
Case-control analysis of single-cell RNA-seq studies
Petukhov V, Rydbirk R, Igolkina A, Mei S, Kharchenko P, Khodosevich K
viktor.s.petuhov@ya.ru
Description of the problem
Two conditions, multiple samples per condition, multiple cells per sample



Case samples

Control samples
Description of the problem
What cell types are affected and how?
Prepare for further experiments:
- Which subtypes should we focus on?
- Which genes per subtype should we investigate further?
Questions to existing data:
- Do some cell types changed their expression in a similar way?
- Which genes changed their expression in a similar way?
- All other patterns in expression changes we can think of
Existing solutions
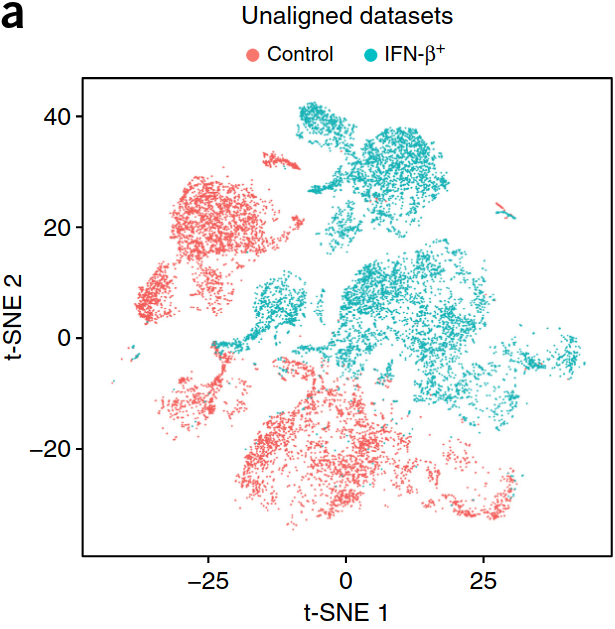
Existing solutions
Align samples
scVI, Conos, ..., Seurat
See the review from the Theis lab
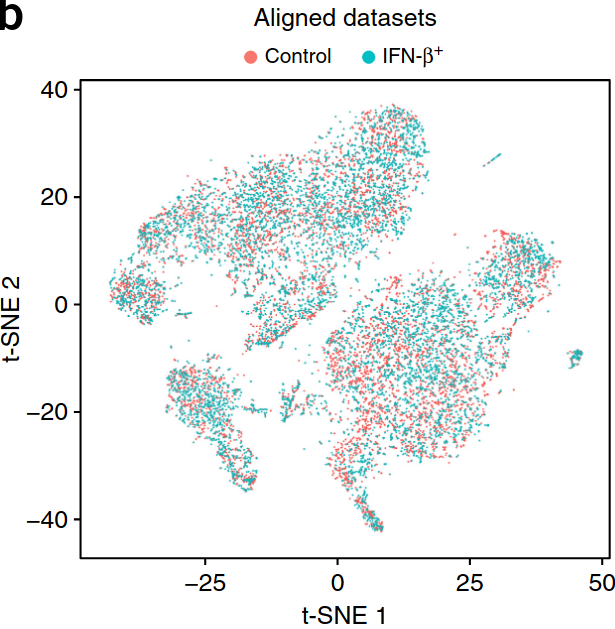
Existing solutions
Align samples
Perform joint annotation
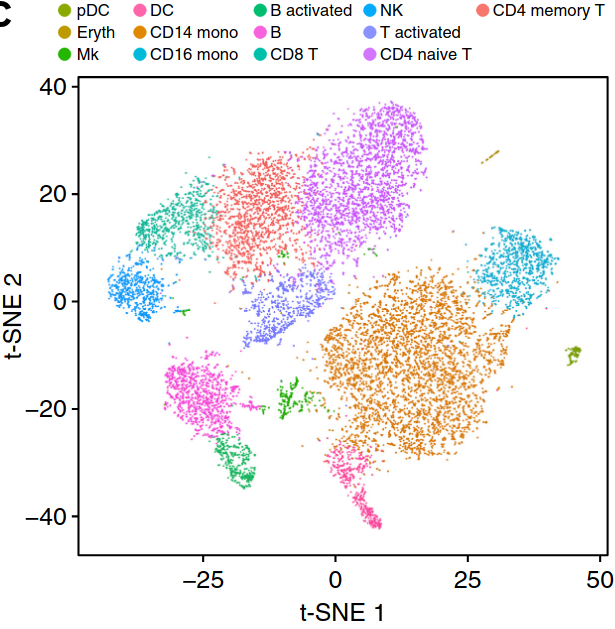
Existing solutions
Align samples
Perform joint annotation
Run differential expression
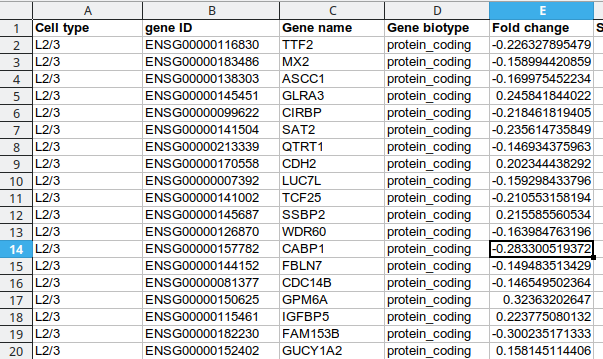
Existing solutions
Align samples
Perform joint annotation
Run differential expression
Run Gene Ontology analysis

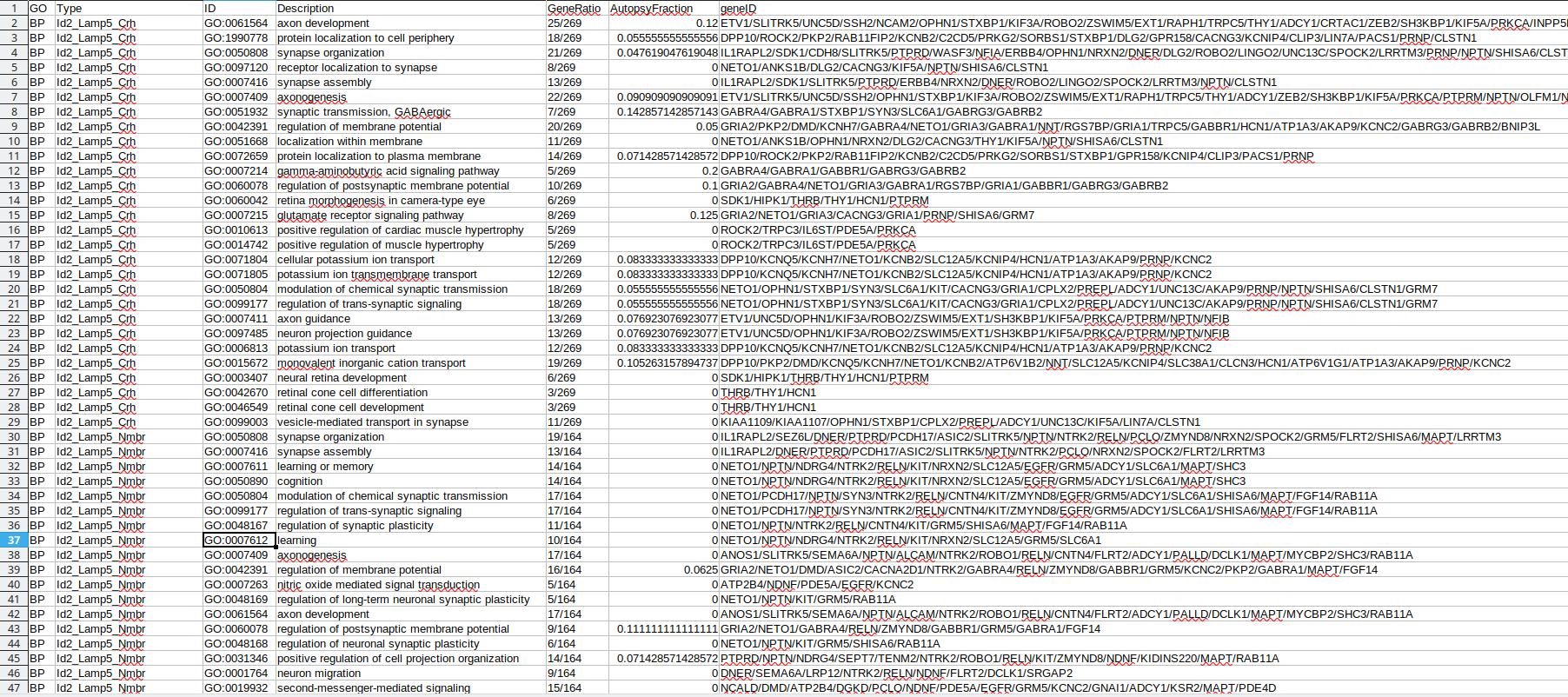
Existing solutions
Align samples
Perform joint annotation
Run differential expression
Run Gene Ontology analysis
Compare cell type proportions
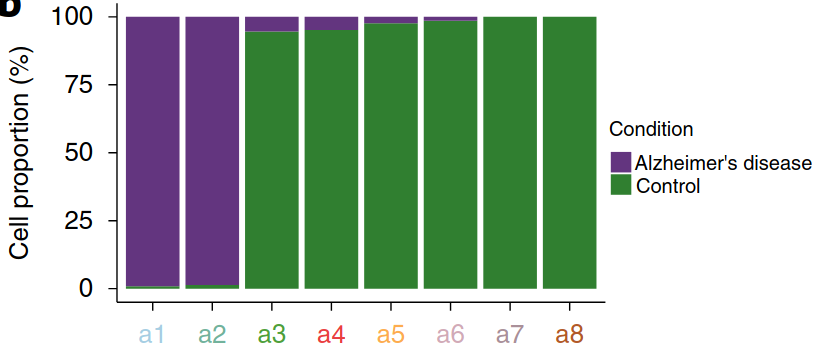
Existing solutions
Align samples
Perform joint annotation
Run differential expression
Run Gene Ontology analysis
Compare cell type proportions
Why it is not enough
- What cell types are the most affected?
- How exactly are they affected?
The main questions:
Why it is not enough
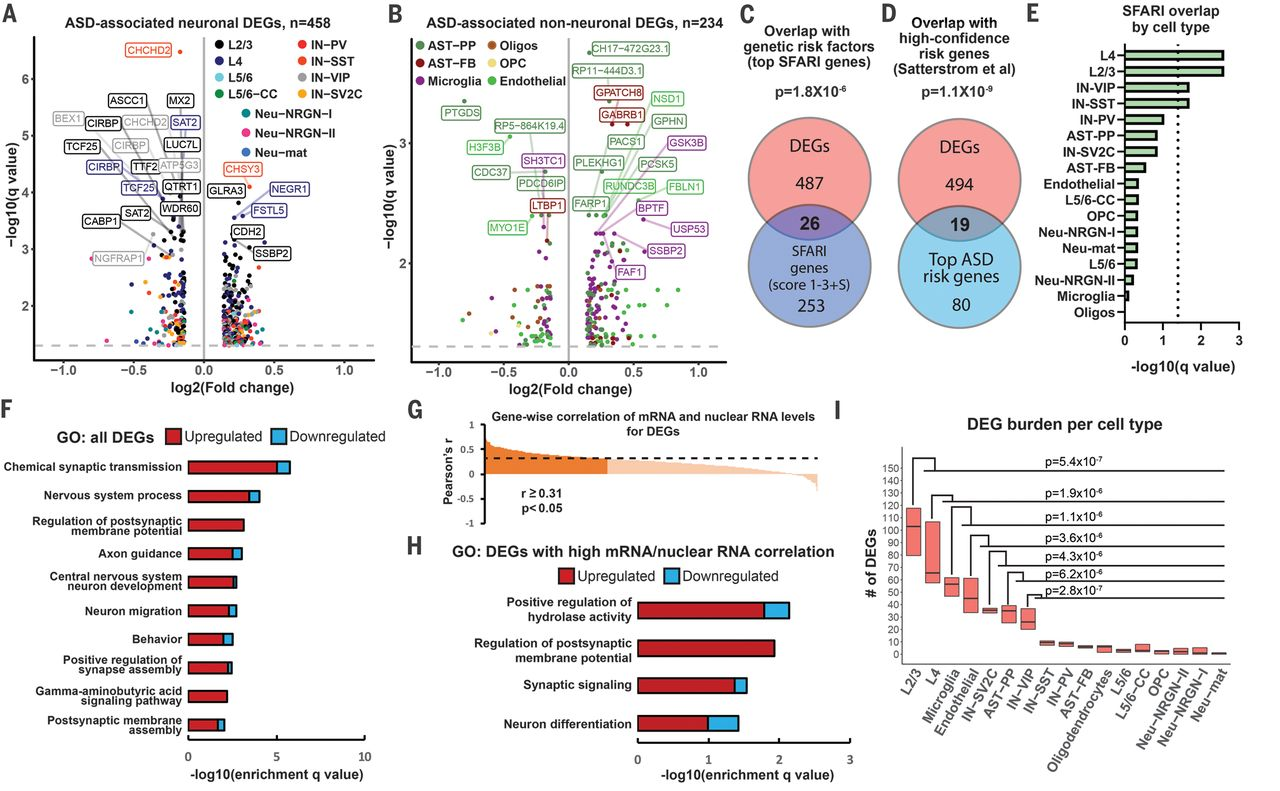
Number of DE genes is not an answer
- What cell types are the most affected?
- How exactly they are affected?
The main questions:

Why it is not enough
There are up to 1000 significant DE genes per type and 2795 unique DE genes in total
- What cell types are the most affected?
- How exactly they are affected?
The main questions:
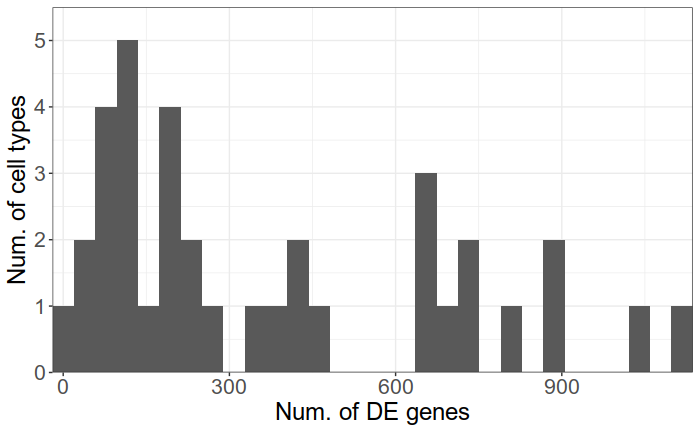
*Epilepsy data from Khodosevich lab
Why it is not enough
There are up to 1000 significant DE genes per type and 2795 unique DE genes in total
- What cell types are the most affected?
- How exactly they are affected?
The main questions:

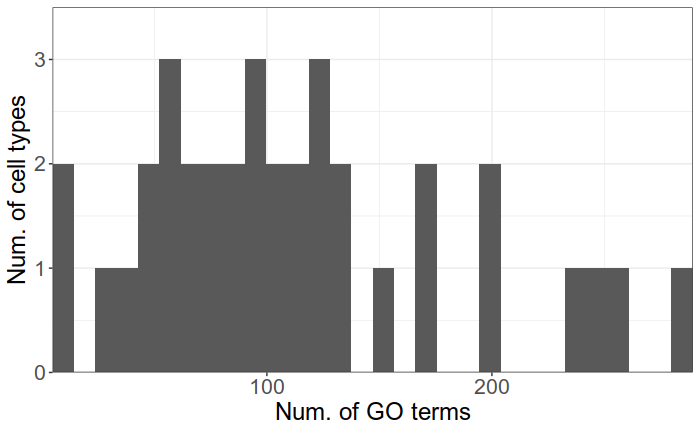
There are up to 300 significant GO terms per type and 796 unique terms in total
Why it is not enough
DE depends on the depth and quality of the annotation
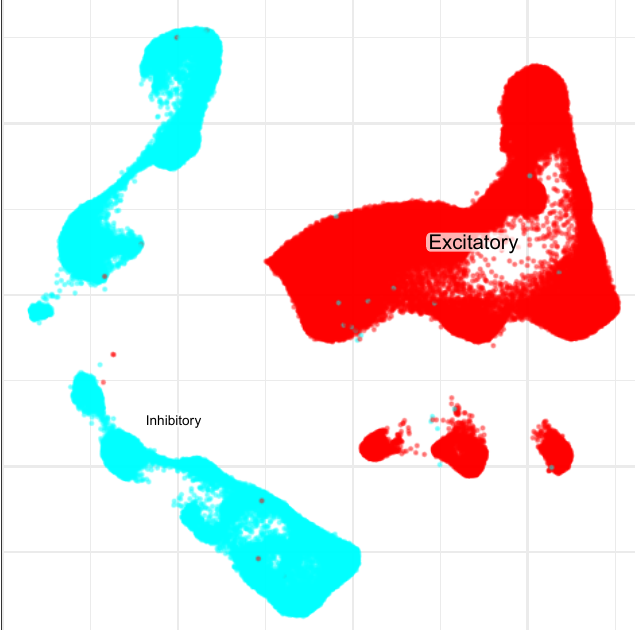
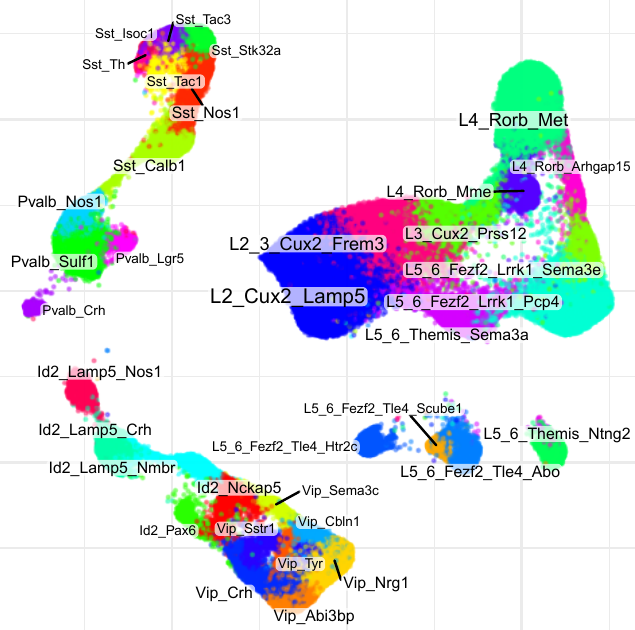
Compositional analysis


Gene expression analysis

Case-control analysis of single-cell studies: a fresh approach
What can we possibly do?
What can we possibly do?
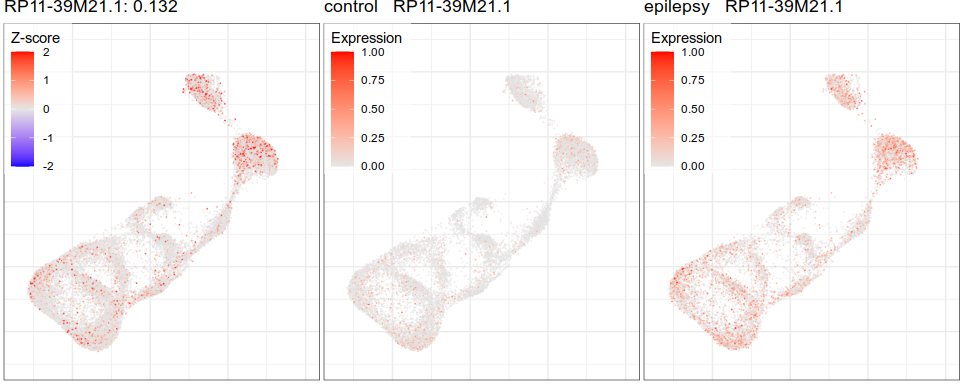
Gene expression analysis

Compositional analysis


Cluster-based
Cluster-free
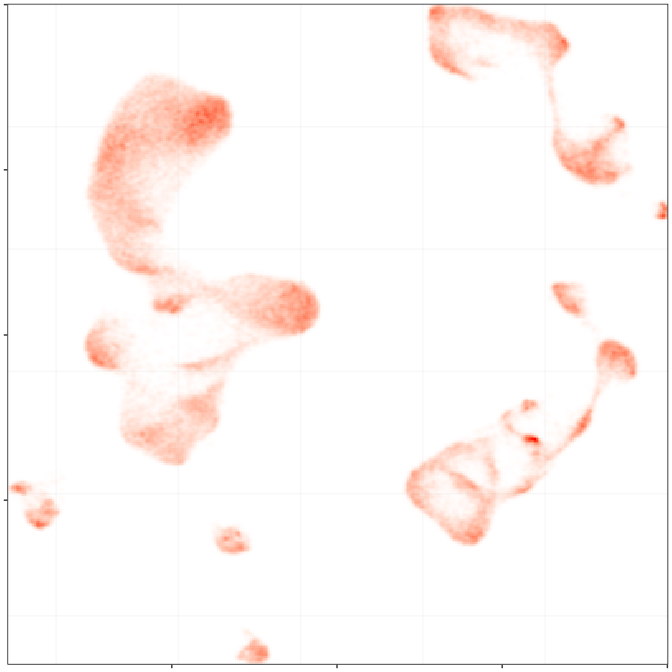
control
epilepsy
Case-control analysis of single-cell studies: a fresh approach
Compositional analysis
Gene expression analysis
Cluster-based
Cluster-free
To be improved
To be improved
Ready
Proof of concept
What is done?
Case-control analysis of single-cell studies: a fresh approach
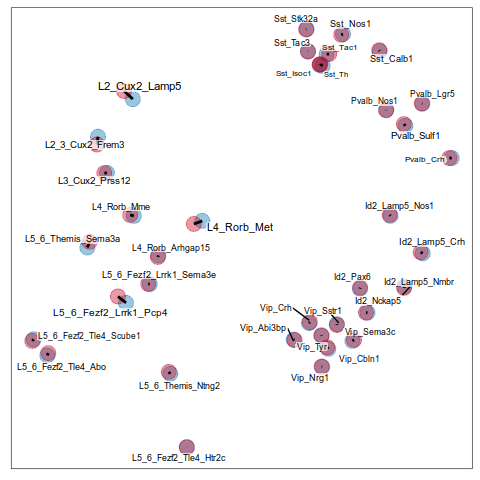
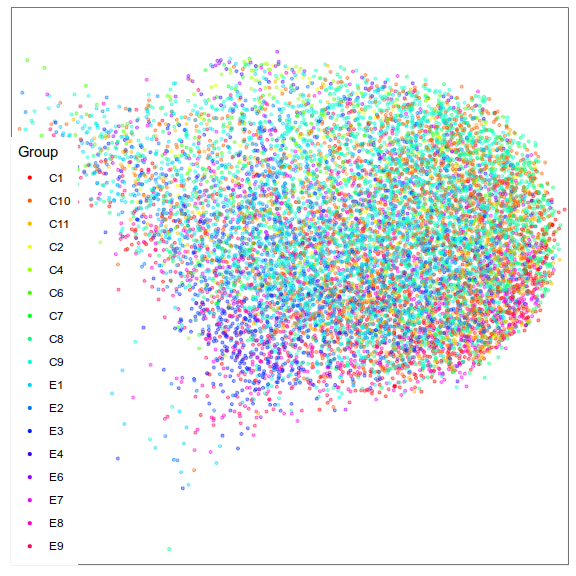
Composition analysis, cluster-based
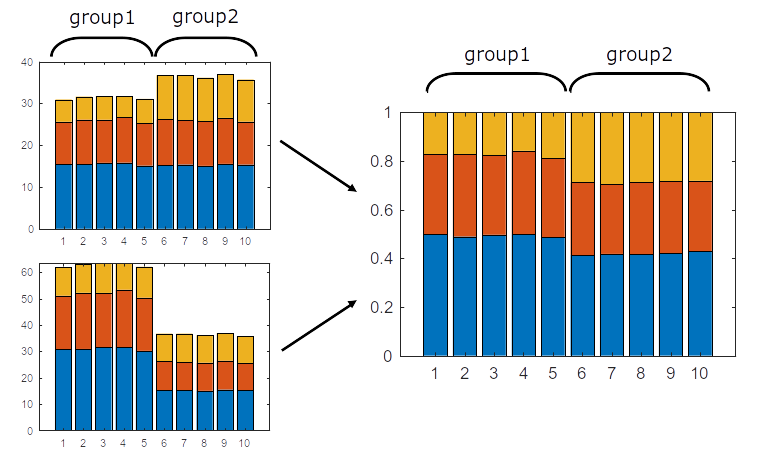
Ideally, replace this plot with 1 increase vs 2 decrease
Problem: changes are not independent
Composition analysis, cluster-based
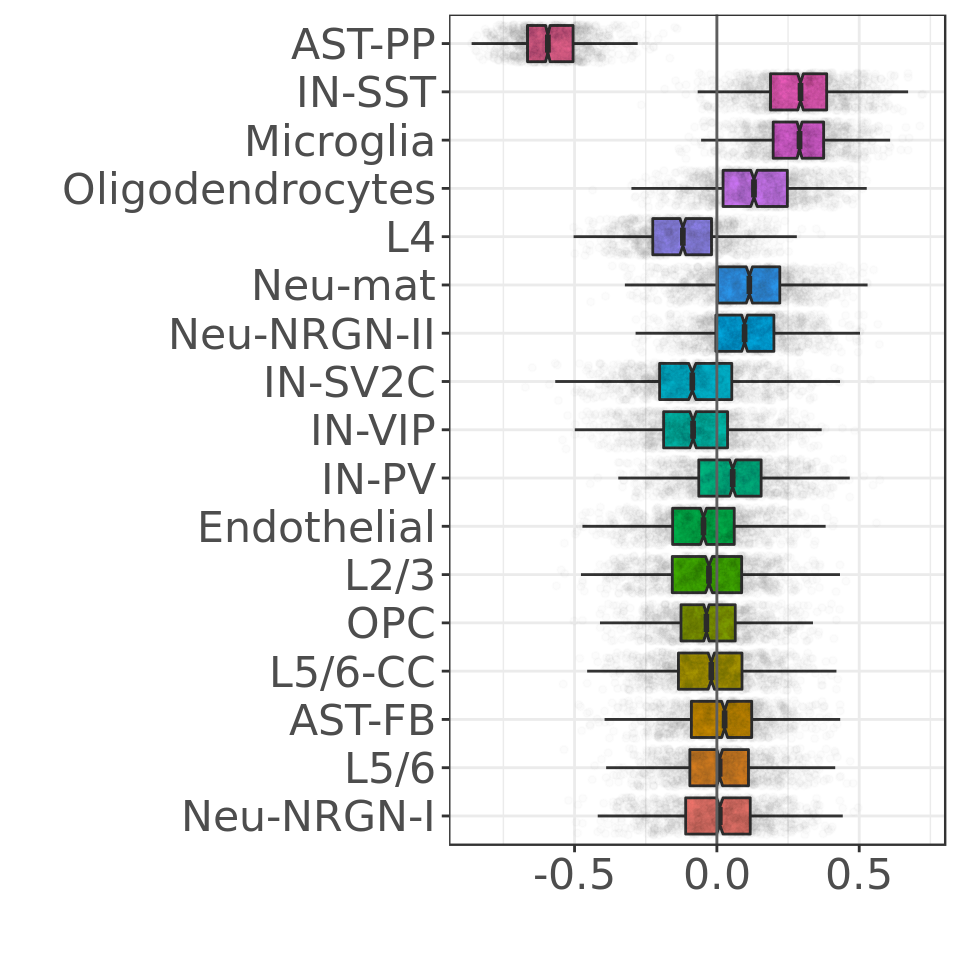
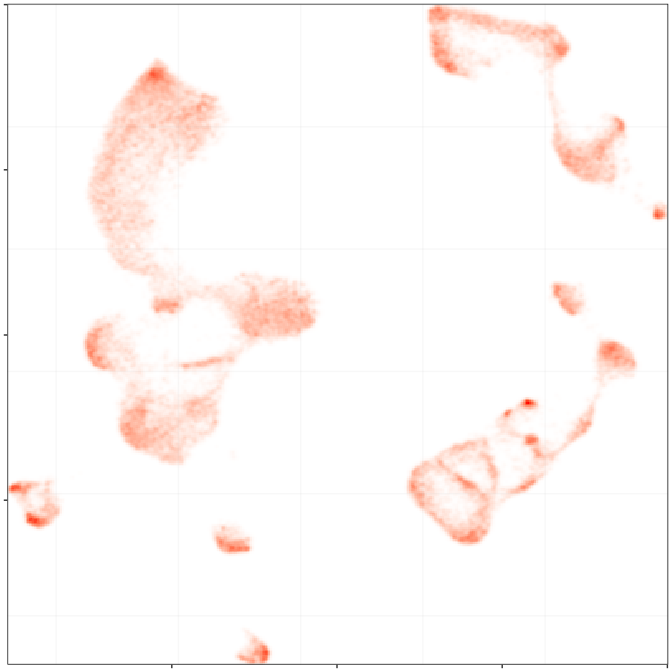
Composition analysis, cluster-free
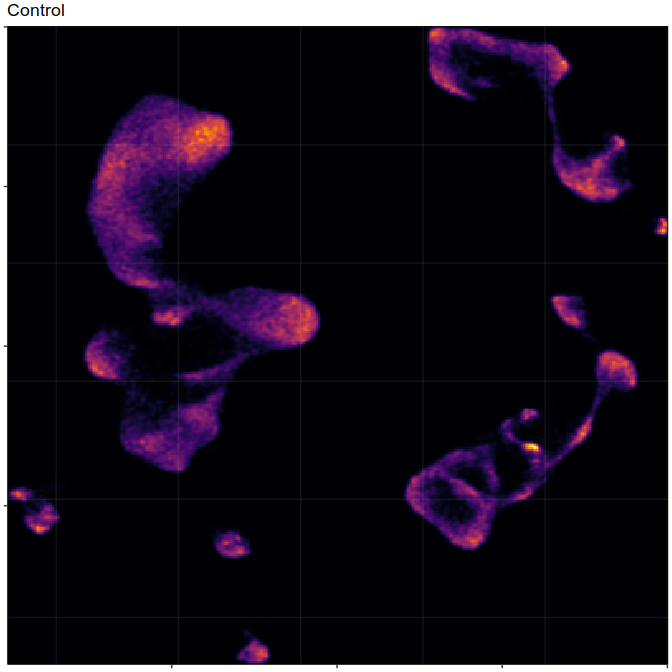
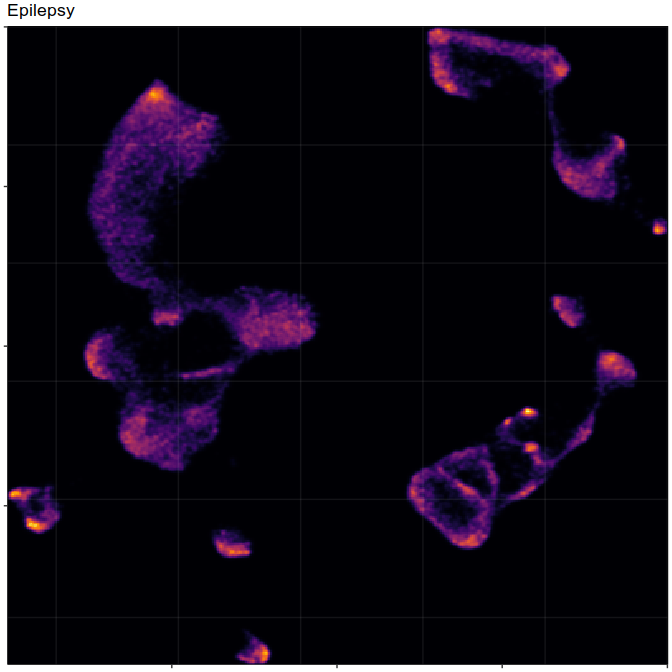
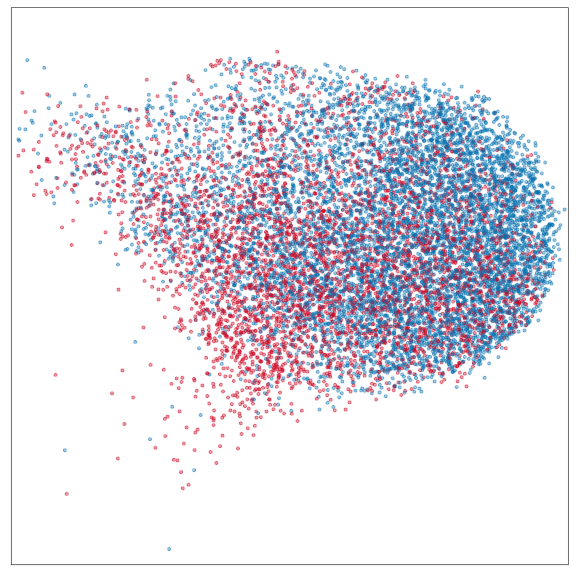
Control
Epilepsy
Embedding densities
Composition analysis, cluster-free

Control
Epilepsy
Graph densities
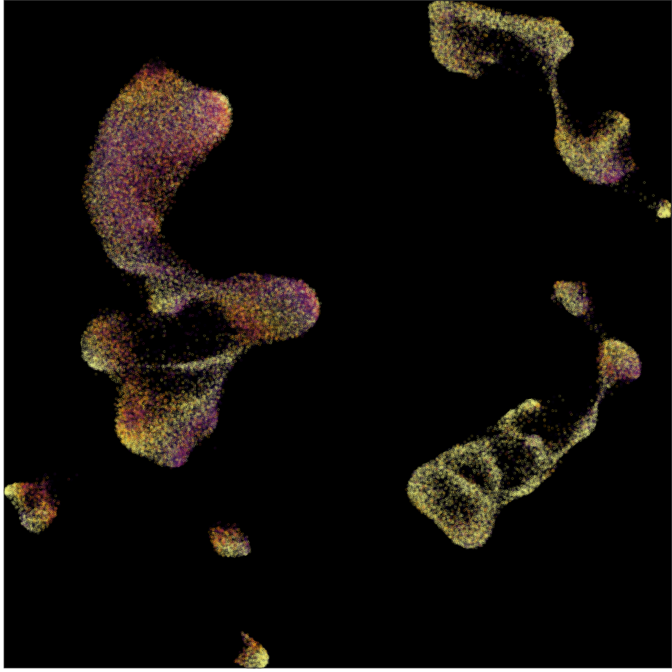
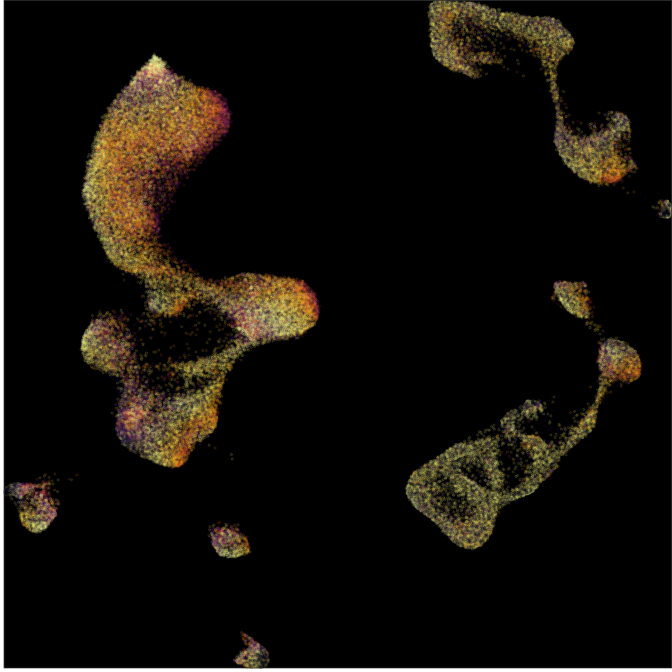
WARNING: preliminary results
Expression analysis, cluster-based
What cell types are affected the most?
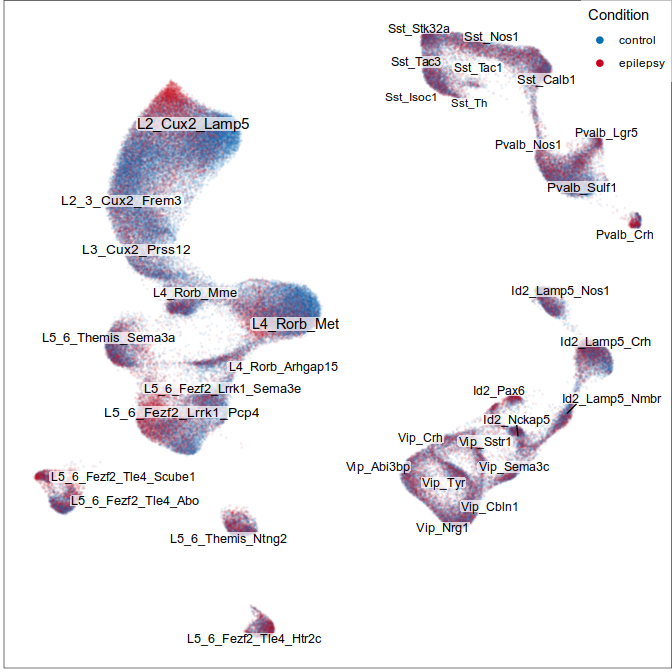
Expression analysis, cluster-based
What cell types are affected the most?


Expression analysis, cluster-based
What cell types are affected the most?
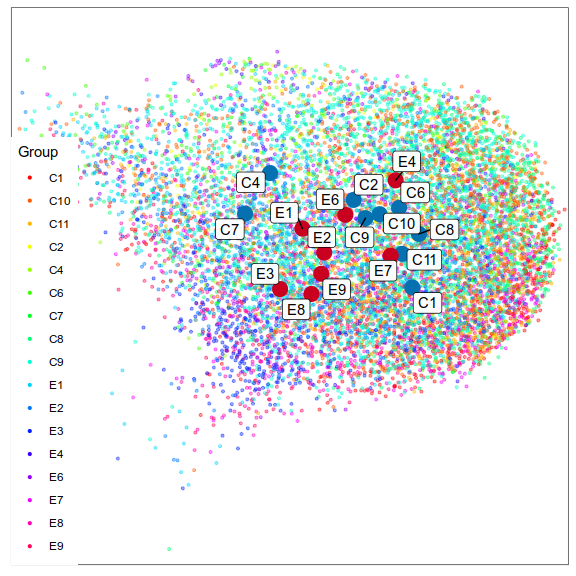
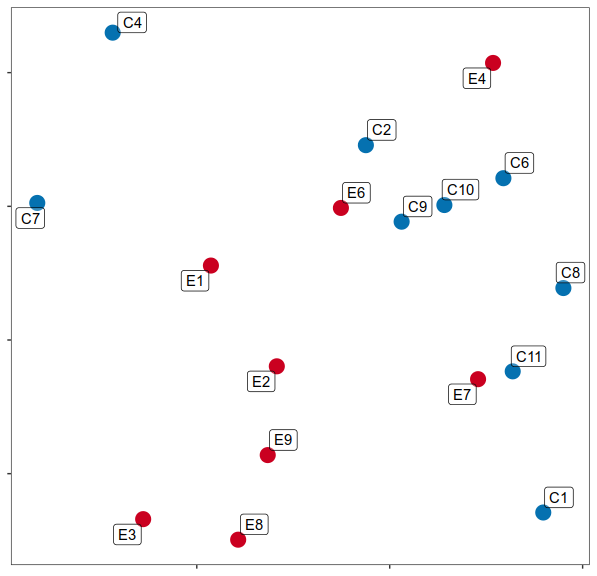
Expression analysis, cluster-based
What cell types are affected the most?

Expression analysis, cluster-based
What cell types are affected the most?
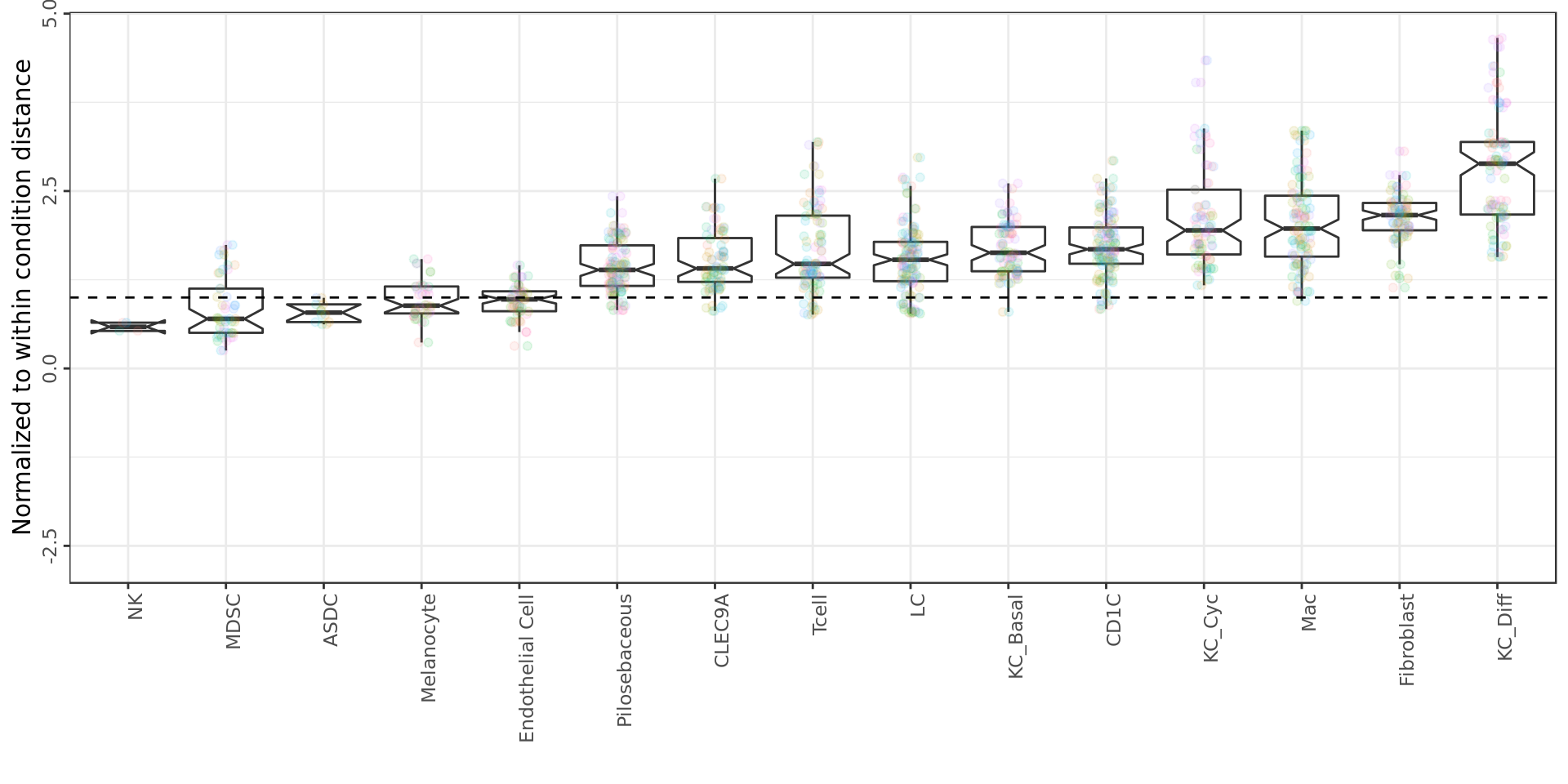
Expression analysis, cluster-based
Can we trust differential expression?
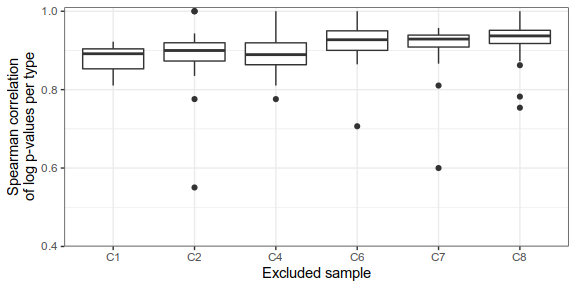
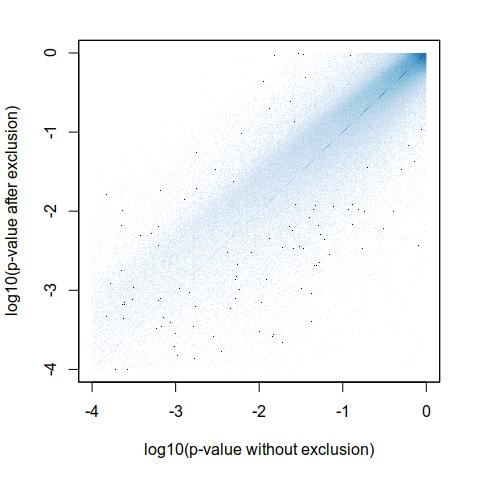
Expression analysis, cluster-based
Can we trust differential expression?
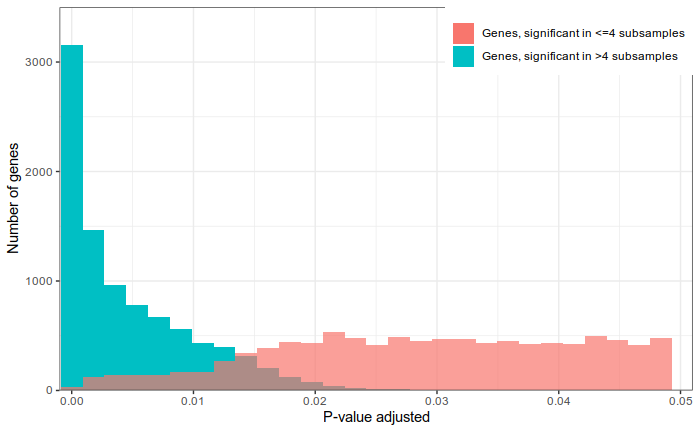

False discoveries

Signal
Expression analysis, cluster-based
Can we trust differential expression?
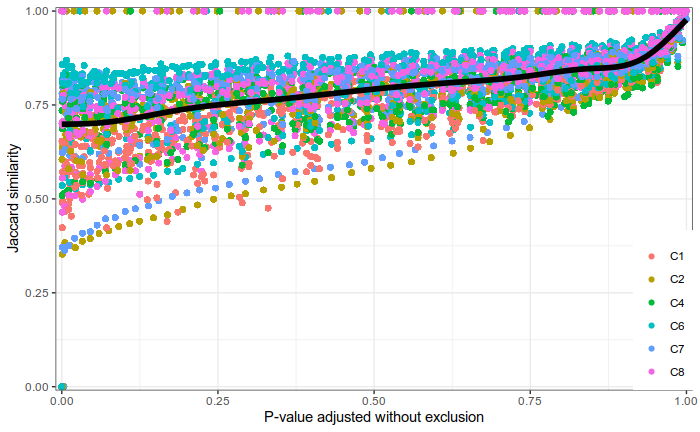
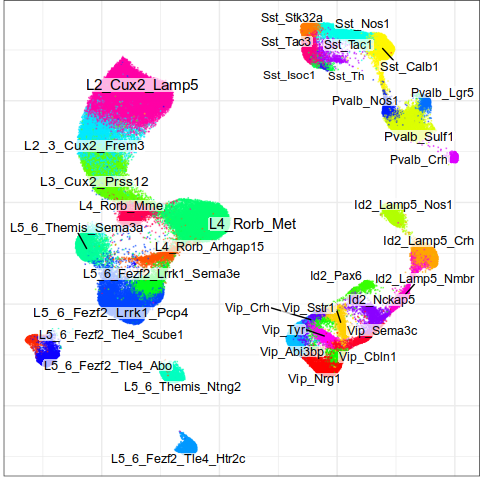
Expression analysis, cluster-free
Differential expression on single cells

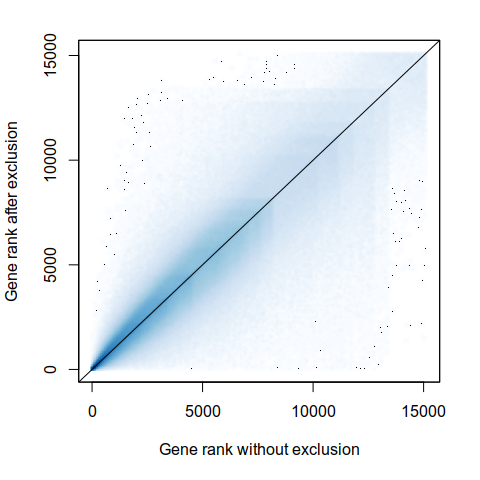
Expression analysis, cluster-free
Differential expression on single cells


Expression analysis, cluster-free
Differential expression on single cells
control
epilepsy
Expression analysis, cluster-free
Differential expression on single cells




control

epilepsy
Differential expression
Expression analysis, cluster-free
Differential expression on single cells
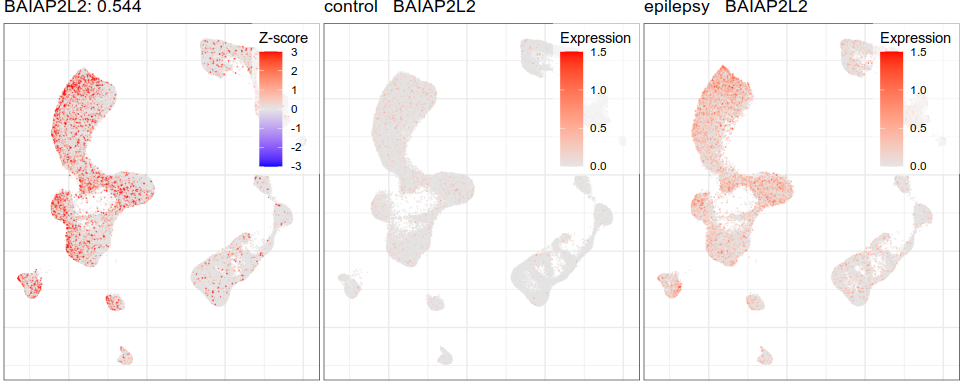
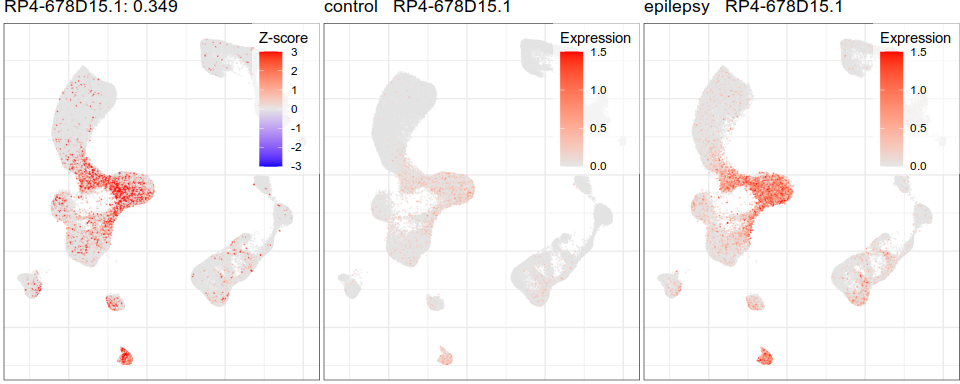
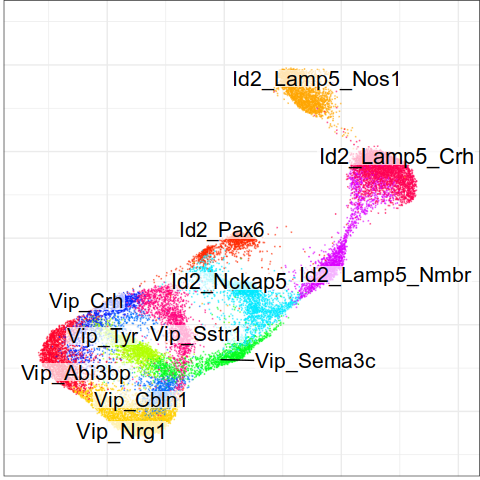
Expression analysis, cluster-free
Differential expression on single cells

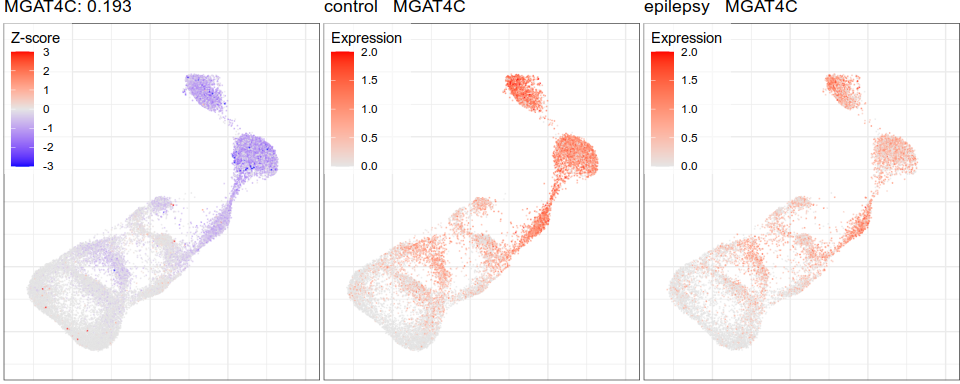


Expression analysis, cluster-free
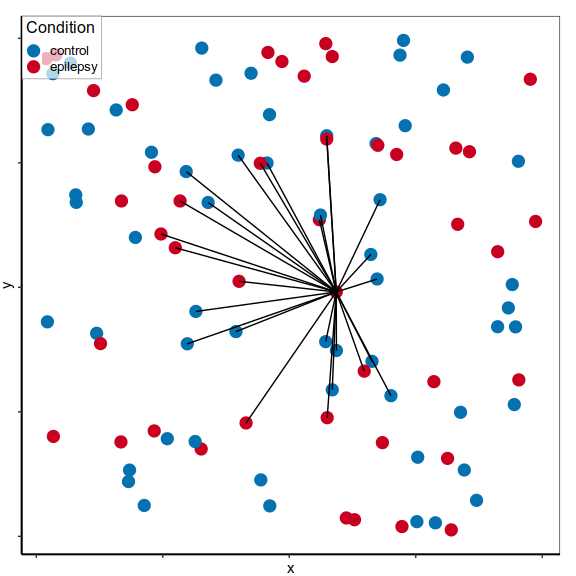
Expression distances
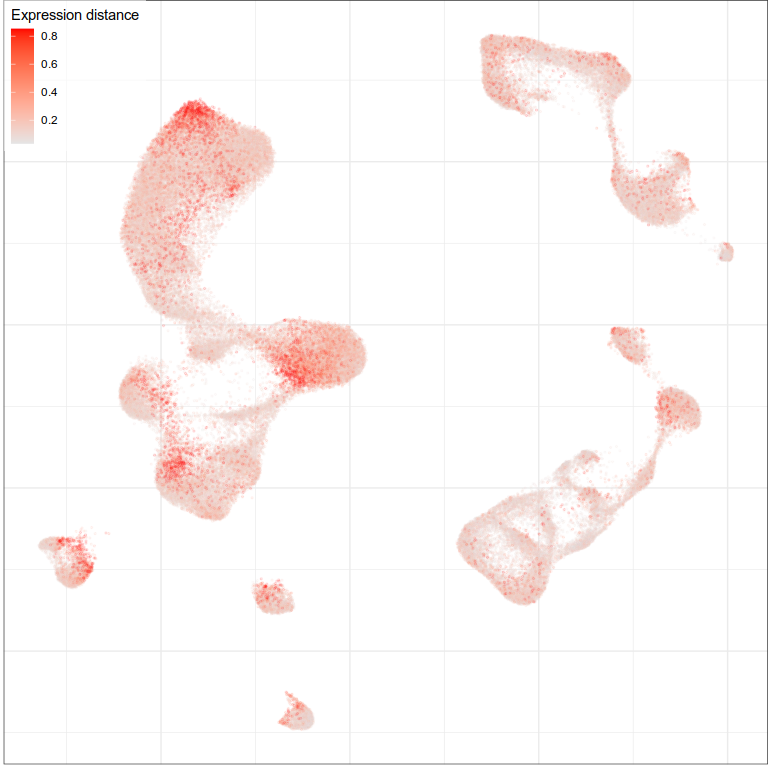


Expression analysis, cluster-free
Expression distances
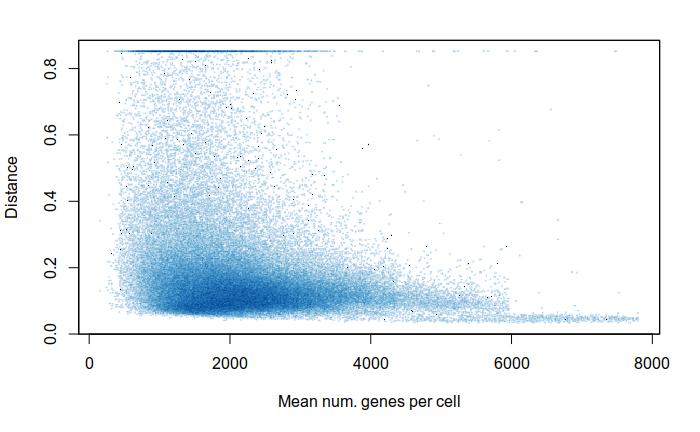


Expression analysis, cluster-free

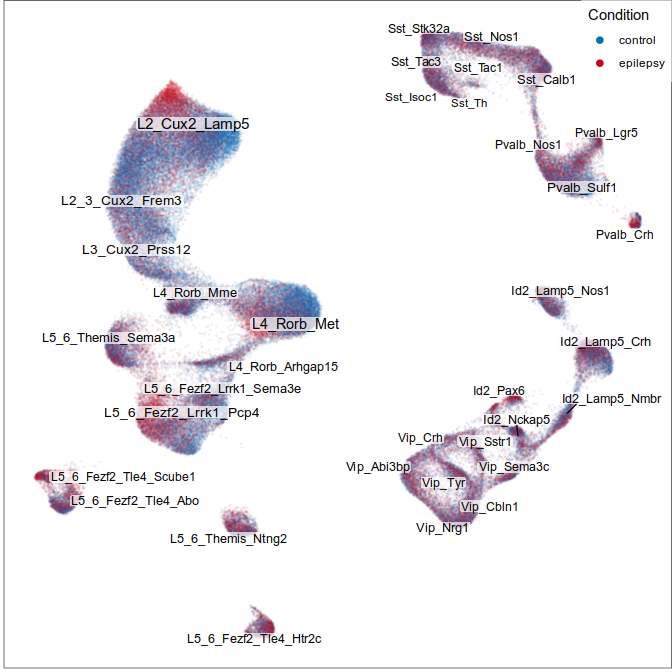
Visualization of joint expression change
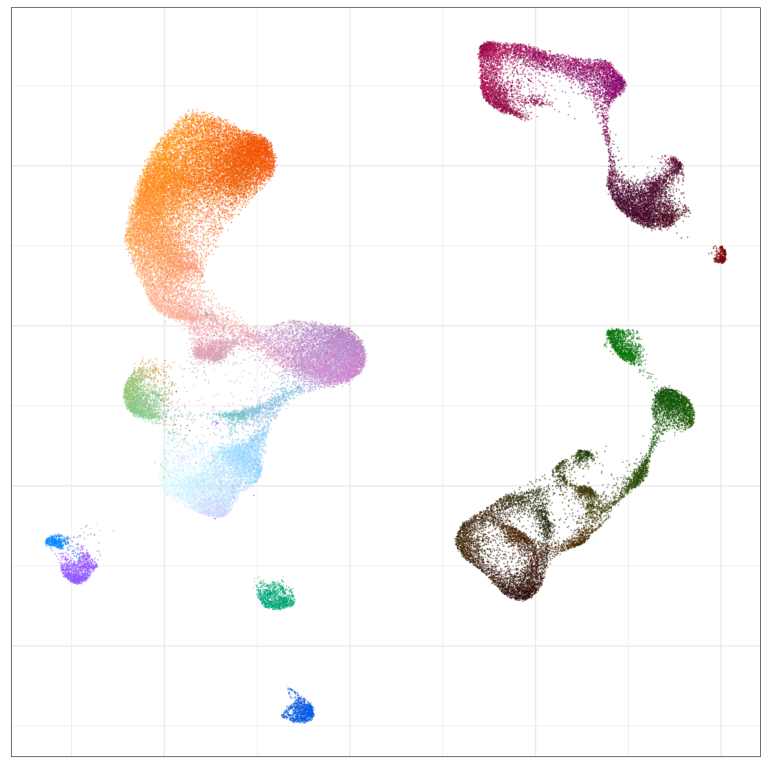
WARNING: very preliminary results
Expression analysis, cluster-free

Visualization of joint expression change
WARNING: very preliminary results
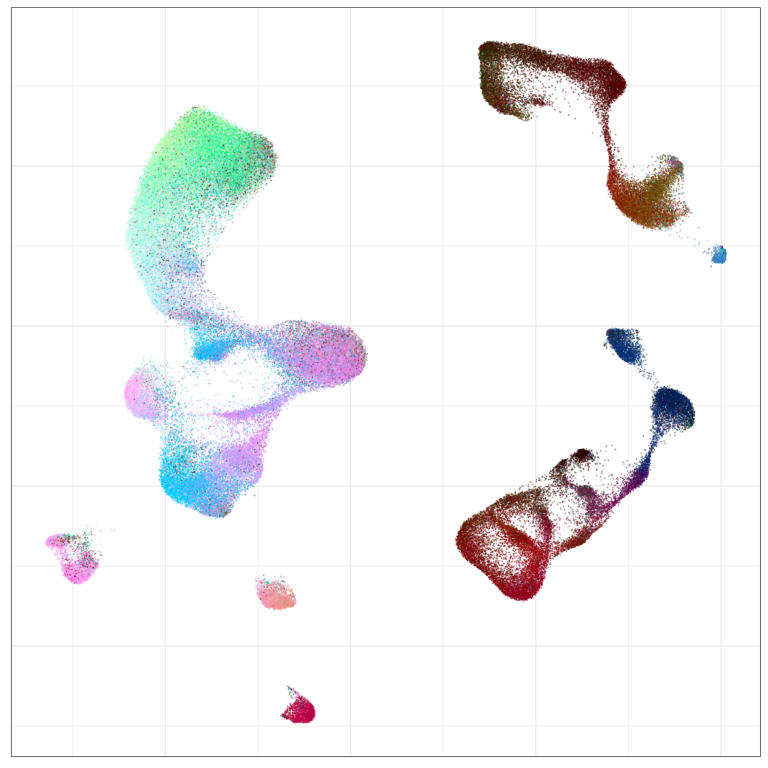
Expression analysis, cluster-free

Visualization of joint expression change
WARNING: very preliminary results

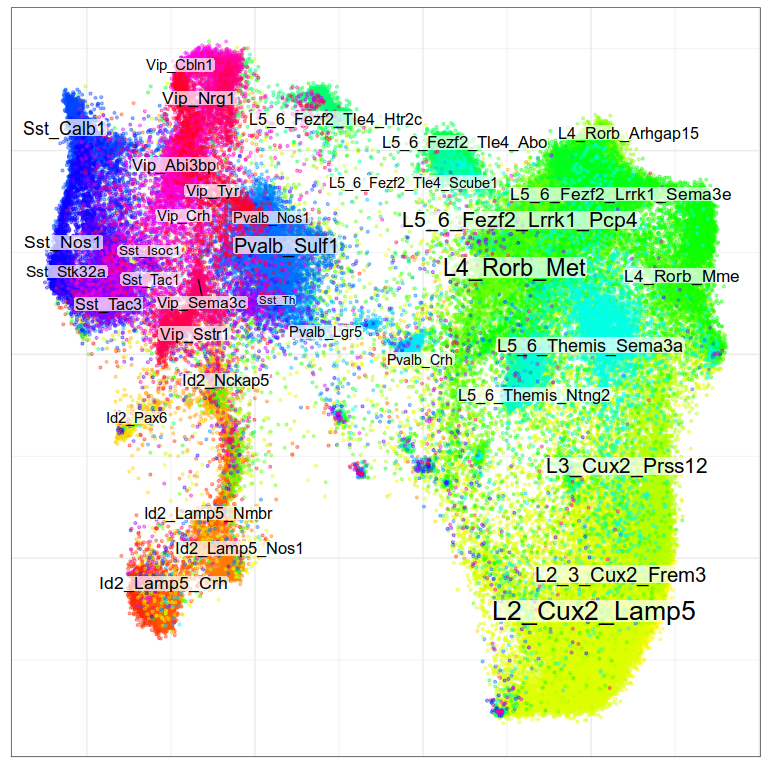

Gene expression analysis

Compositional analysis


Cluster-based
Cluster-free


control
epilepsy
Summary
Thank you!


Konstantin Khodosevich lab
Peter Kharchenko
- Rasmus Rydbirk
- Anna Igolkina
- Shenglin Mei
Co-authors
viktor.s.petuhov@ya.ru