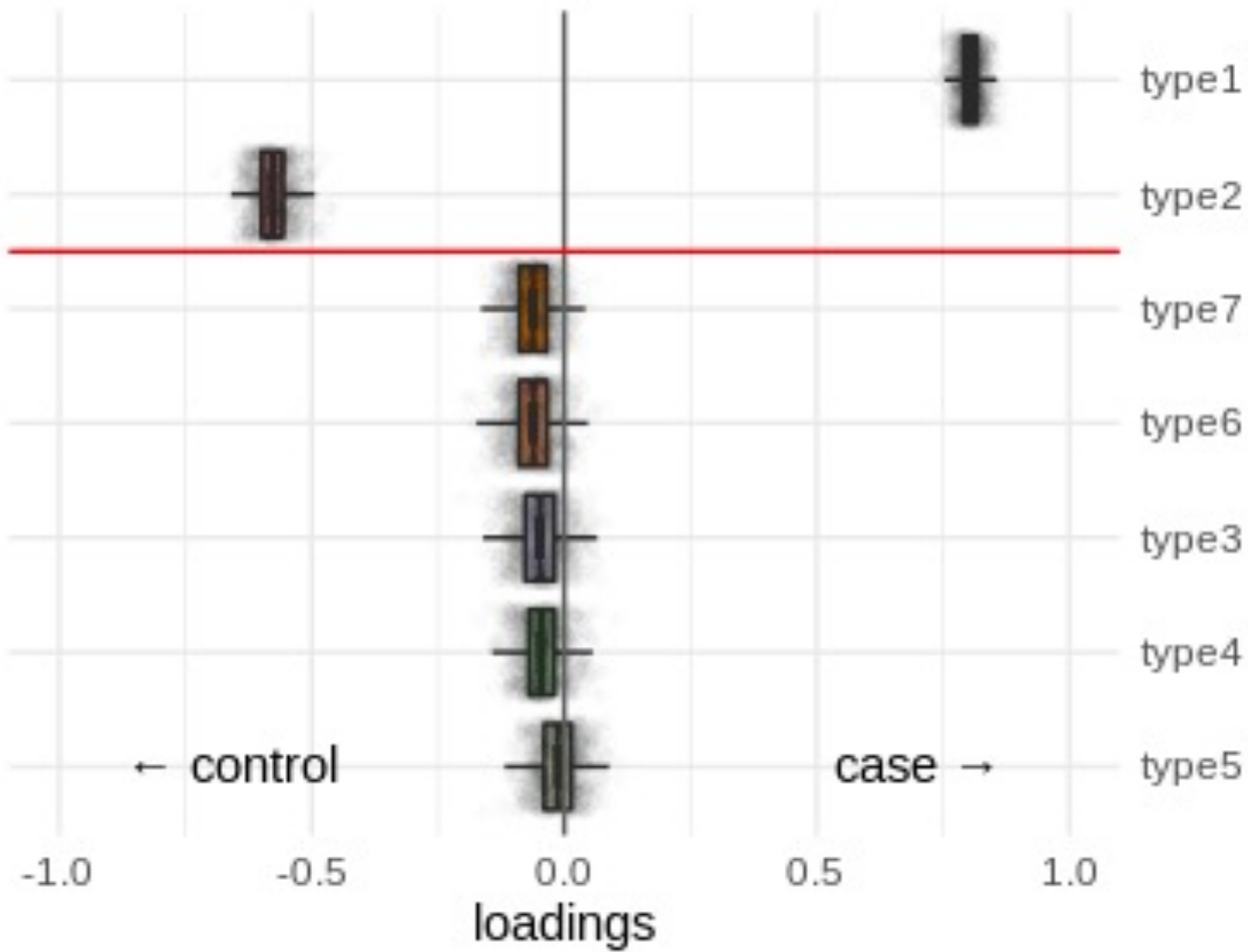
Compositional analysis: cluster-based
*: CoDA significance
*: proportion significance
Compositional analysis: cluster-based
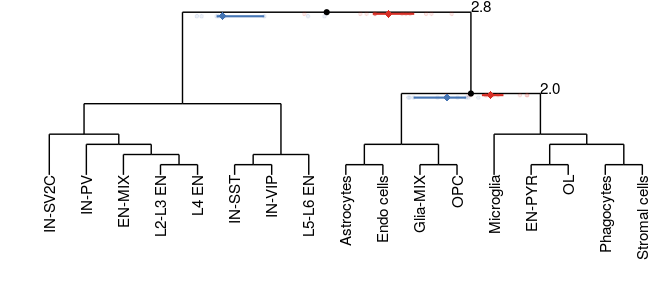
Neurons
Glia

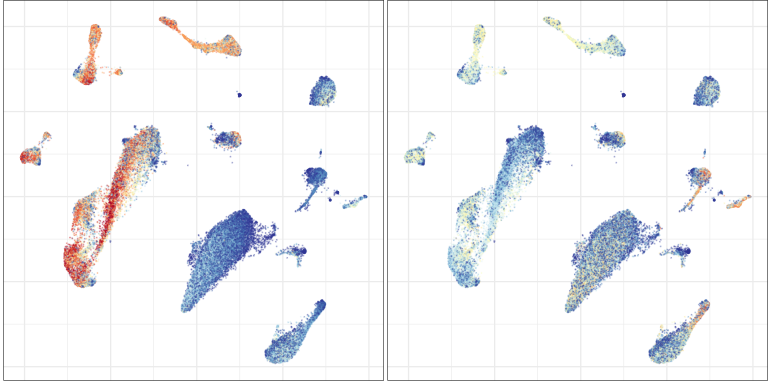
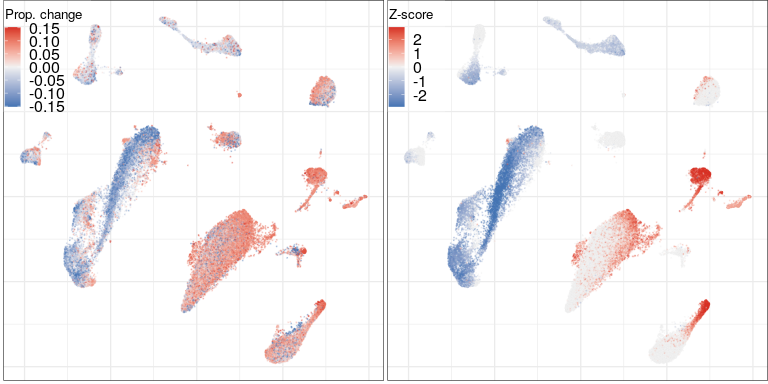
Compositional analysis: cluster-free
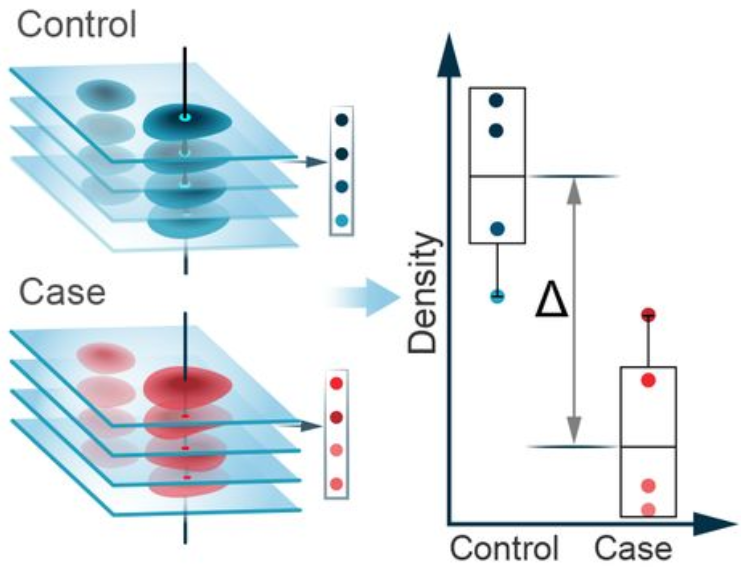
Compositional analysis: cluster-free


Control
Multiple sclerosis
Compositional analysis: cluster-free
Effect size
Significance
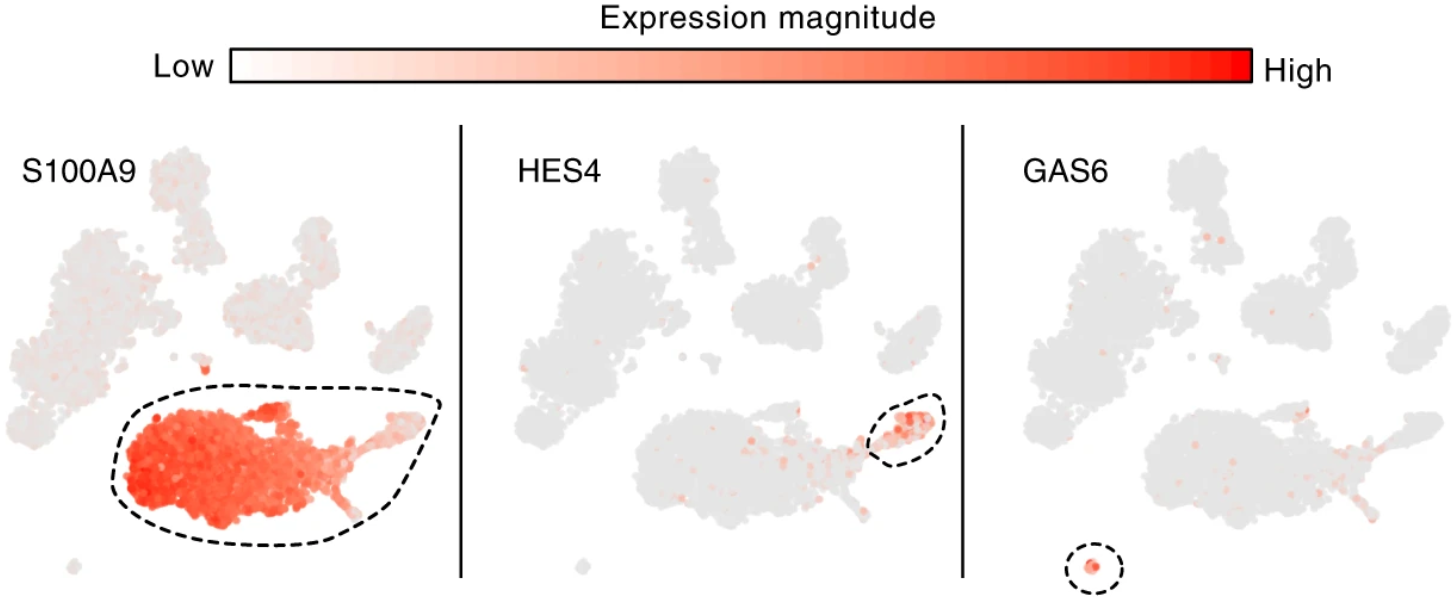
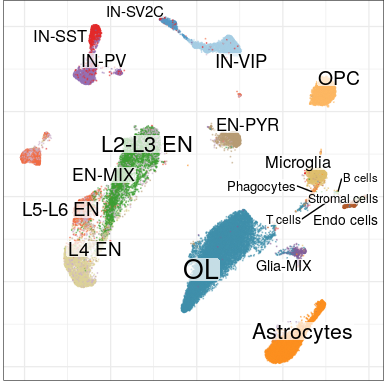
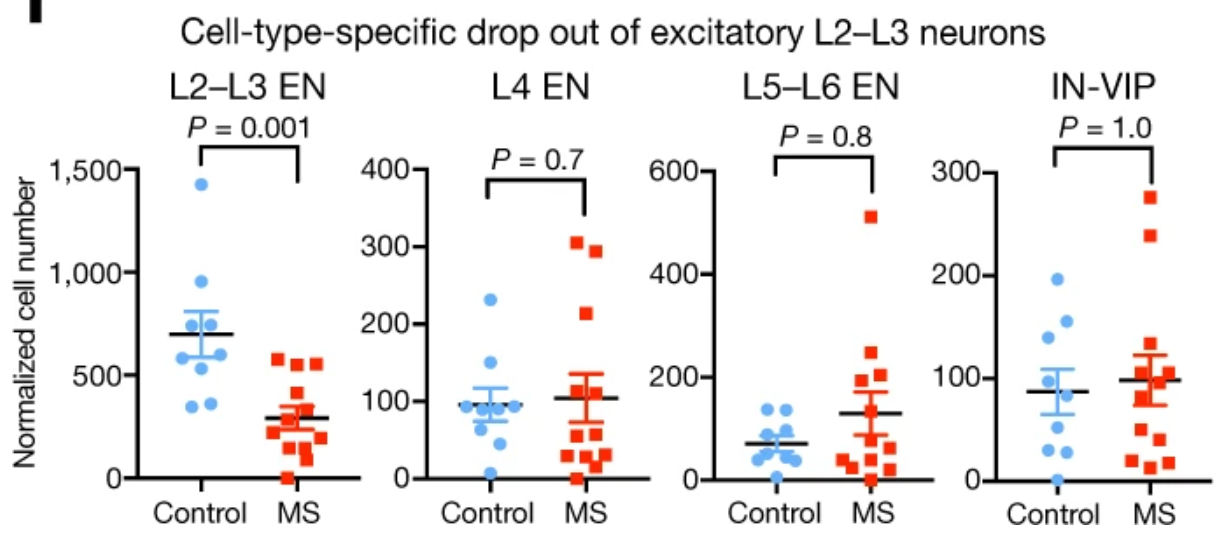
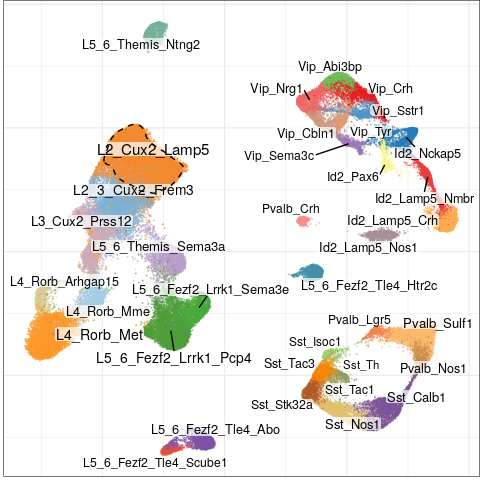
Expression analysis: cluster-based

EN L2-L3
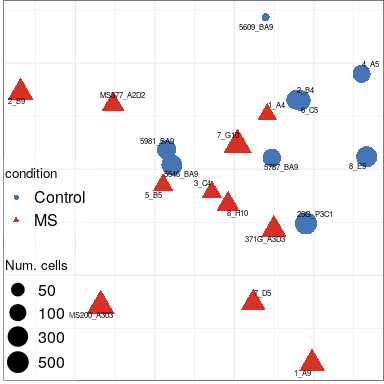
Expression analysis: sample structure

EN L2-L3
EN L2-L3
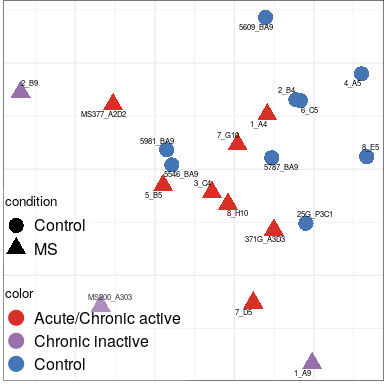
Expression analysis: sample structure
Color by batch
Aggregated across all cell types
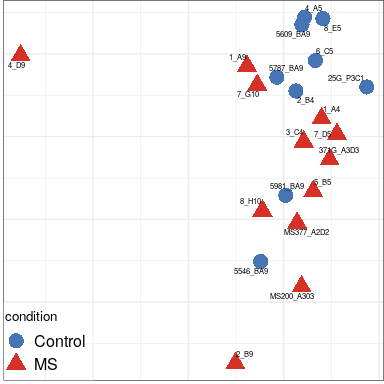
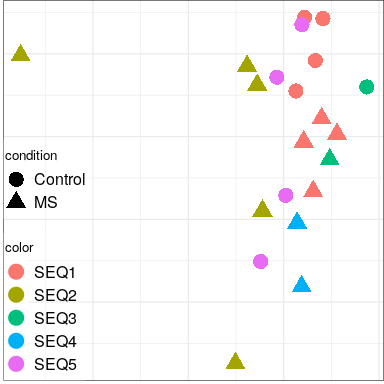
Expression analysis: sample structure
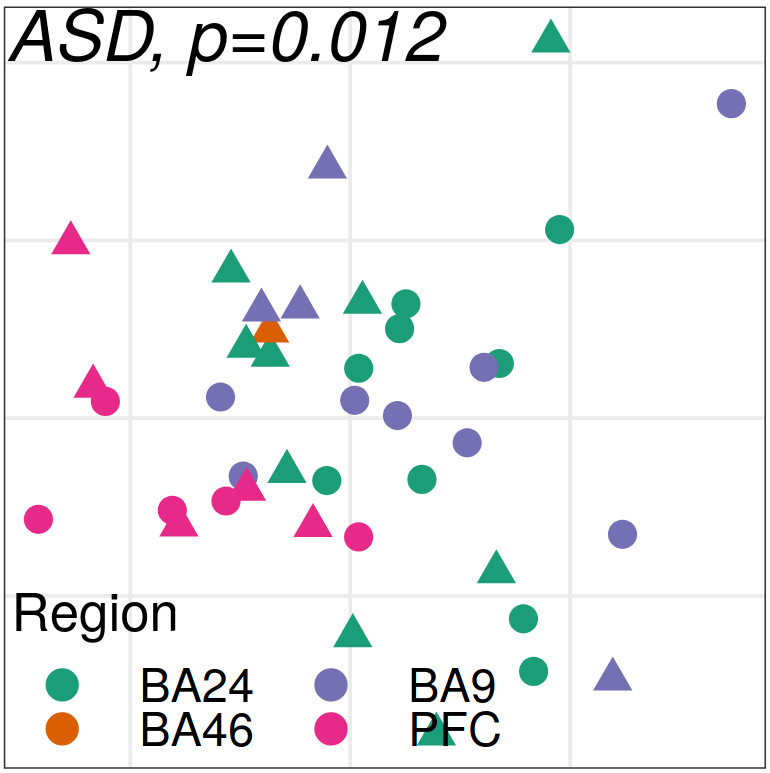
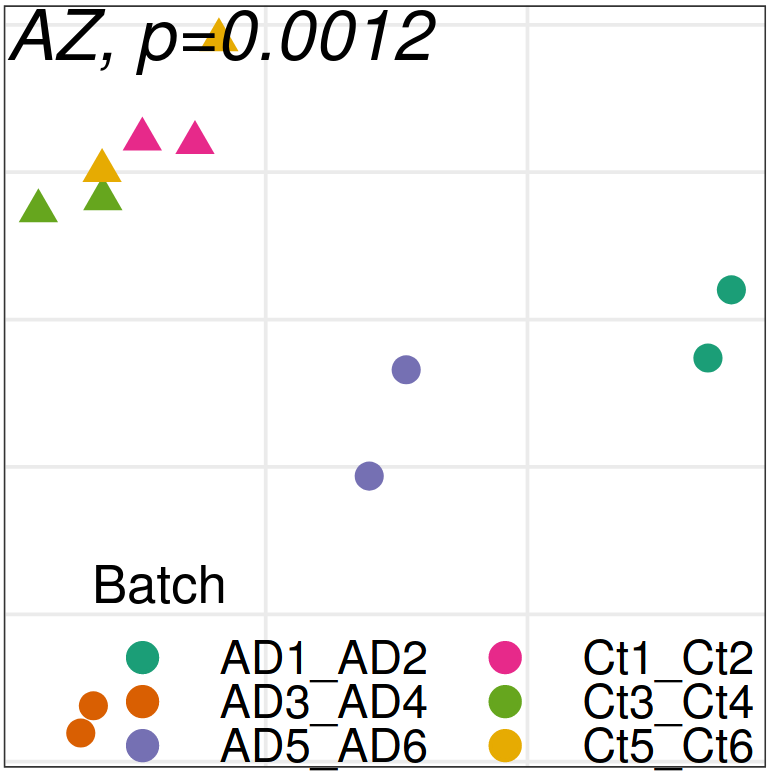
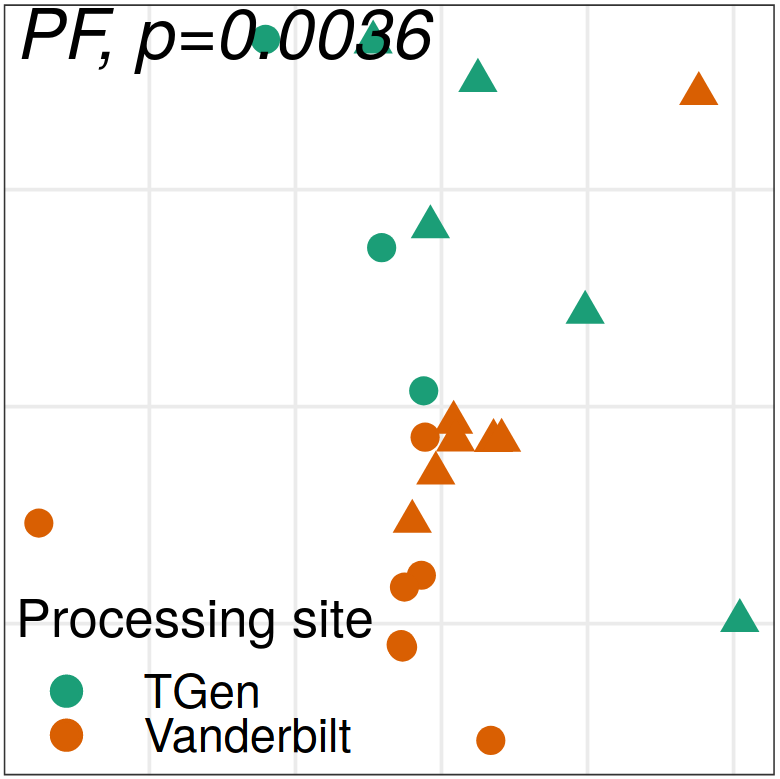
Expression analysis: expression distance
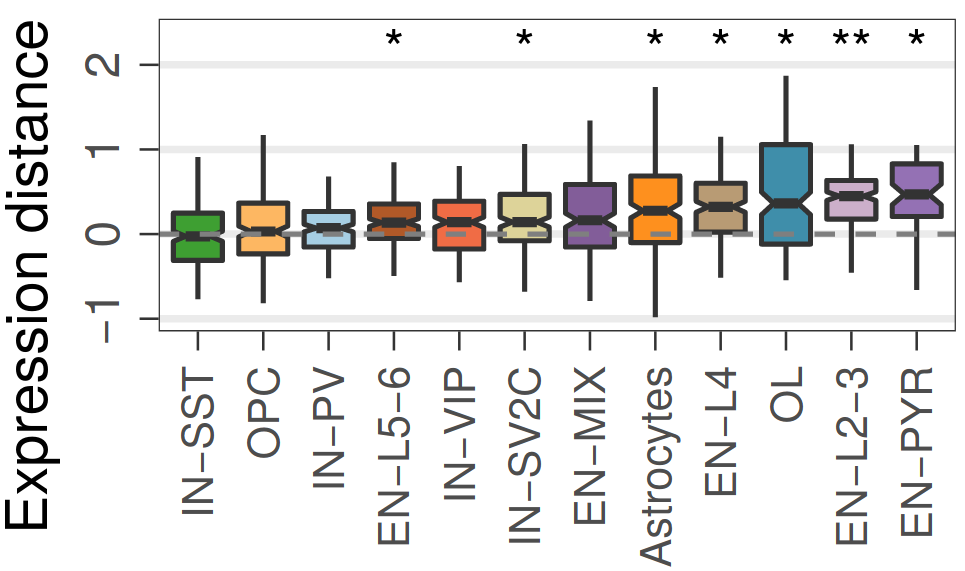
separation
EN L2-L3

Expression analysis, cluster-free
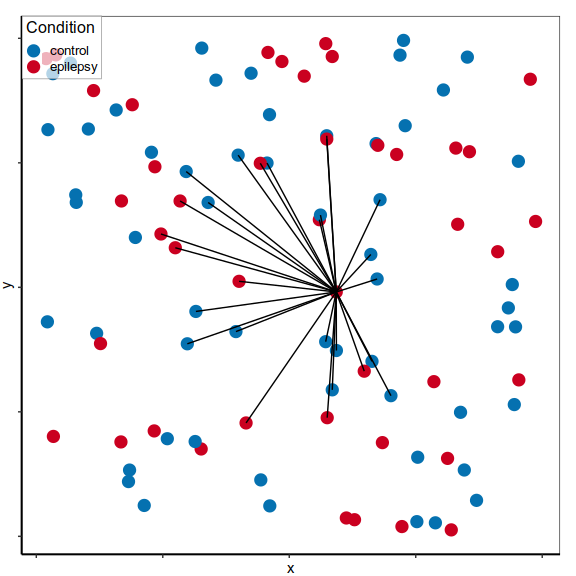
Differential expression on single cells

Expression analysis, cluster-free
Differential expression on single cells


Expression analysis, cluster-free
Differential expression on single cells
control
epilepsy
Expression analysis, cluster-free
Differential expression on single cells




control

epilepsy
Differential expression
Expression analysis, cluster-free
Differential expression on single cells

PDE10A
MS
Control
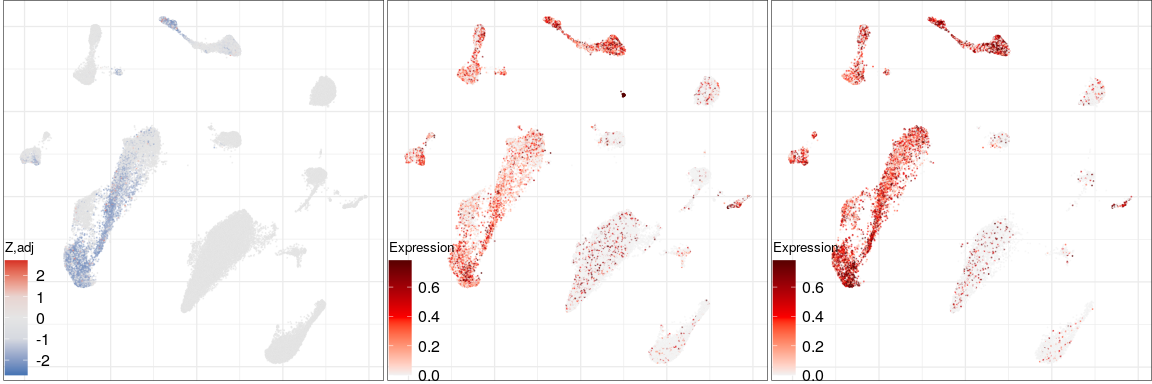
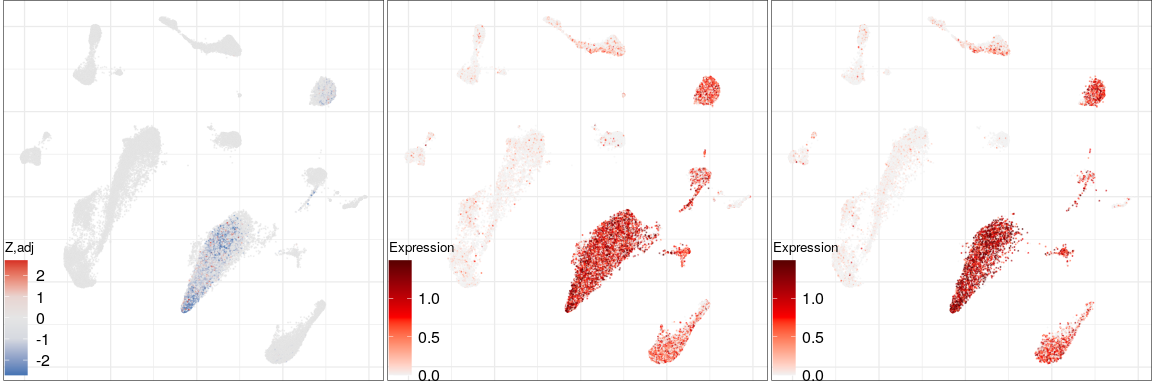
NCKAP5
MS
Control
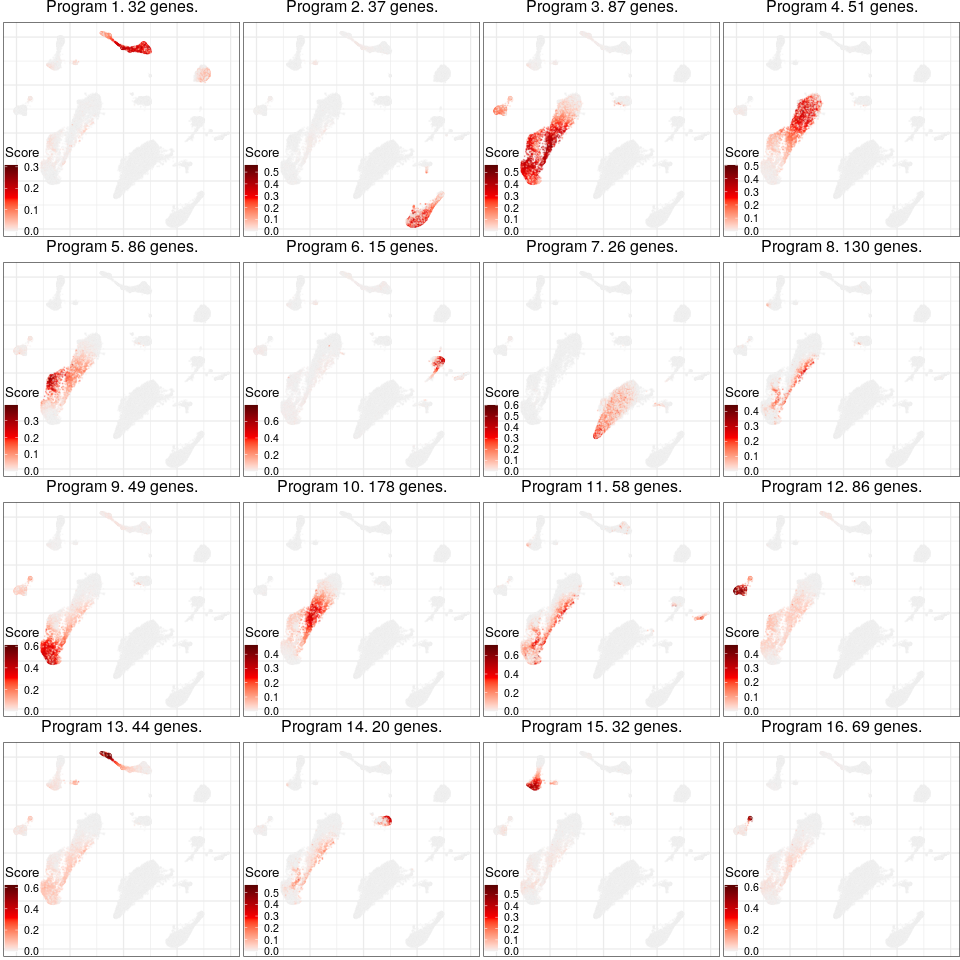
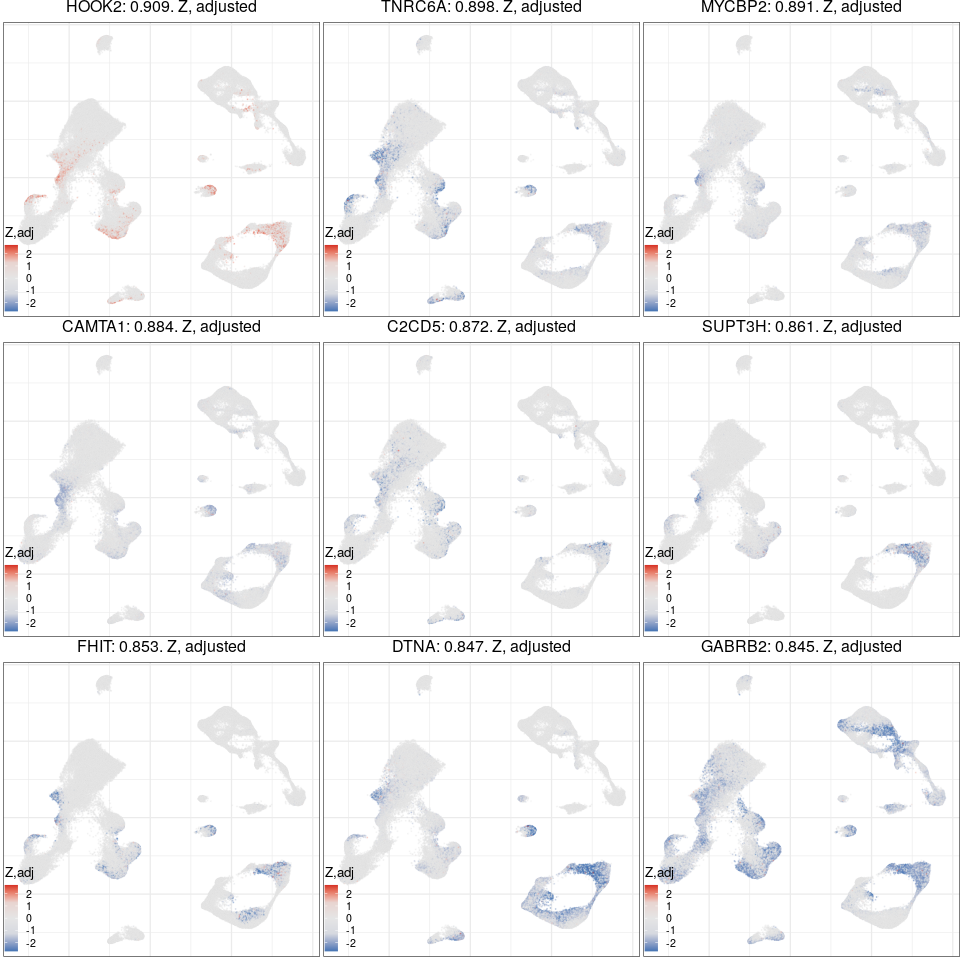
Expression analysis, cluster-free
Gene programs

Science reproducibility
>70% of publications had
major flows, compromising main results
Science reproducibility


>70% of publications had
major flows, compromising main results
Science reproducibility
- Publish your code
- Double-check your results
- Don't get too excited

Gene expression analysis

Compositional analysis


Cluster-based
Cluster-free

Control
Multiple sclerosis
Summary

Full analysis of the Epilepsy dataset
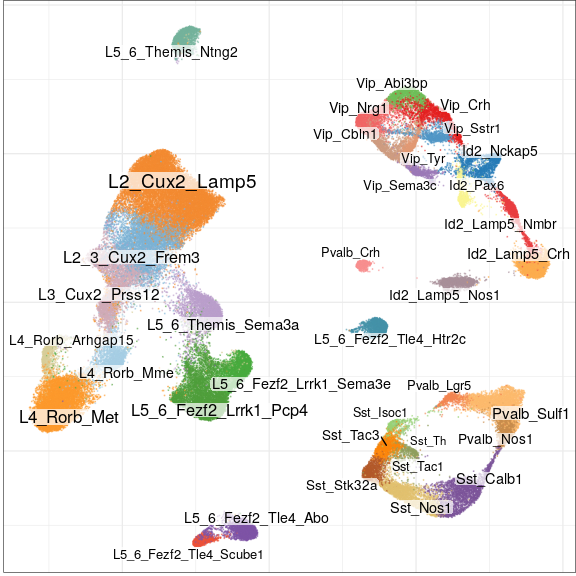
- Temporal Cortex
- 9 control vs 10 epilepsy samples
- 117k nuclei
Epilepsy
Cacoa
Overview
State
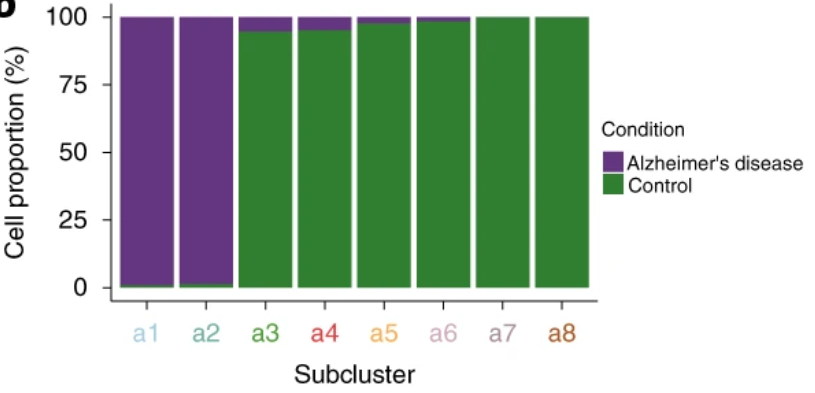
Full analysis of the Epilepsy dataset

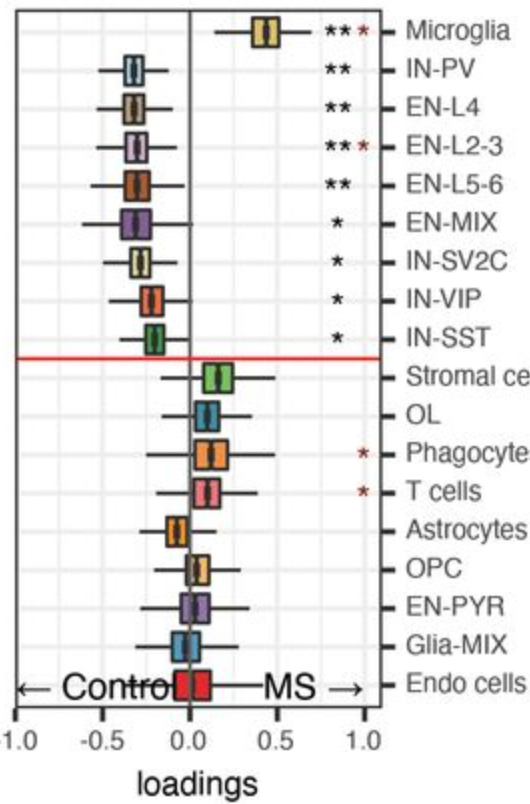
Compositional analysis
No significance!
Full analysis of the Epilepsy dataset


Compositional analysis, cluster-free
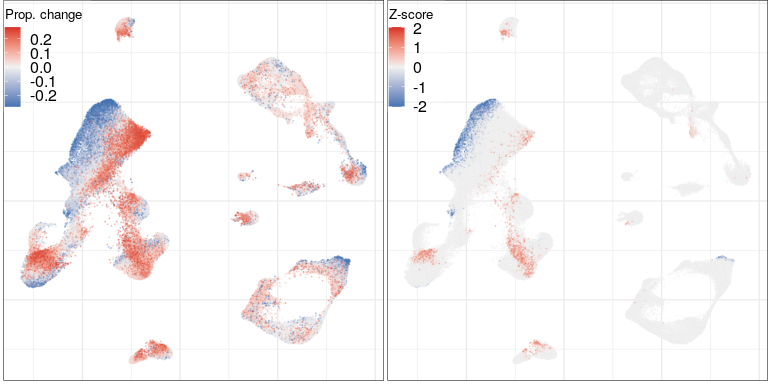
Effect size
Significance
Full analysis of the Epilepsy dataset
Expression analysis: sample embeddings
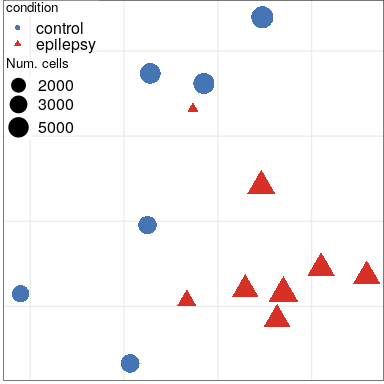

Condition
Full analysis of the Epilepsy dataset

Expression analysis: sample embeddings

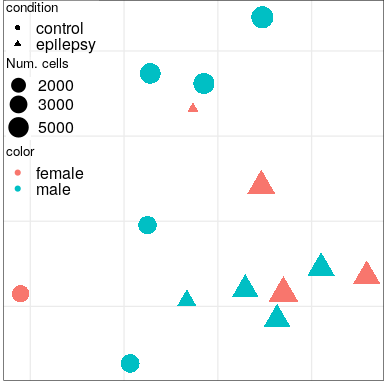
Condition
Sex
Full analysis of the Epilepsy dataset

Expression analysis: sample embeddings


Condition
Sex
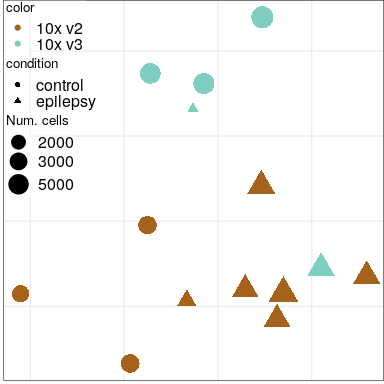
Protocol
Full analysis of the Epilepsy dataset

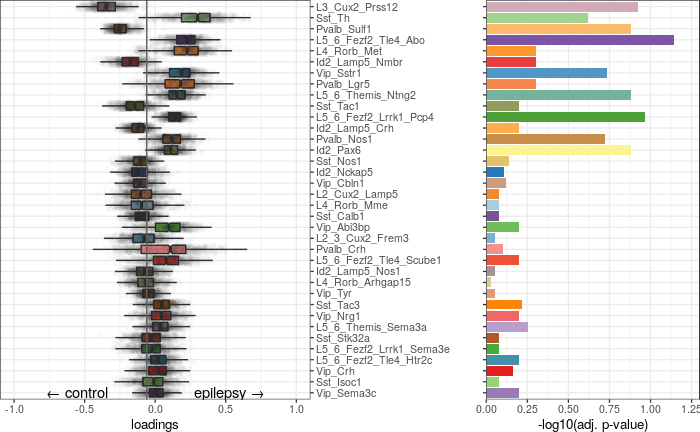
Expression analysis: shift magnitudes
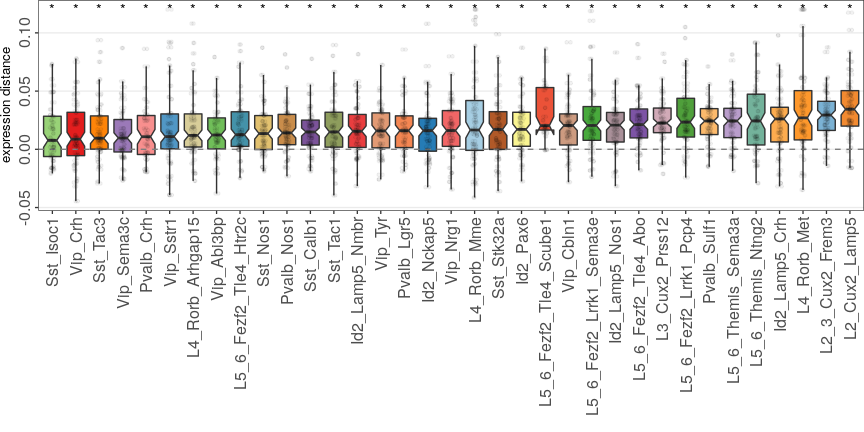
Full analysis of the Epilepsy dataset

Expression analysis: shift magnitudes


Full analysis of the Epilepsy dataset
Expression analysis: cluster-free DE
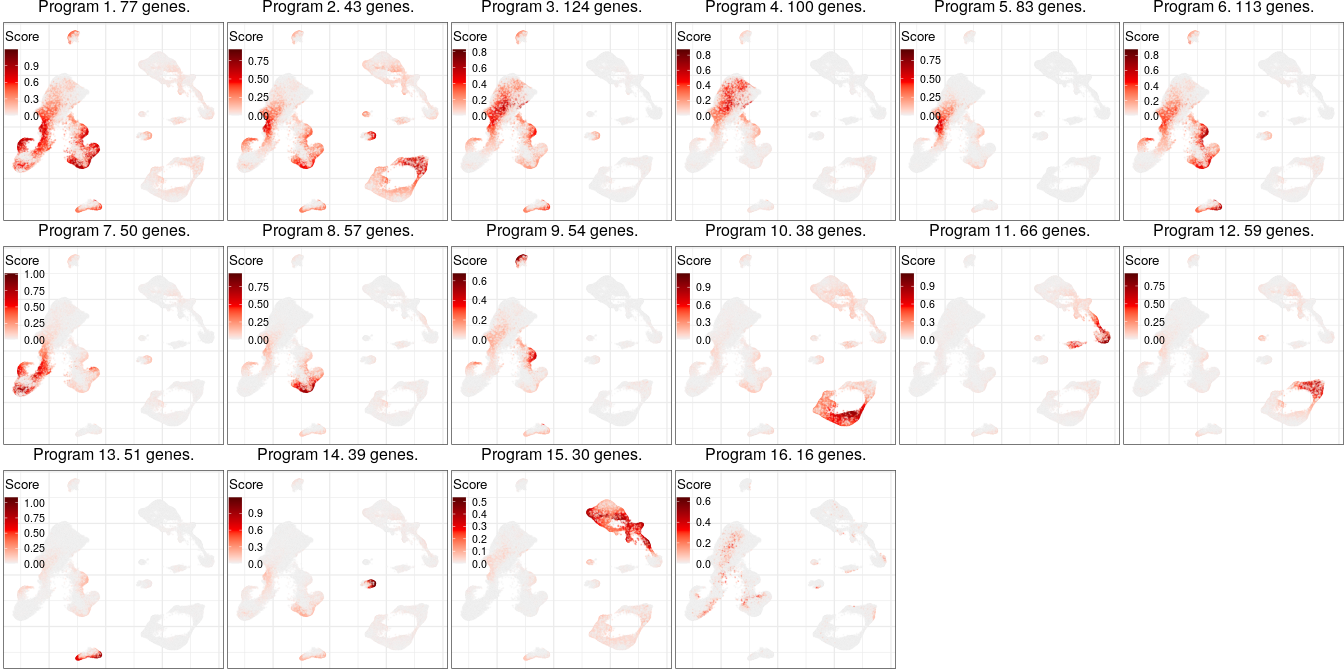

Full analysis of the Epilepsy dataset
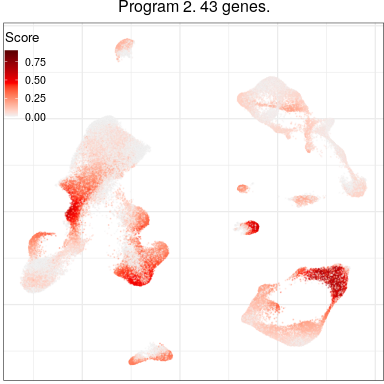
Programs for excitatory neurons

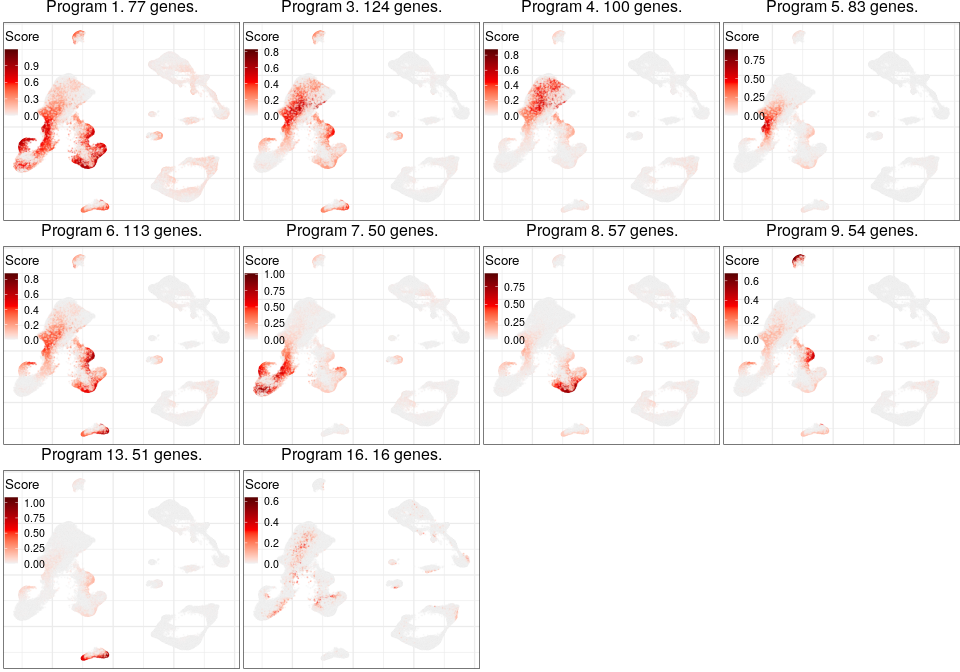
Full analysis of the Epilepsy dataset
Global excitatory program

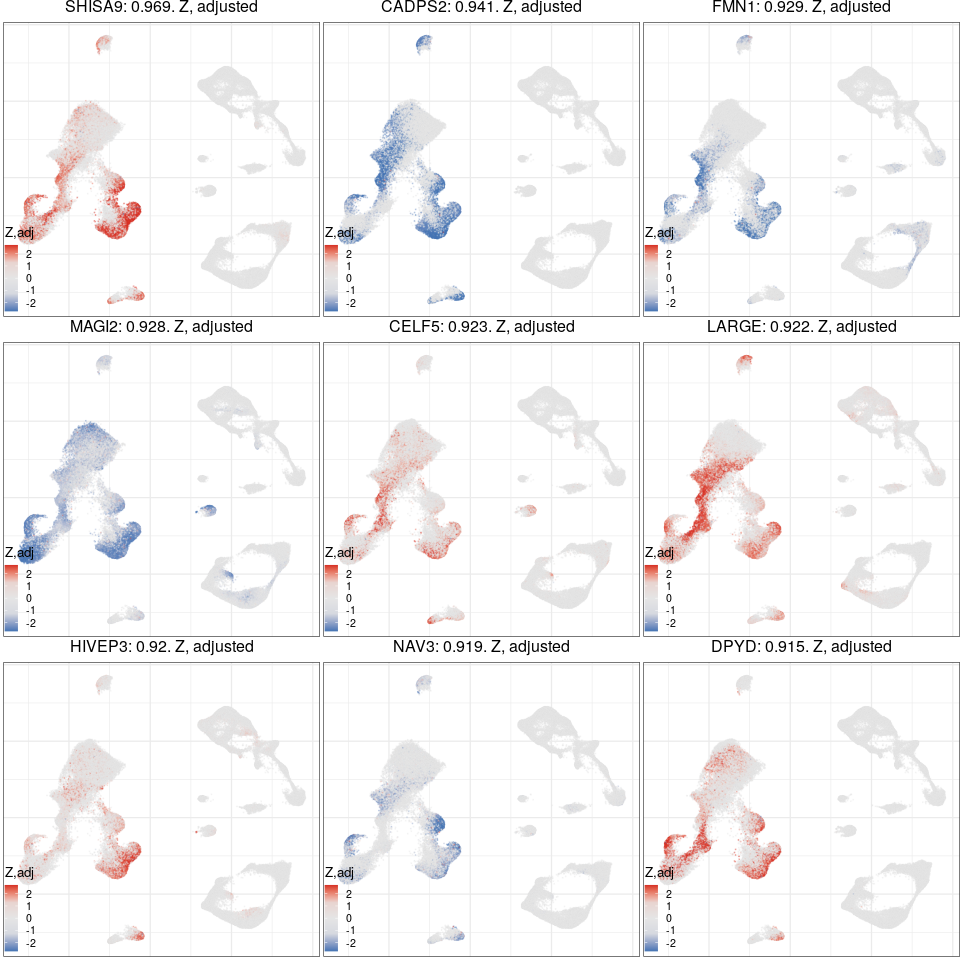
Full analysis of the Epilepsy dataset
Programs for inhibitory neurons

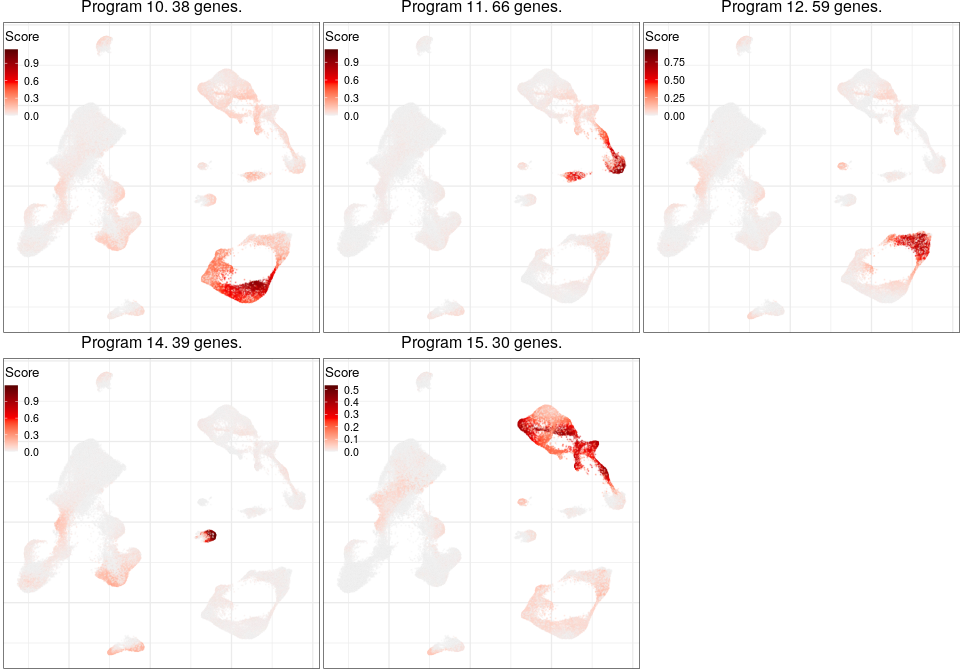
Full analysis of the Epilepsy dataset
Joint program

Full analysis of the Epilepsy dataset
Expression analysis: local cluster-free DE
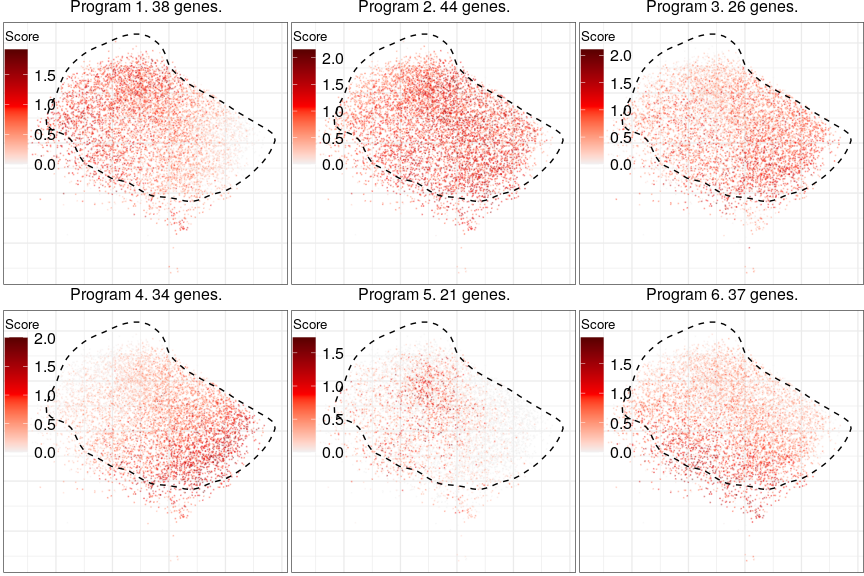
L2_Cux2_Lamp5 programs
Full analysis of the Epilepsy dataset
Expression analysis: local cluster-free DE


L2_Cux2_Lamp5 programs
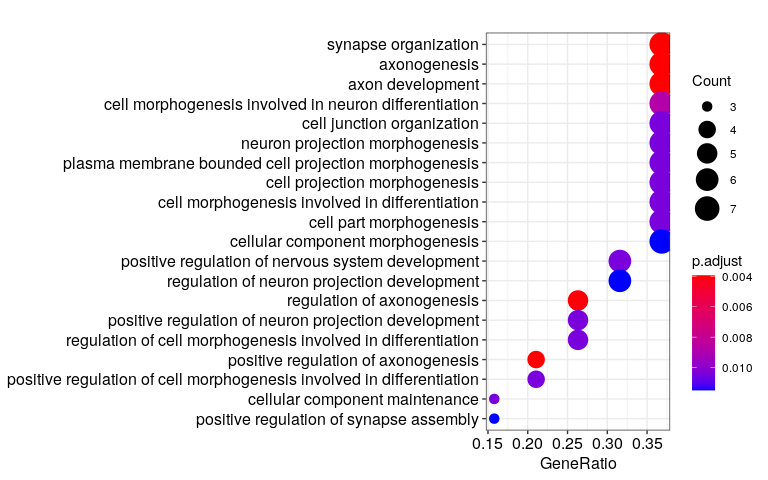
Full analysis of the Epilepsy dataset
Expression analysis: local cluster-free DE



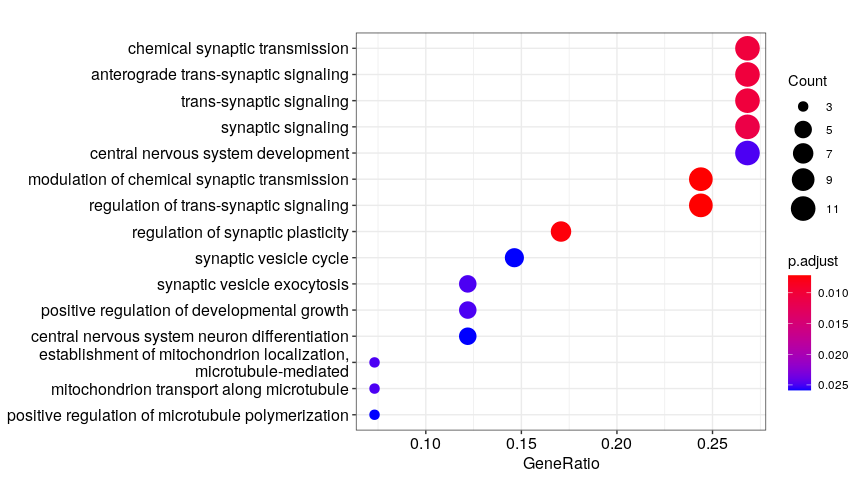
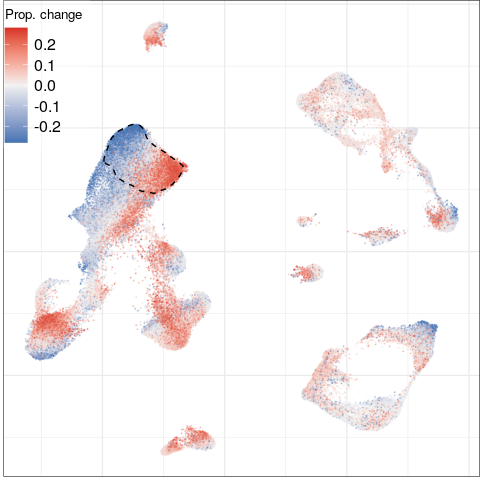
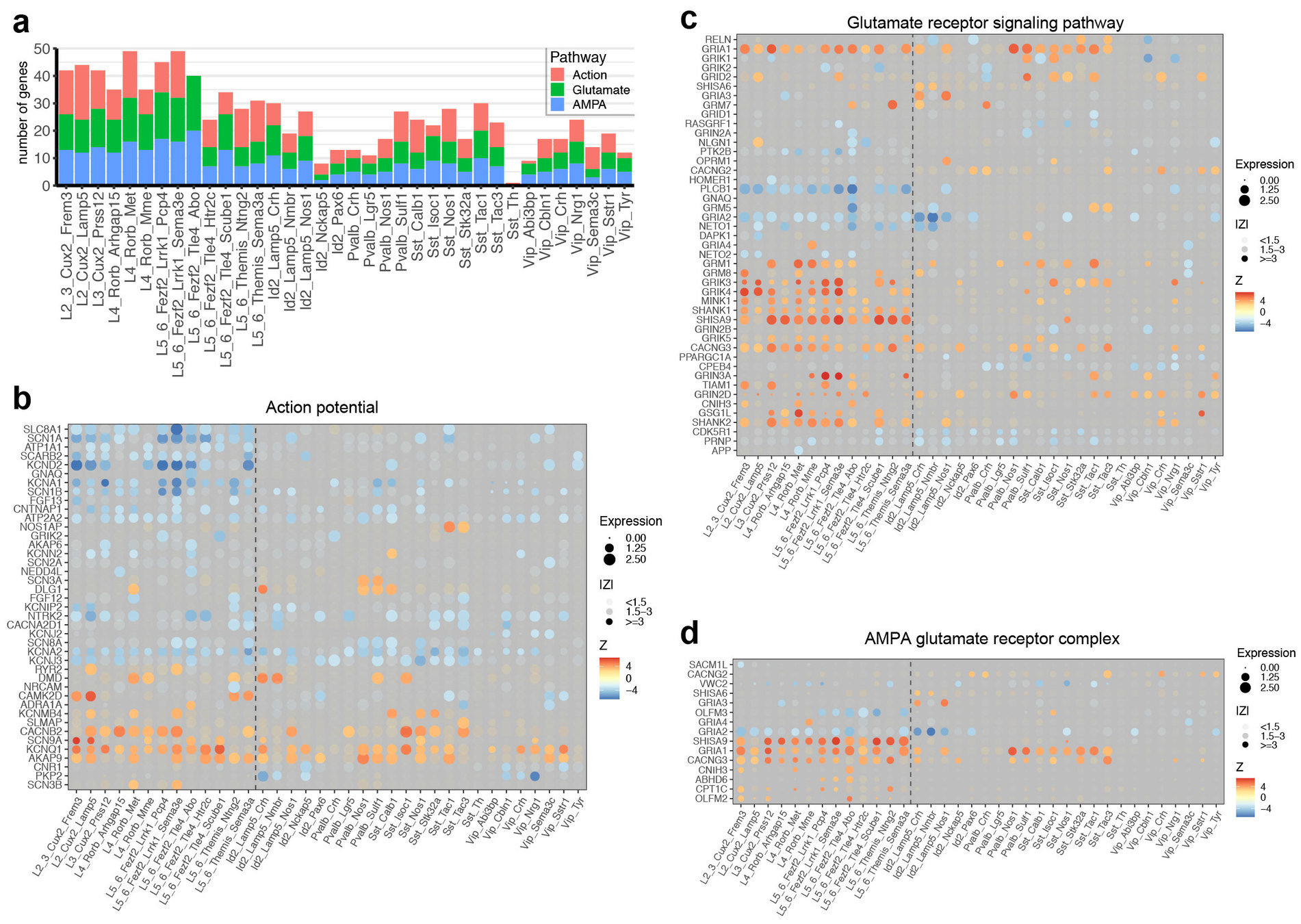
Step 3.1: global structure of changes


developmental processes, neural circuit re-organization and neurotransmission
ion transport and glutamate signaling
protein transport to axons/dendrites
cell adhesion, ion transport and synaptic plasticity
regulation of neuronal morphogenesis
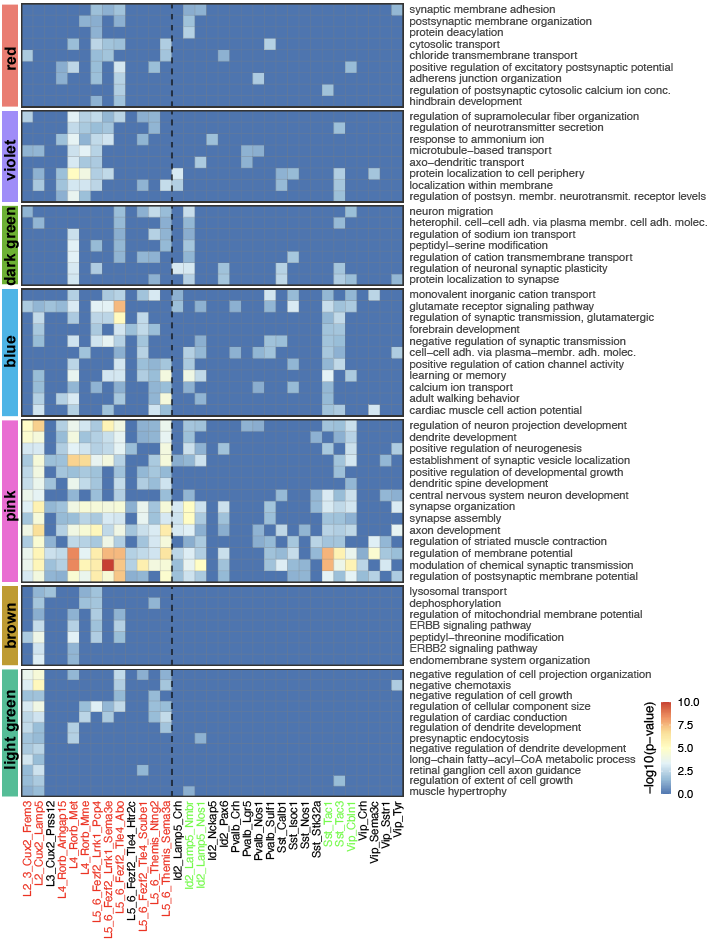
Step 3.2: local structure of changes


Step 3.2: local structure of changes

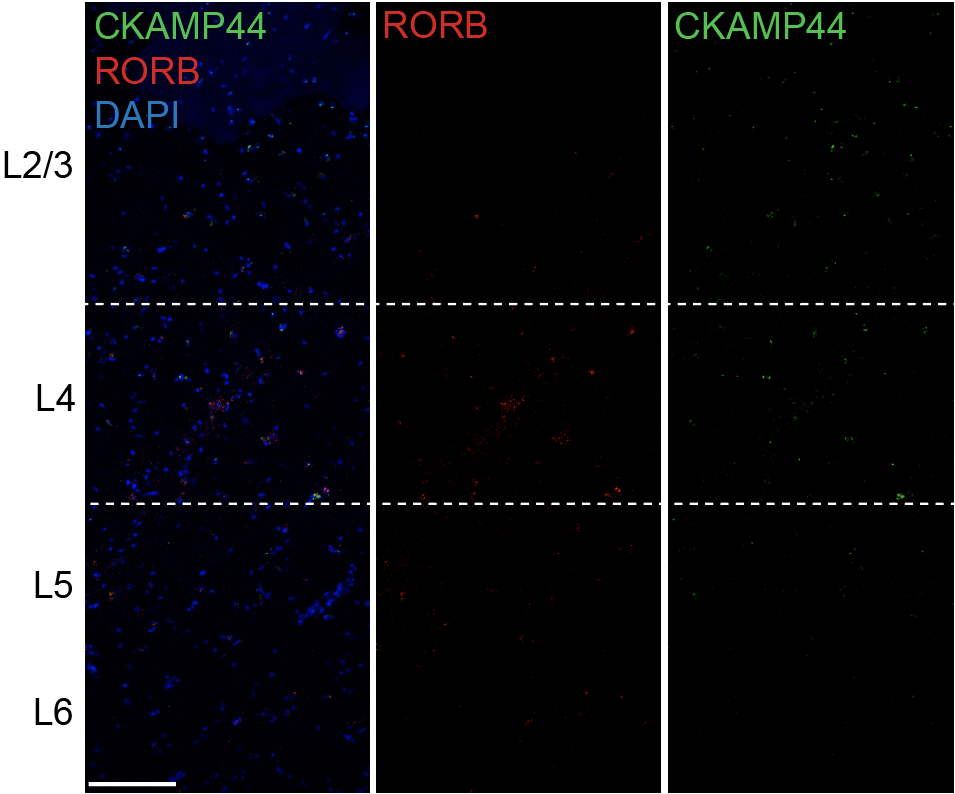

Control
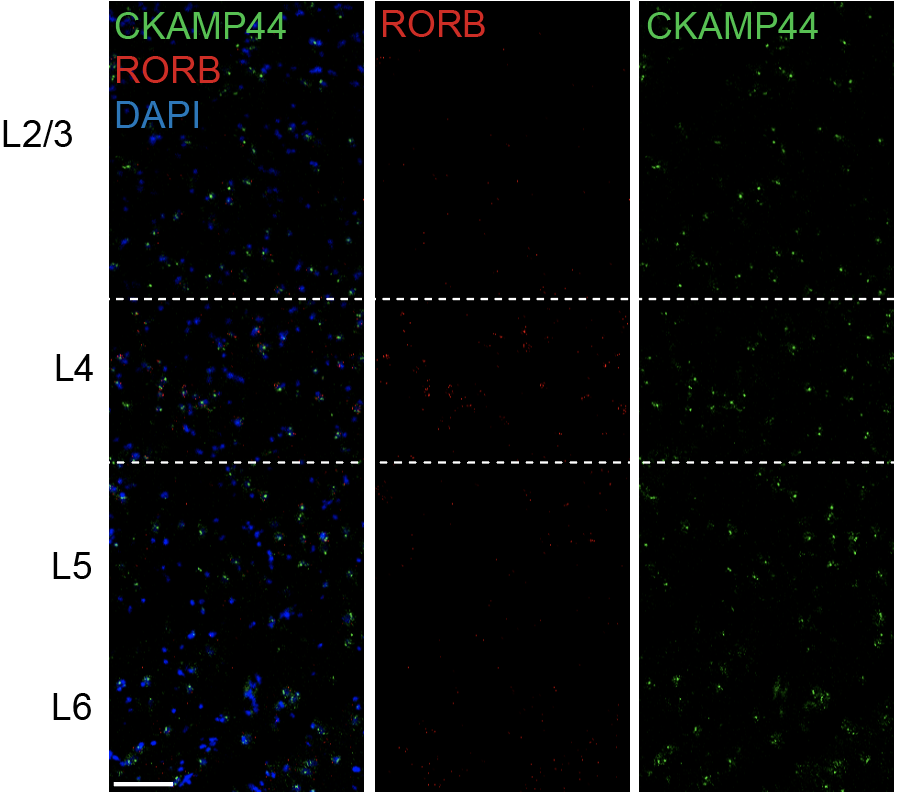

Epilepsy
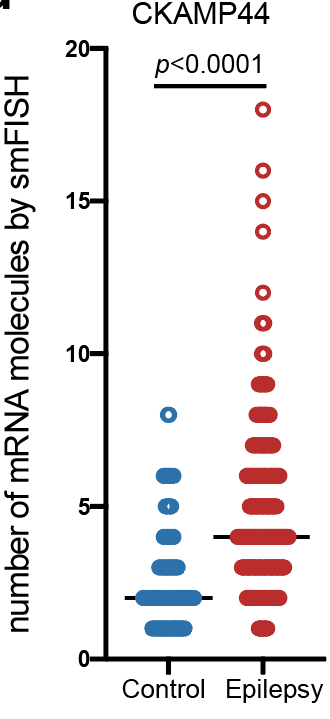
Step 3.2: local structure of changes

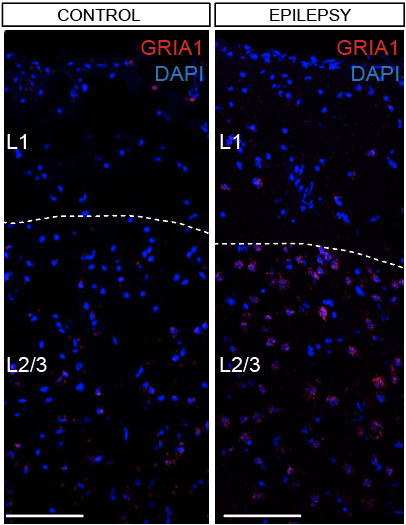
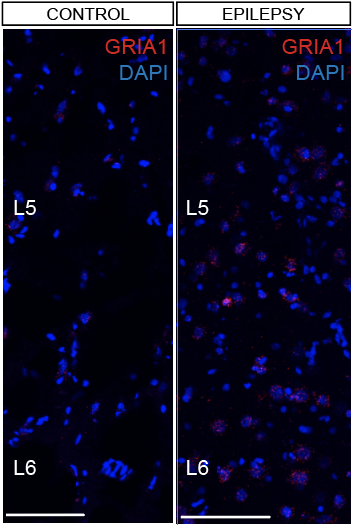
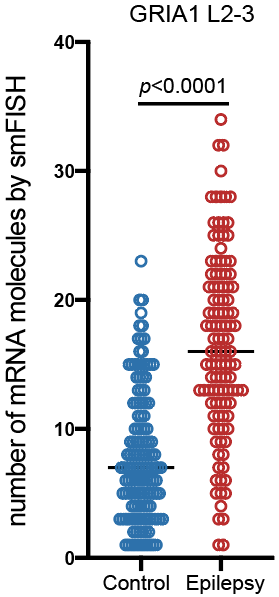
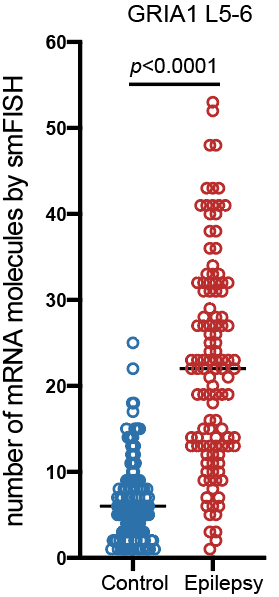
Step 3.2: local structure of changes

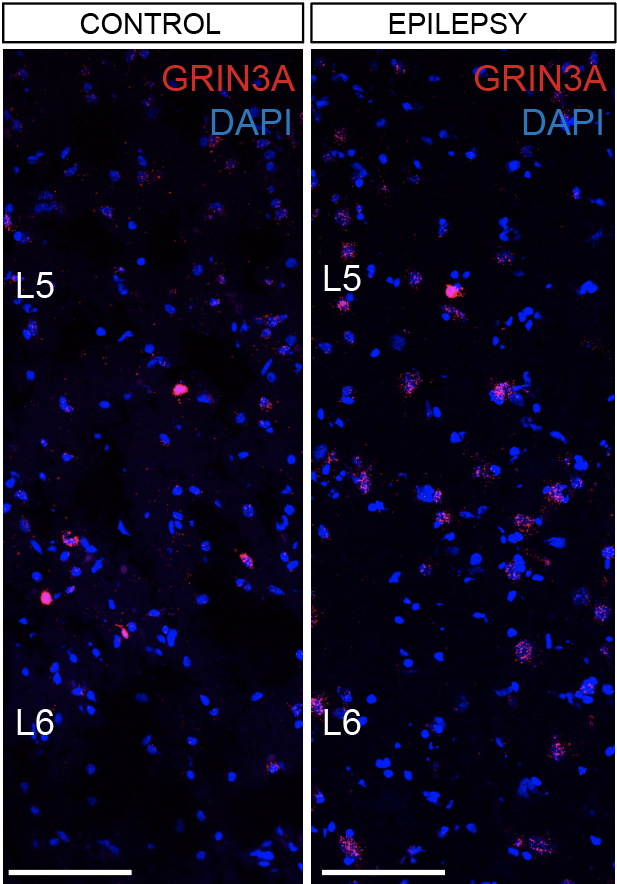
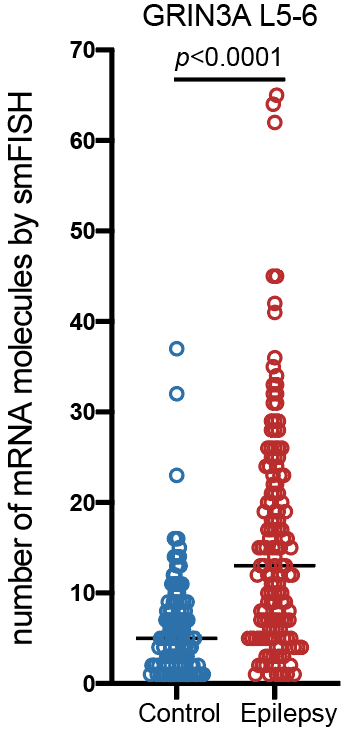
Epilepsy
Cacoa
Overview
State
Acknowledgements

Khodosevich Lab

Jonathan Mitchel

Ruslan Soldatov

Shenglin Mei

Evan Biederstedt
Navneet Vasistha
Konstantin Khodosevich
Rasmus Rydbirk
Irina Korshunova

Diego González

Katarina Dragicevic

Mykhailo Batiuk

Anna Igolkina

Peter Kharchenko
Thank you for your attention!
Petukhov Viktor
viktor.petukhov@pm.me
Petukhov Viktor
viktor.petukhov@pm.me
Let's collaborate!
- Bioinformatics & statistics
- Analysis of scRNA-seq data
- Anti-ageing & cell rejuvenation
- Case-control analysis of multi-omics data