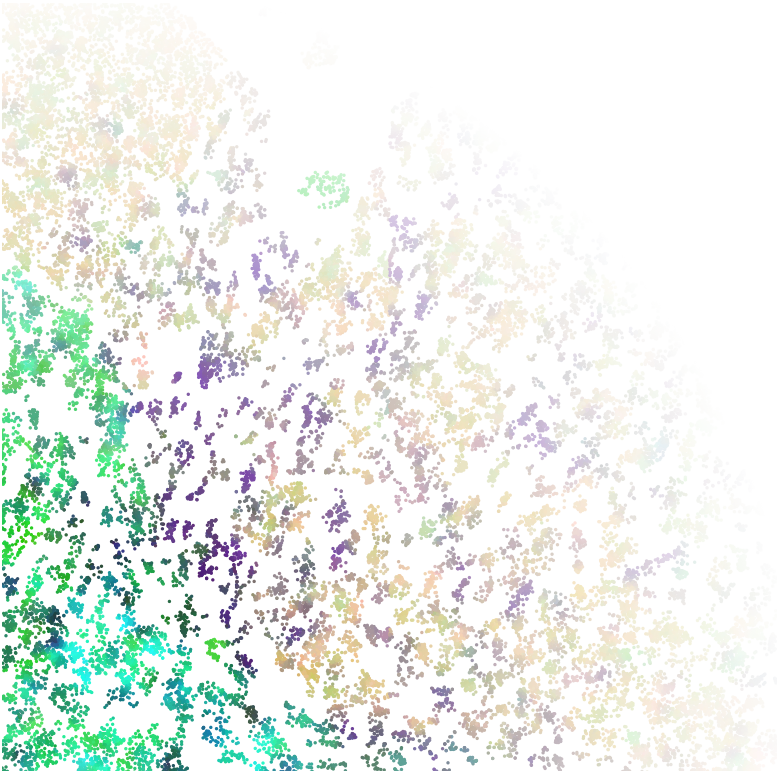
Boundary detection
V. Petukhov, P. Kharchenko
Goal

Estimate
boundaries
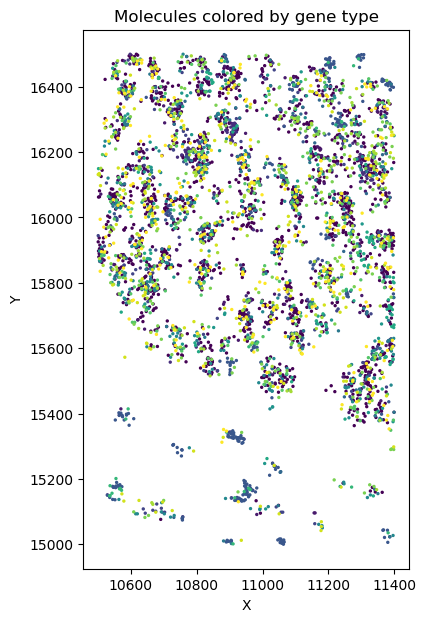
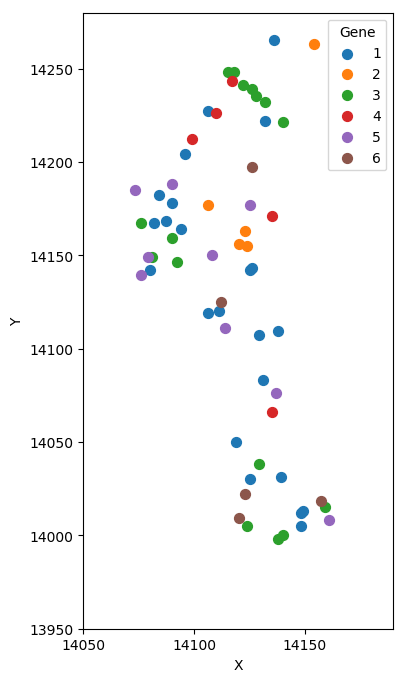
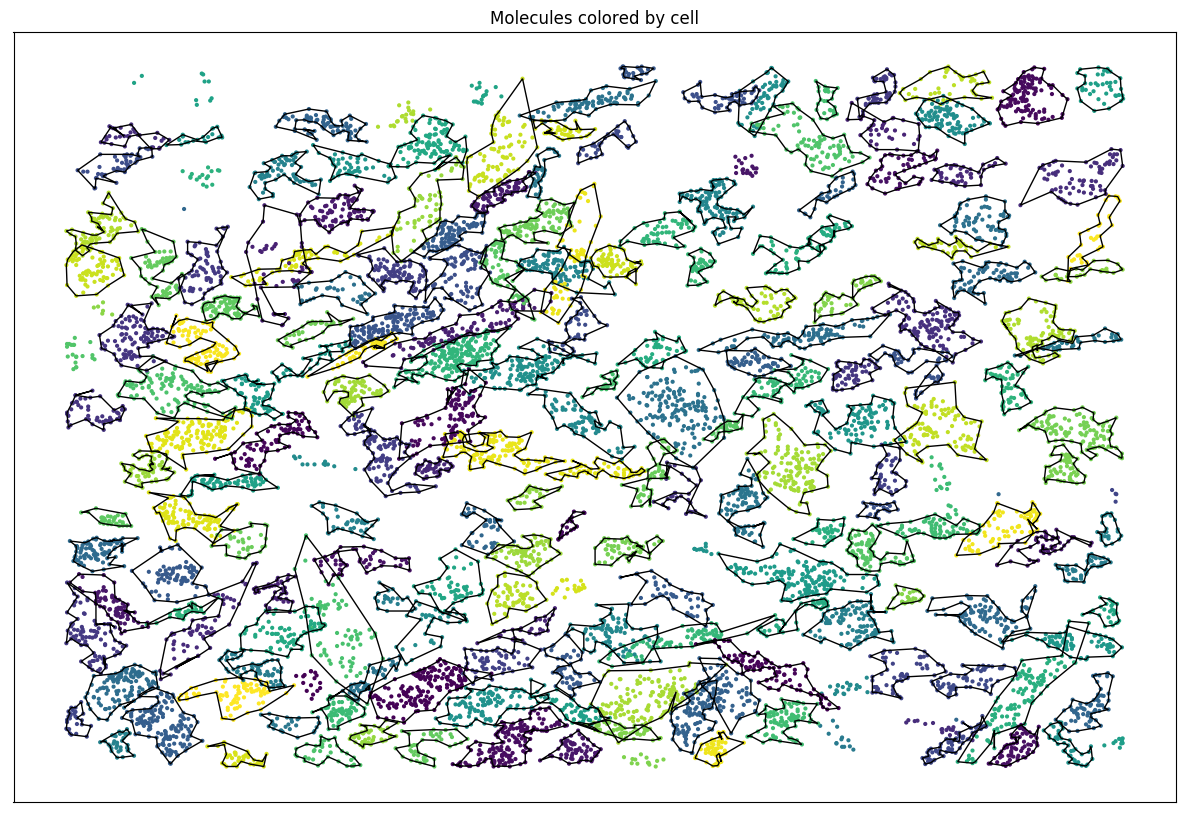
Set of molecules
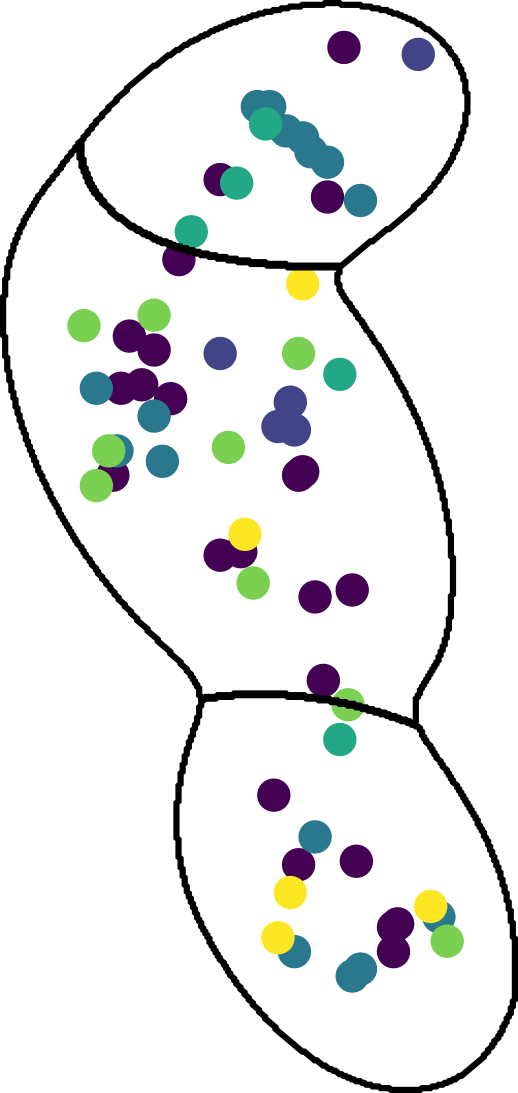


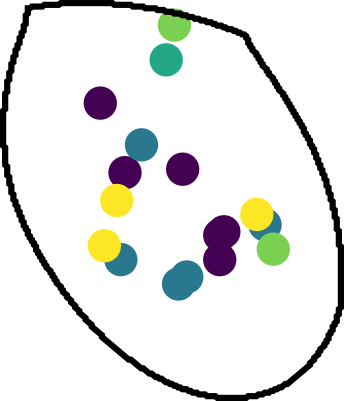
Split molecules by cells
Basic idea
| X | Y | Gene |
|---|---|---|
| ... | ... | ... |
We know:

What is the source?
Gene 1: 20%
Gene 2: 80%
Gene 1: 80%
Gene 2: 20%
Basic idea
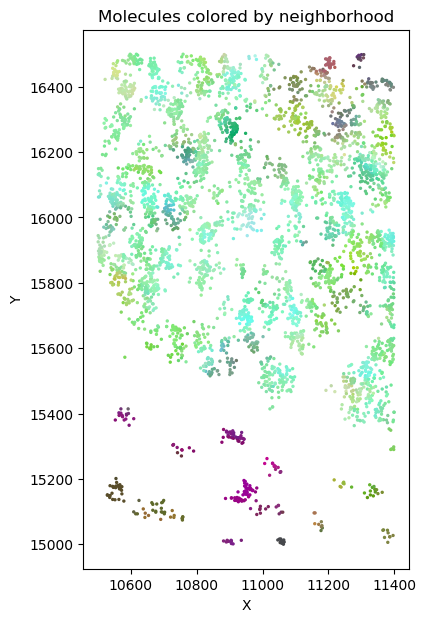

| Gene 1 | ... | Gene k |
|---|---|---|
| N1 | Nk |
Gene vector
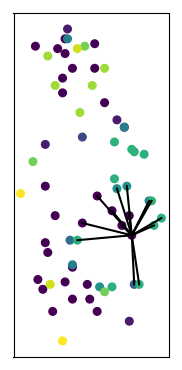
Embedding to 3d space
k nearest
neighbors
Model description
Global scale
Cell type
Shape / size
Position of center

scRNA-seq data
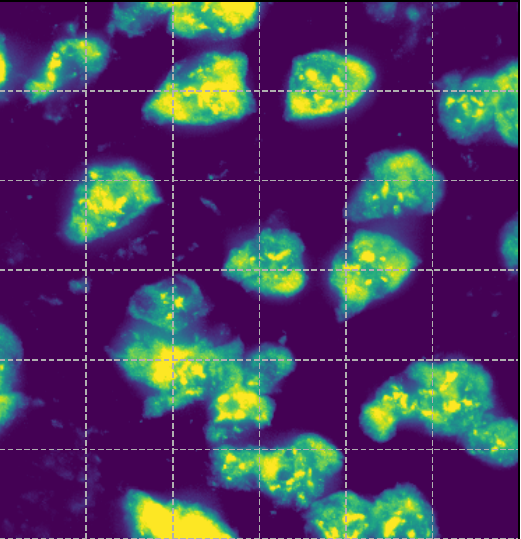
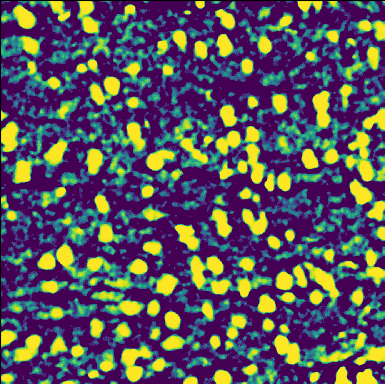



Composition
Nuclei stains
Membrane stains
Molecules
Boundaries
Model description




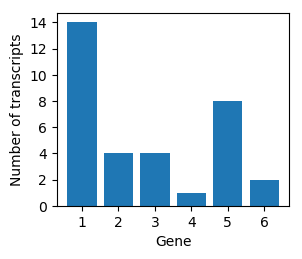
Transcript composition
Center position, ellipsoid shape
2D Normal distribution
Multinomial distribution
| 0.43 | 0.12 | 0.12 | 0.03 | 0.24 | 0.06 |
| 10 | 20 |
| 3.5 | -1.5 |
| -1.5 | 3.5 |
Position
Shape
Composition
Cell is a distribution:
Goal v2.0
Separate probability distributions from a mixture model
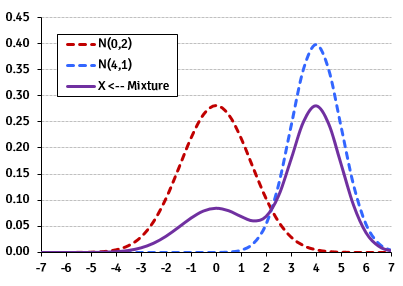
EM on spatial data
Algorithm v0.01
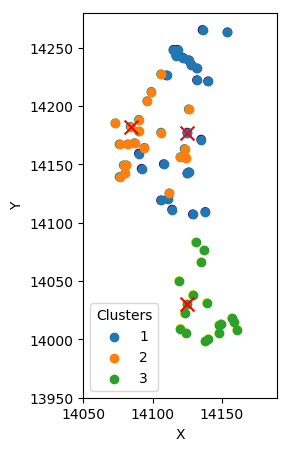
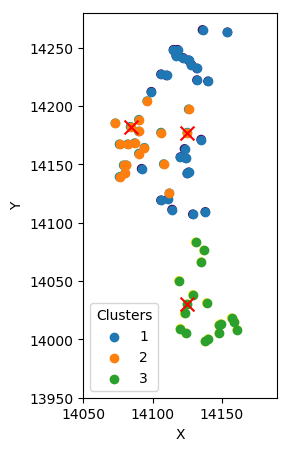
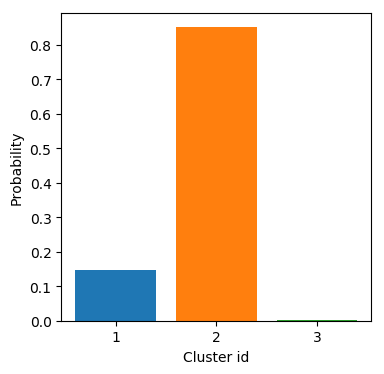
Initial approximation
Expect
Maximize

EM on spatial data
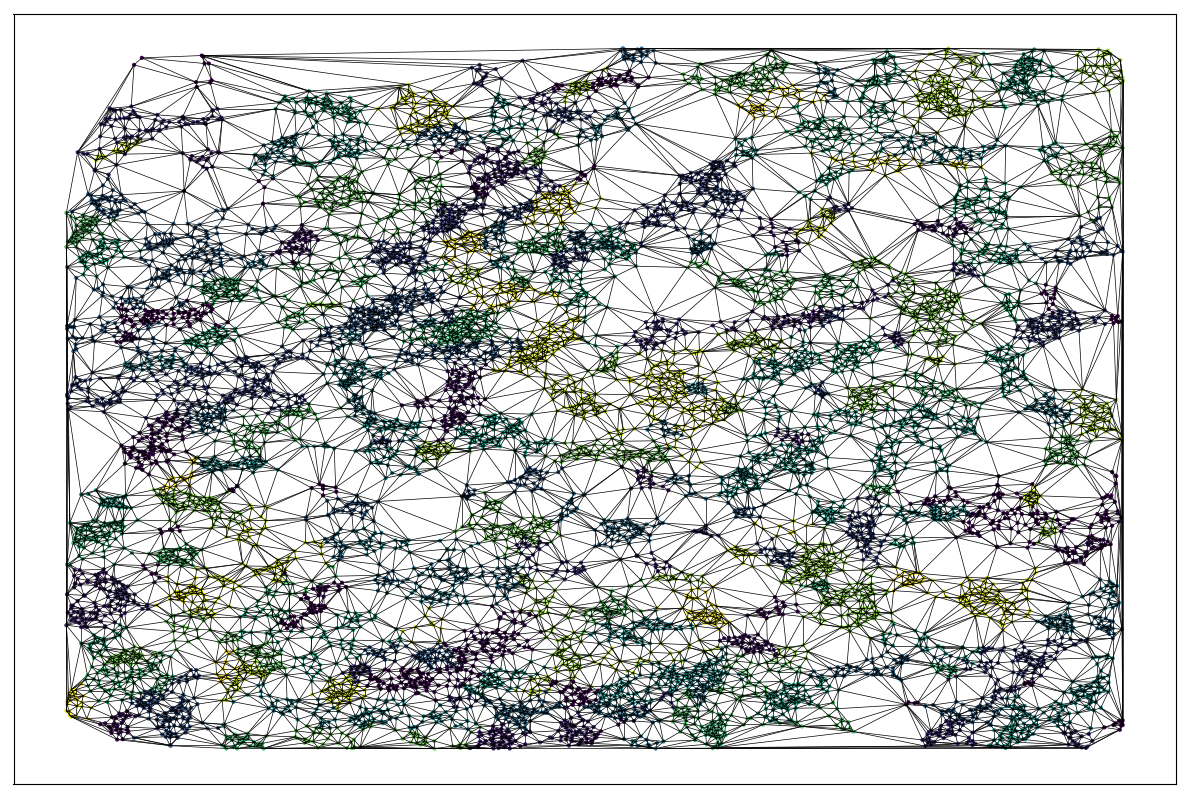
Problem: spatial constraints
EM on spatial data
Solution: Graph Cut Optimization
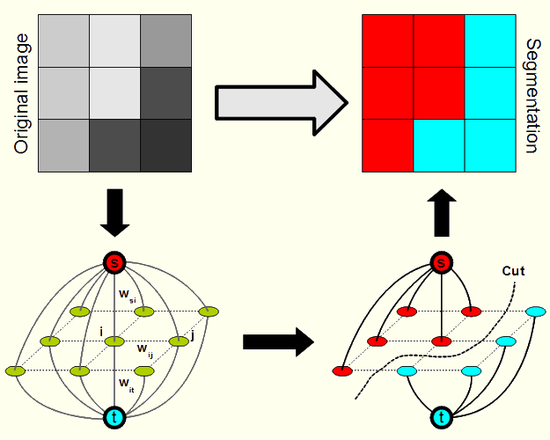
EM on spatial data
Algorithm v0.02




Initial approximation
Expect
Maximize

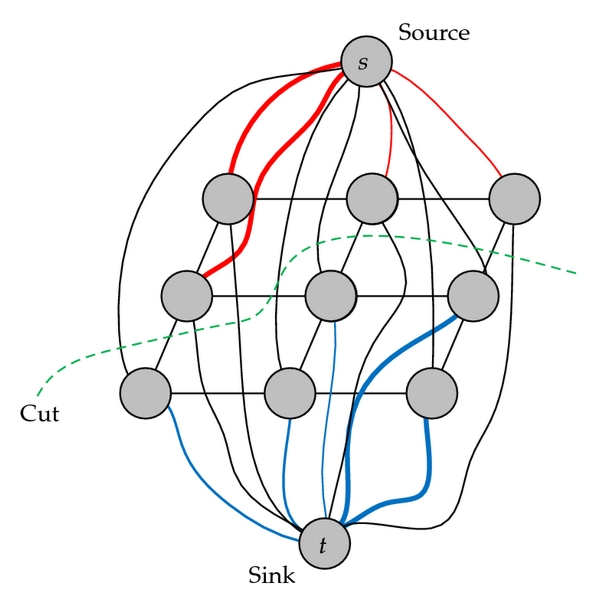
GCO
EM on spatial data
Algorithm v0.02: Results
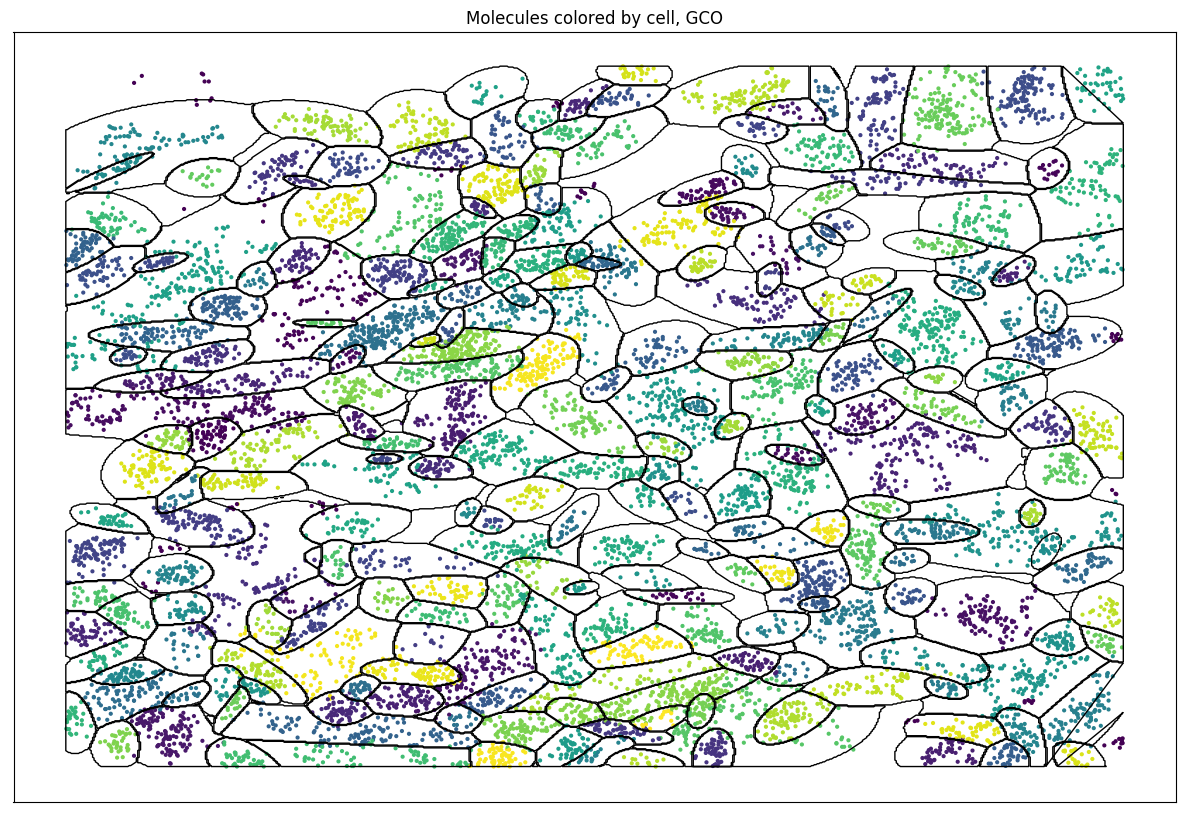
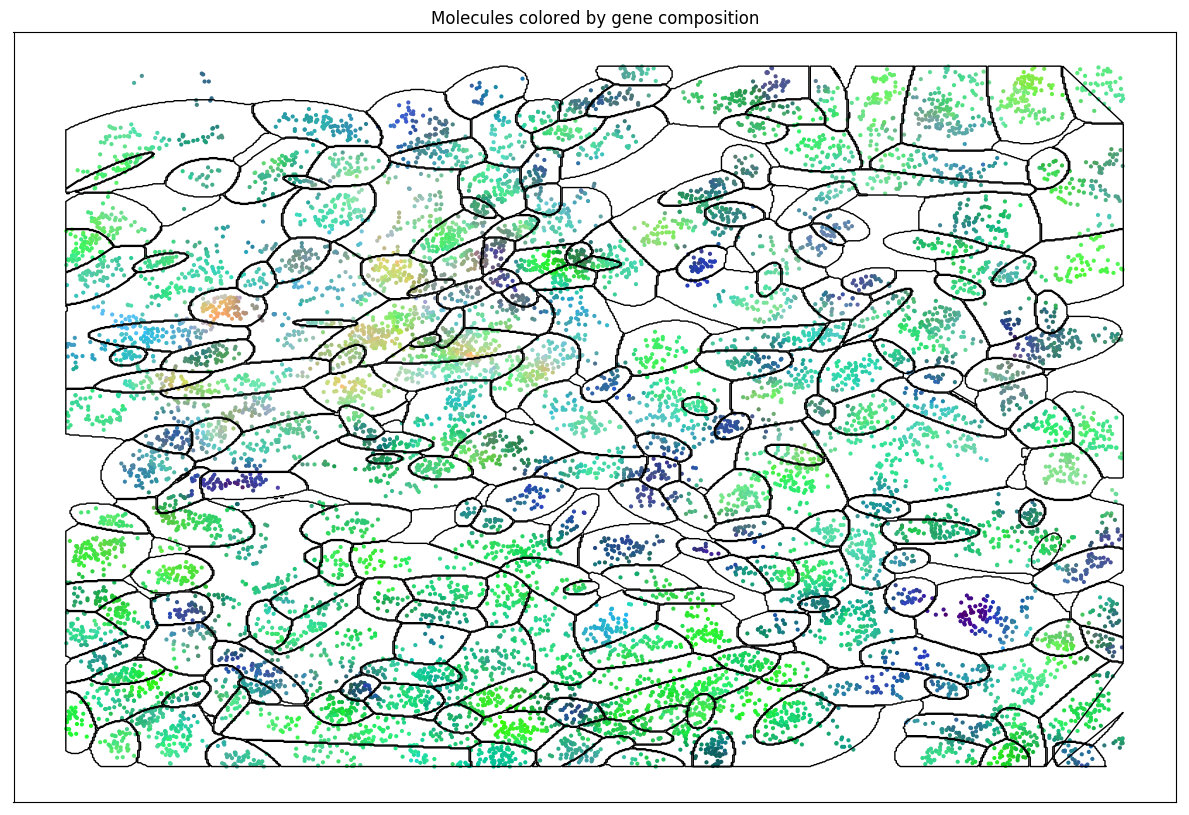
EM on spatial data
Algorithm v0.1: Stochastic EM
Expect
Sample
Maximize

- Convergence to global optimum
- Doesn't require specification of number of clusters
EM on spatial data
Algorithm v0.1: Stochastic EM
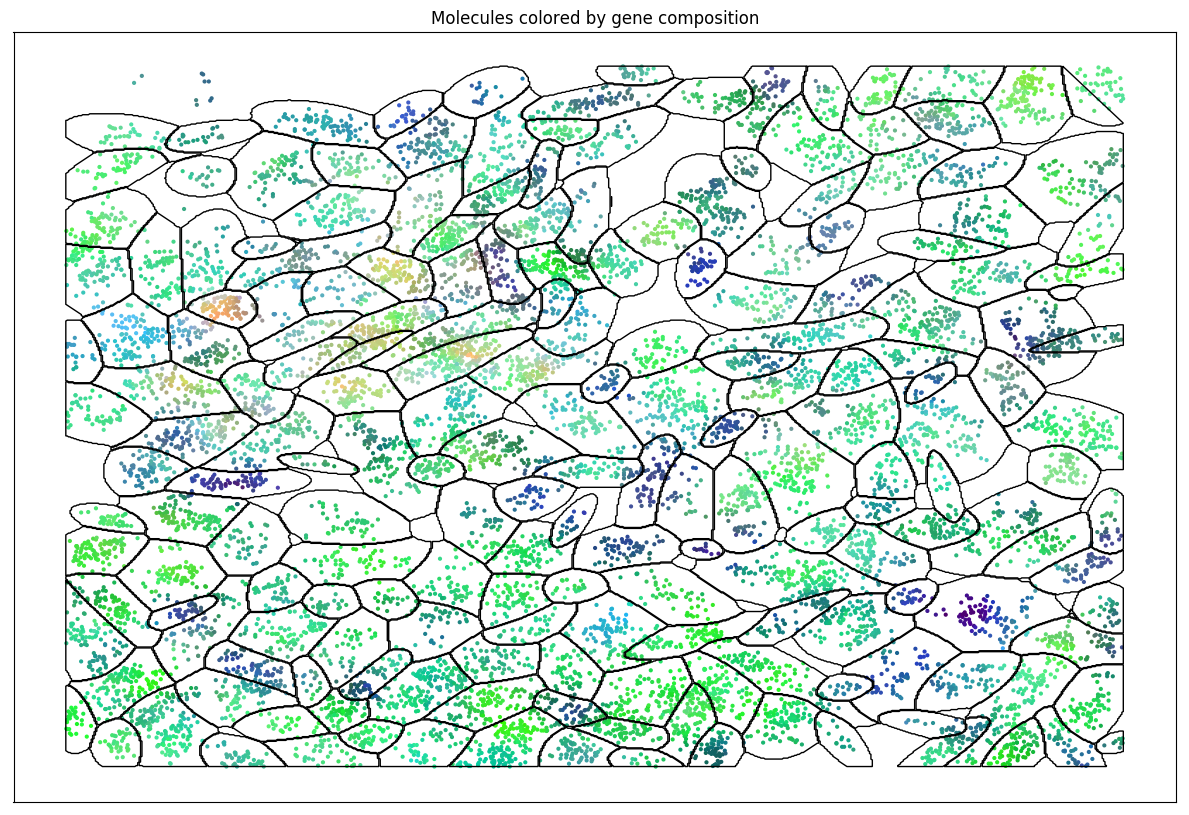
Hierarchical Bayesian Models
Composition prior
Expect
Sample
Maximize
