Case-control analysis of single-cell RNA-seq studies
Petukhov Viktor
Khodosevich Lab, BRIC
viktor.petukhov@pm.me
Computational methods for single-cell analysis of brain disorders

Co-supervisor
- Computational methods for single-cell biology
- Harvard Medical School
Peter Kharchenko
Background

Main supervisor
- Biology of neurodevelopmental disorders
- scRNA-seq
- University of Copenhagen
Konstantin Khodosevich
Structure
Overview of projects
Biological introduction
Projects: Conos
Projects: Epilepsy
Projects: Cacoa
Future directions
Slide complexity
Overview of projects
in prep.

preprint

package

published

authorship

Biology

Schizophrenia



Neuron Maturation
Macaque Vis Region


brain


Conos
Epilepsy
Cacoa






2019

2020

co-

co-


case-control

Baysor
SpaceTx




2022
spatial

Future work
Cacoa
Epilepsy
Conos
Overview
Introduction
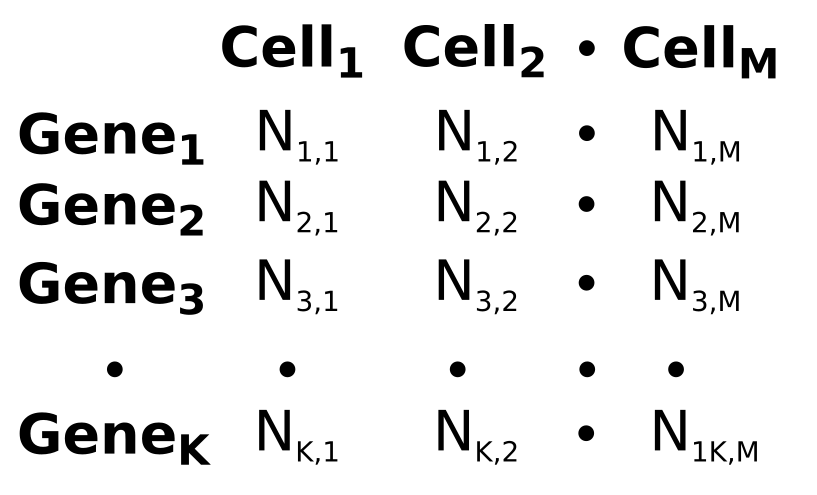
Our data: single-cell RNA-sequencing (2015)
DNA:
RNA molecules
Genes
}
Expression
levels
(3;
1;
5)

Future work
Cacoa
Epilepsy
Conos
Overview
Introduction
Genes
DNA:
(3;
1;
5)
}
Expression
levels







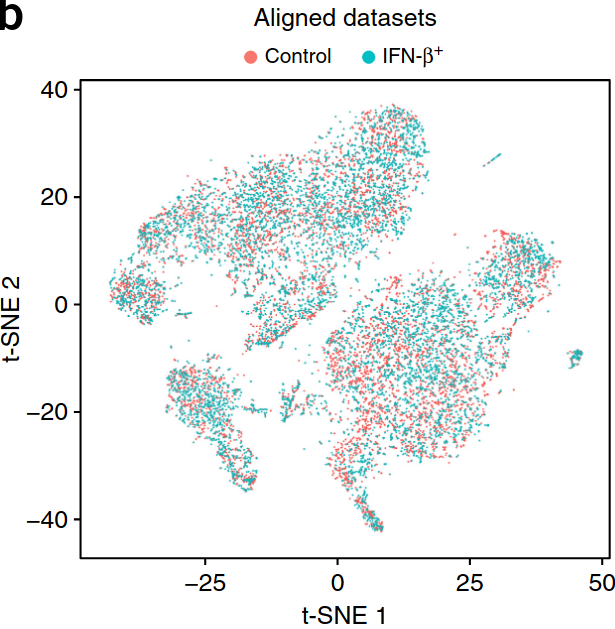
t-SNE 1
t-SNE 2
Our data: single-cell RNA-sequencing (2015)
~10k cells x 20k genes







Genes
DNA:
(3;
1;
5)
}
Expression
levels







































































































t-SNE 1
t-SNE 2
Our data: single-cell RNA-sequencing (2015)




Our data: multiple samples (2018)
Problem: multiple conditions (2020)
Control patients
Case patients










Problem: multiple conditions (2020)
Control patients
Case patients


















































Cells
























Cell
types
Problem: multiple conditions (2020)
Control patients
Case patients










Cell
types
Cells
































































Cells
Genes


Cells
Genes


Cells
Genes


Cells
Genes


Cells
Genes


Cells
Genes


Cells
Genes


Cells
Genes


How to compare?

Conos
Epilepsy
Cacoa



Conos
Epilepsy
Cacoa
Future work
Cacoa
Epilepsy
Conos
Overview
Introduction
Problem: batch-effect on multiple samples


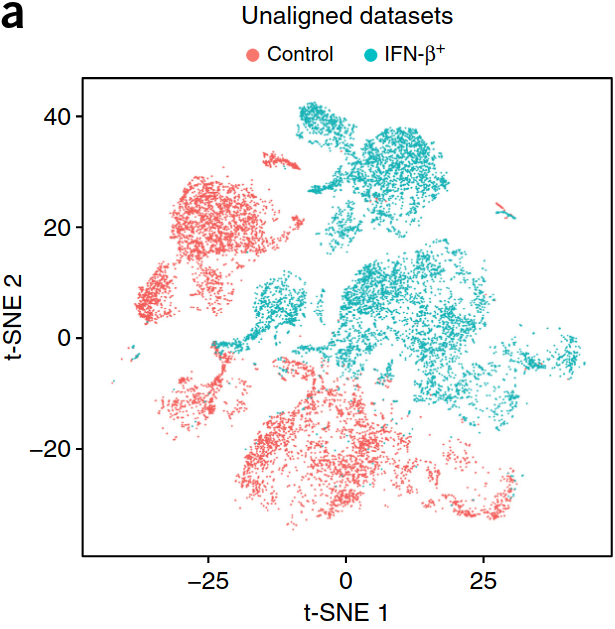
Problem: batch-effect on multiple samples

- Individuals
- Tissues
- Species
Potential sources of differences:
- Treatment
- Environment
- Disease
- Protocol
- Lab
- ...


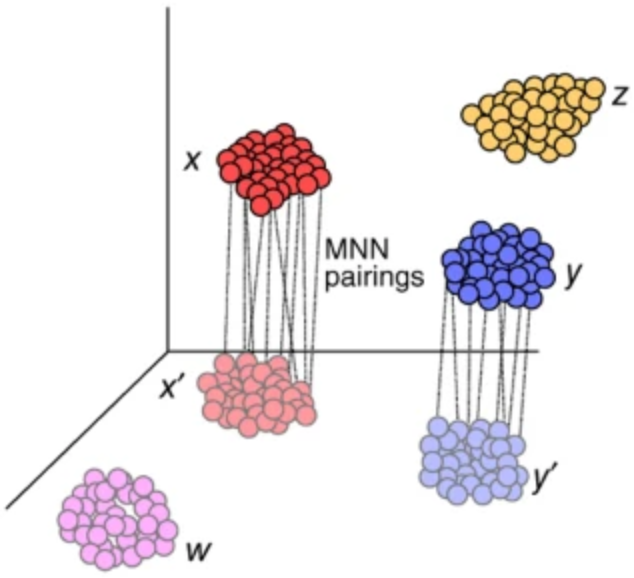
Conos: clustering of networks of samples
Goal: develop a framework to analyze heterogeneous collections of samples without suffering from batch-effect
State of the art (Seurat, mnn): create a joint 'batch-corrected' expression space
Conos: clustering of networks of samples
Goal: develop a framework to analyze heterogeneous collections of samples without suffering from batch-effect
State of the art (Seurat, mnn): create a joint 'batch-corrected' expression space
Problem: joint expression space removes important variation and distorts distributions
Conos: clustering of networks of samples
Problem: joint expression space removes important variation and distorts distributions
Solution: work with a joint topology (graph), not expression space
Conos: clustering of networks of samples
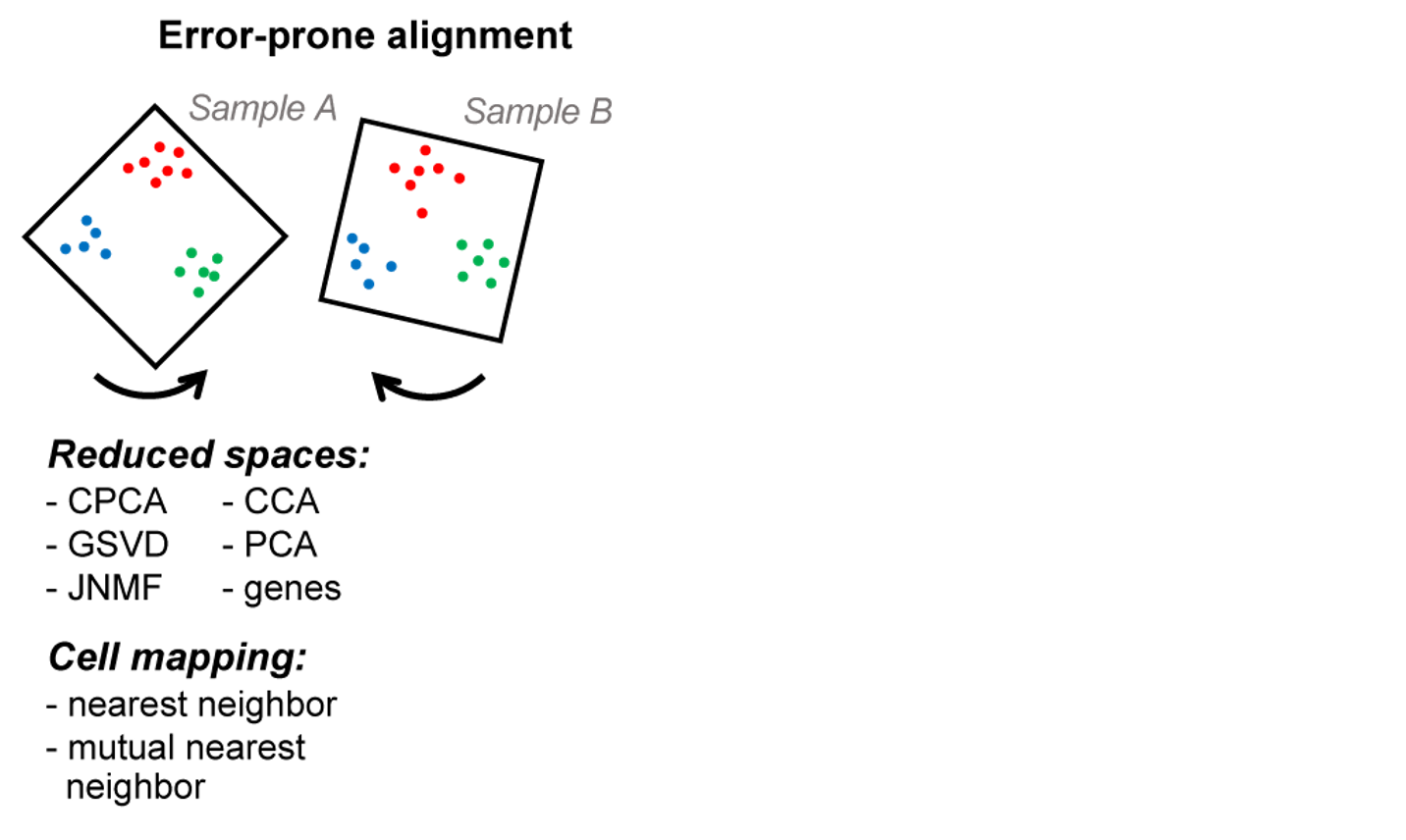
Pairwise alignment
Conos: clustering of networks of samples
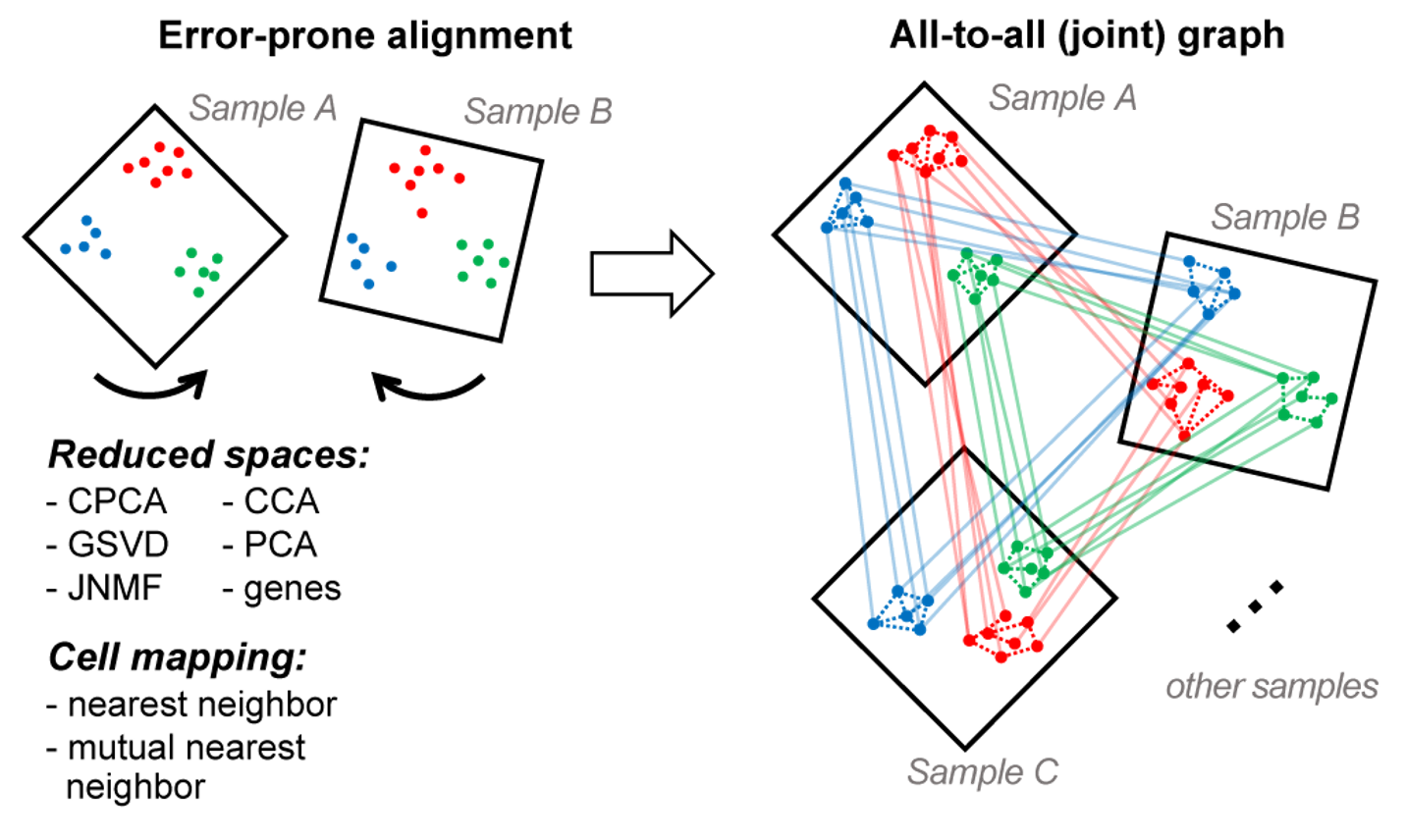
Joint graph

Conos: clustering of networks of samples

Graph analysis
- Clustering
- 2D embedding
- Propagating information between samples
- Other graph processing
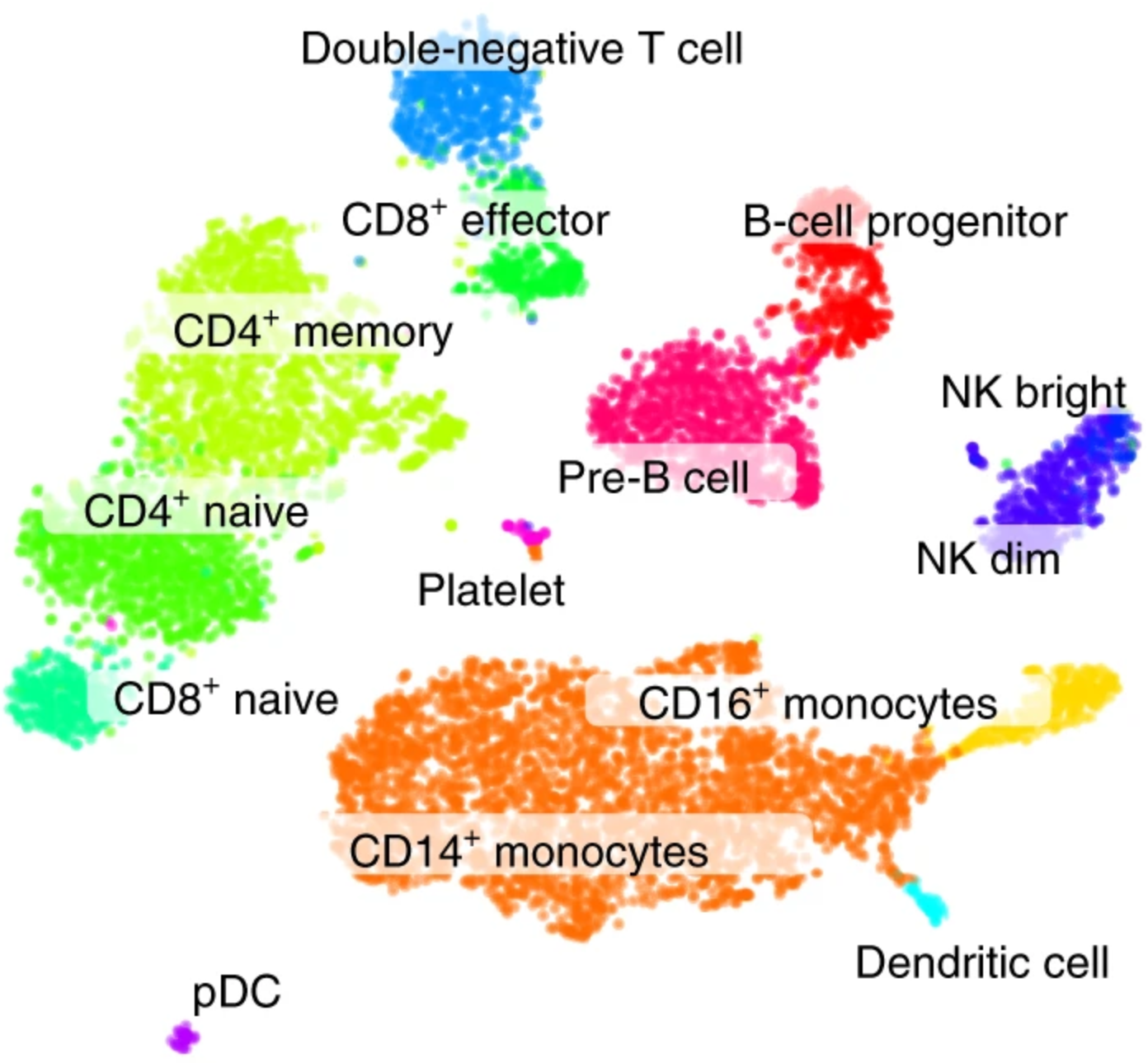
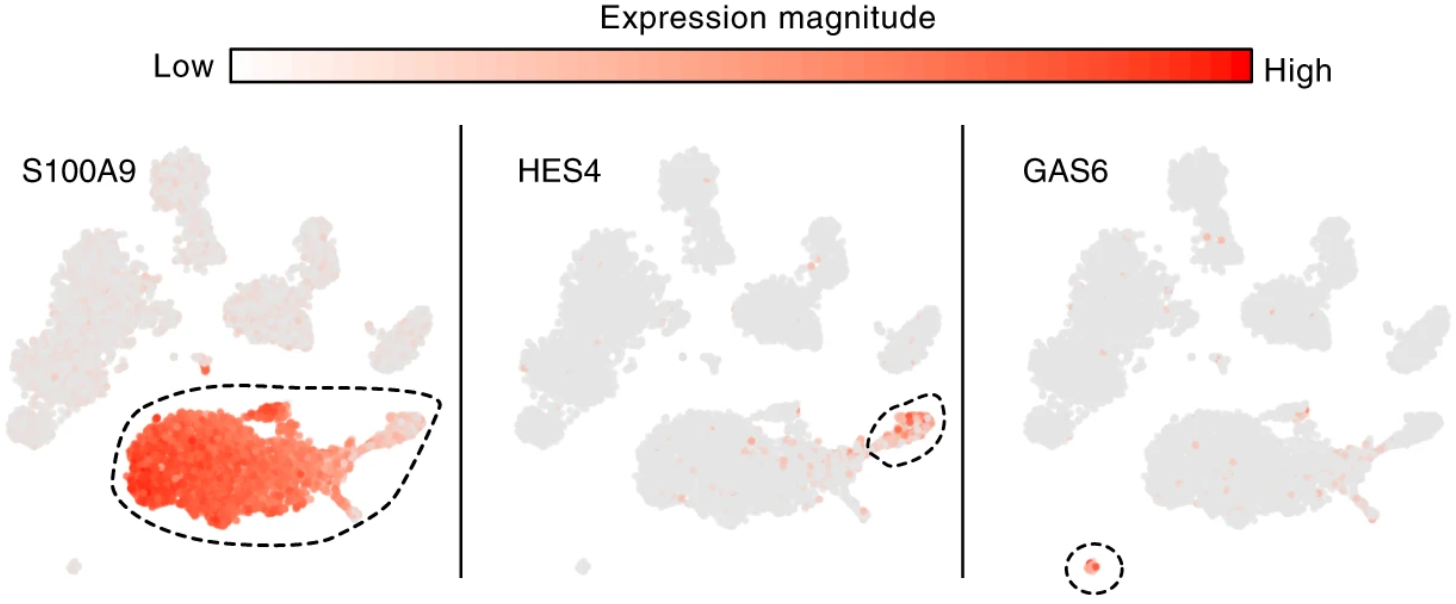
Conos: results
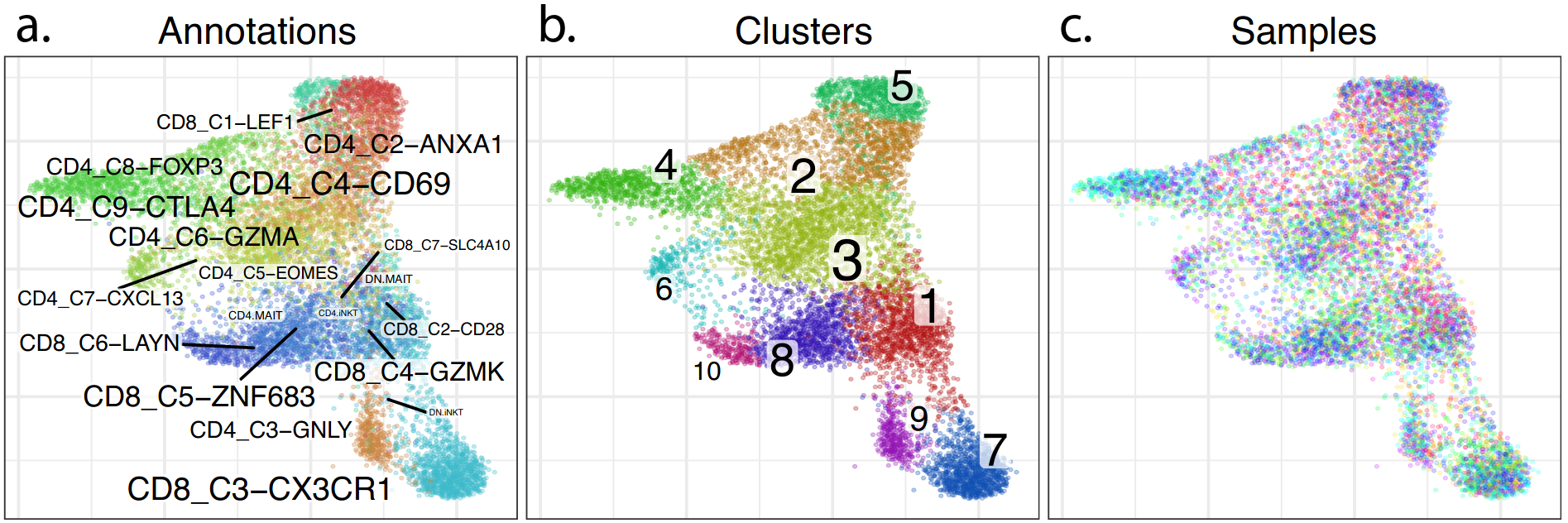
Annotation

Samples

Clusters
Conos: benchmarking
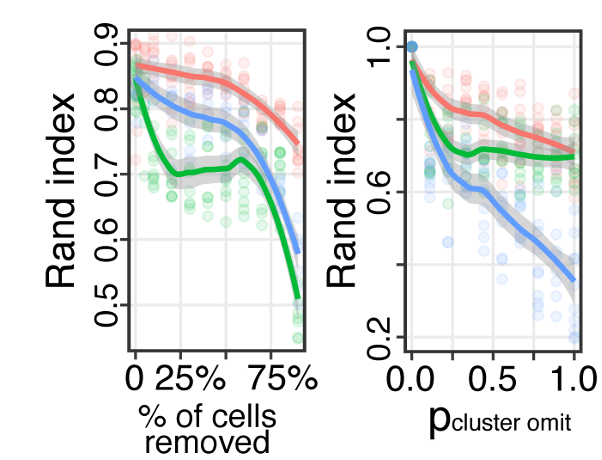

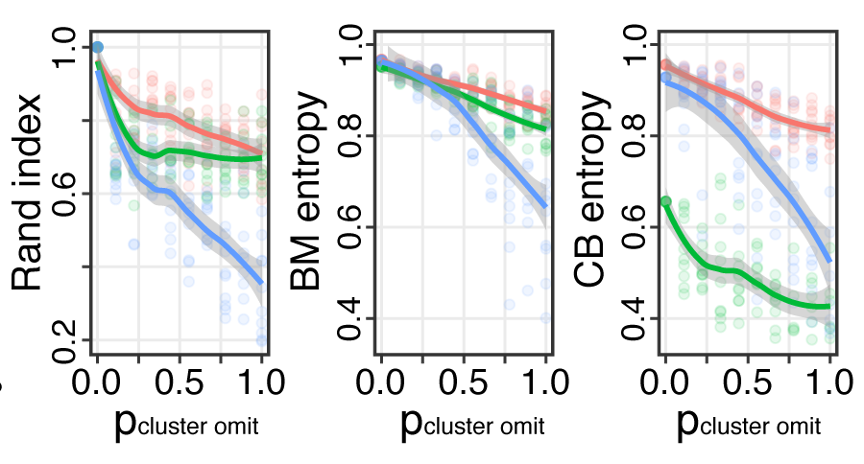

Projects



Conos
Epilepsy
Cacoa
Projects



Conos
Epilepsy
Cacoa
Future work
Cacoa
Epilepsy
Conos
Overview
Introduction





















Our data: multiple samples (2018)
Problem: multiple conditions (2020)
Healthy patients
Epilepsy patients










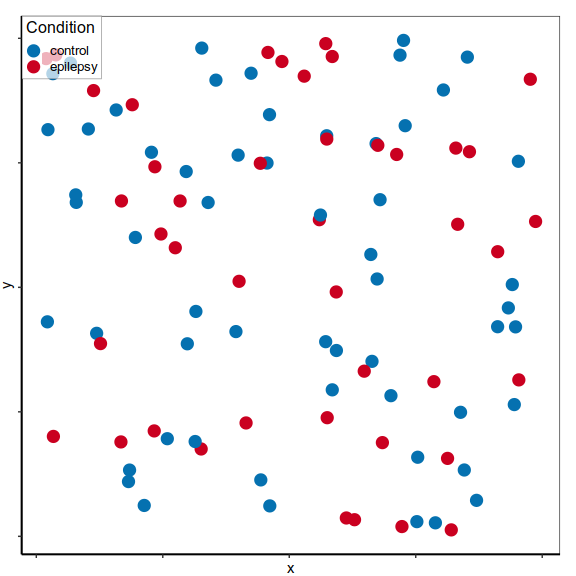
scRNA-seq to study Temporal Lobe Epilepsy
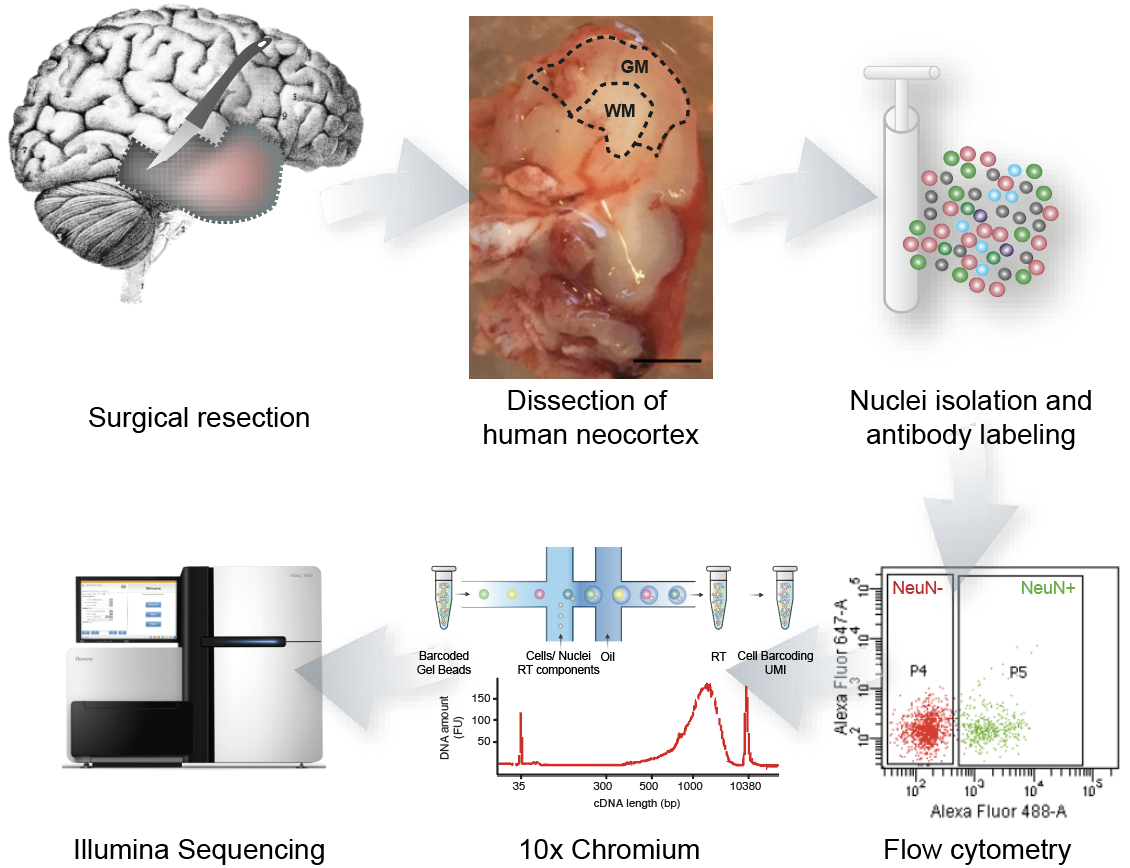
- Temporal Cortex
- 9 control vs 10 epilepsy samples
- 117k nuclei
scRNA-seq to study Temporal Lobe Epilepsy
Goal: investigate molecular mechanisms of Temporal Lobe Epilepsy using single-cell data
State of the art: no scRNA-seq studies were done on Epilepsy data
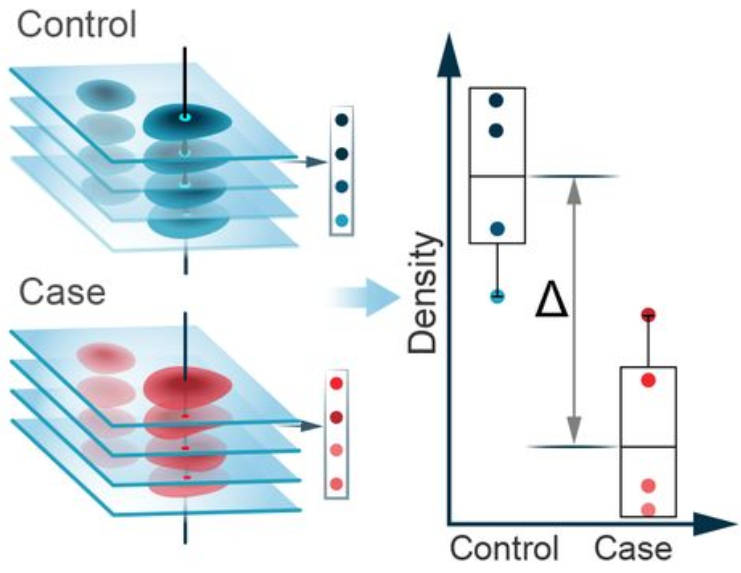
Problem: how to measure changes between conditions?
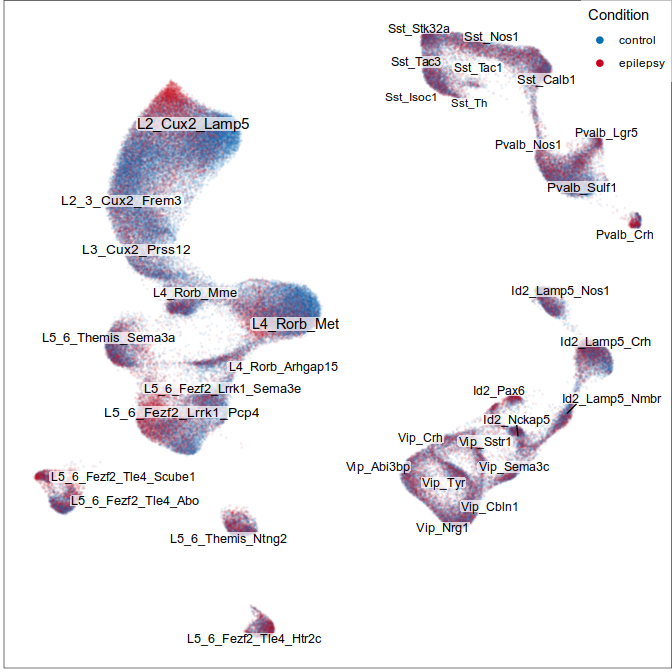
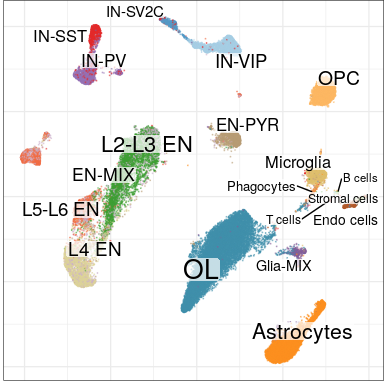
Step 1: align and annotate
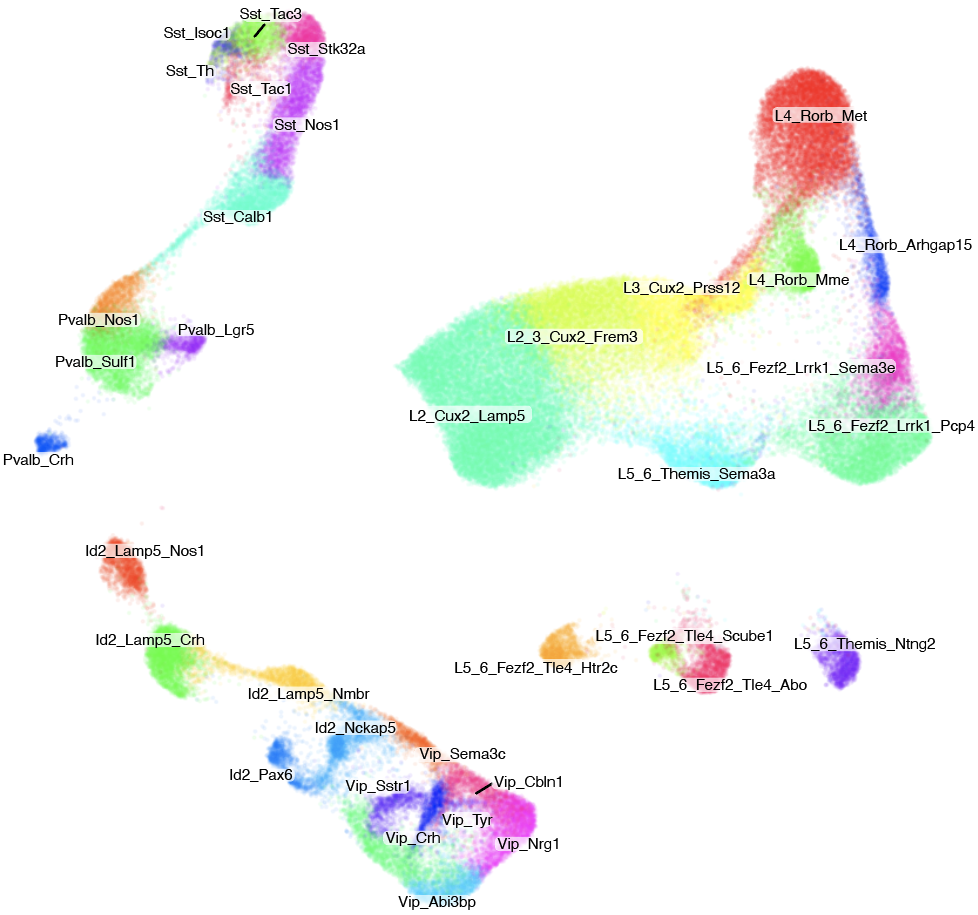
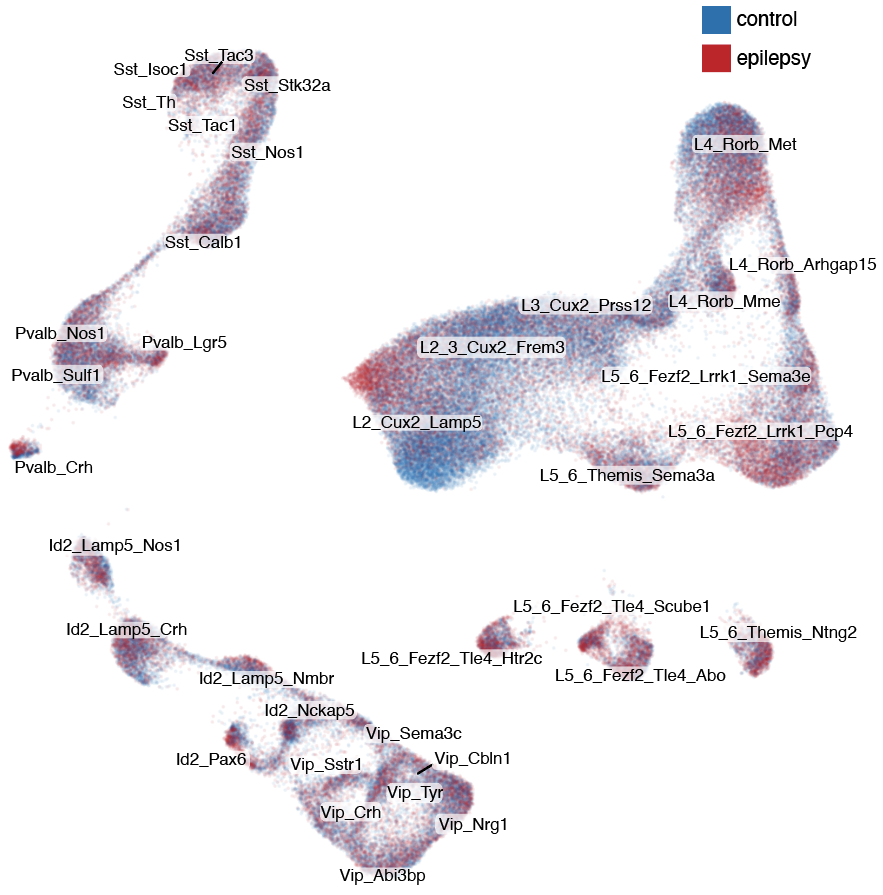
Step 2: identification of changed types
Gene expression analysis

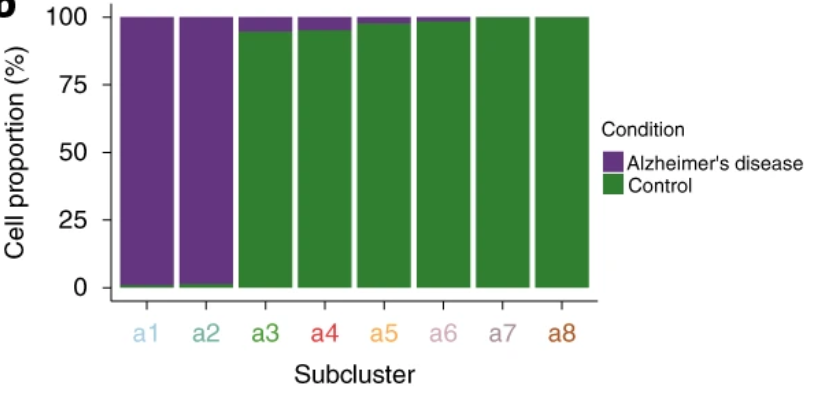
Compositional analysis
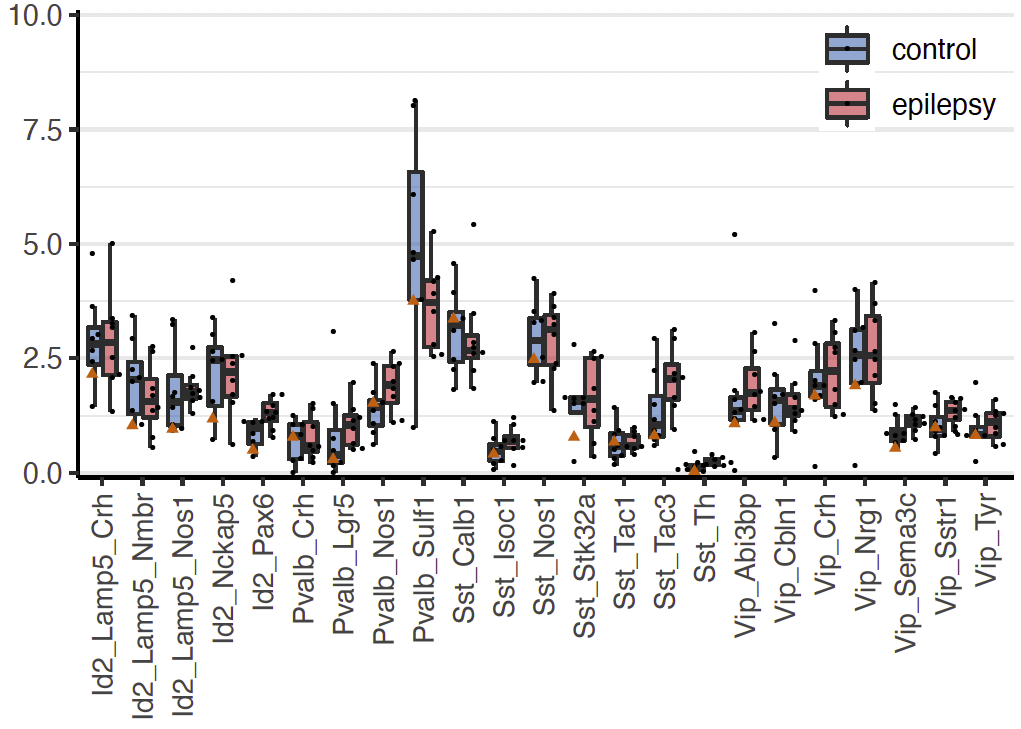
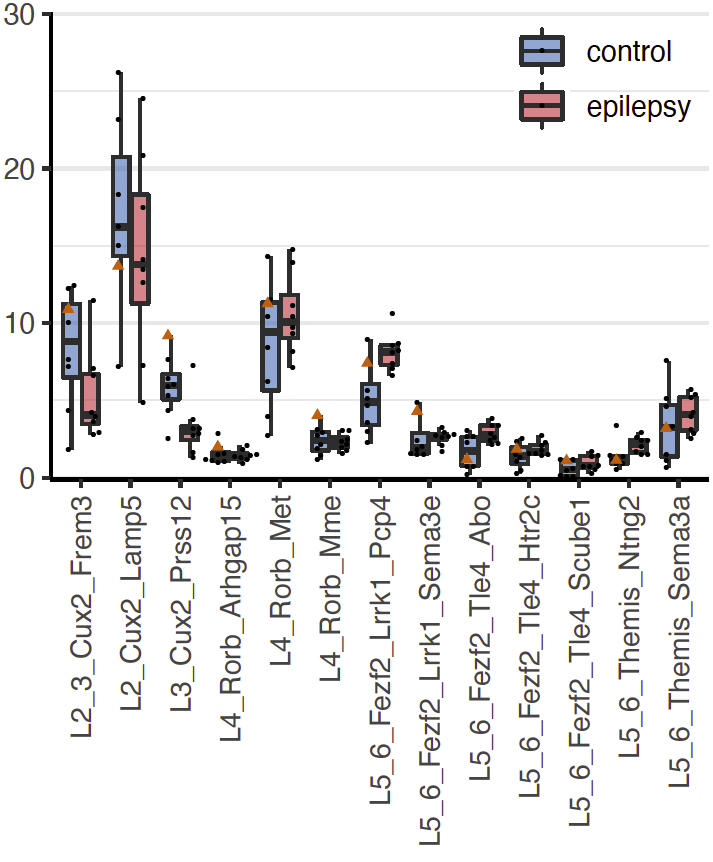
%of nuclei
Step 2.1: compositional analysis
%of nuclei

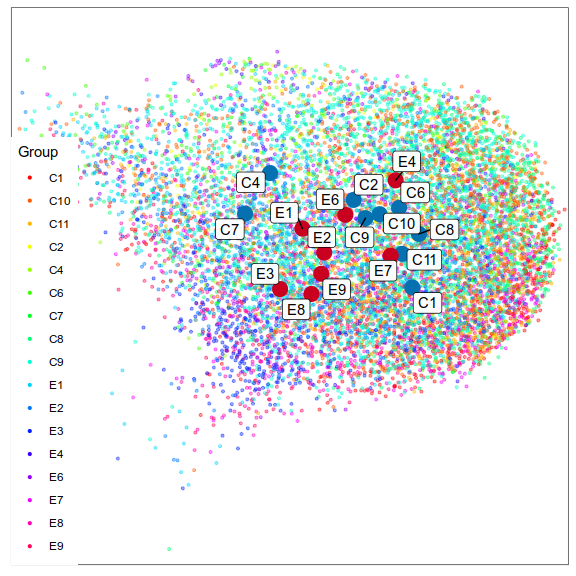
Step 2.2: expression similarity
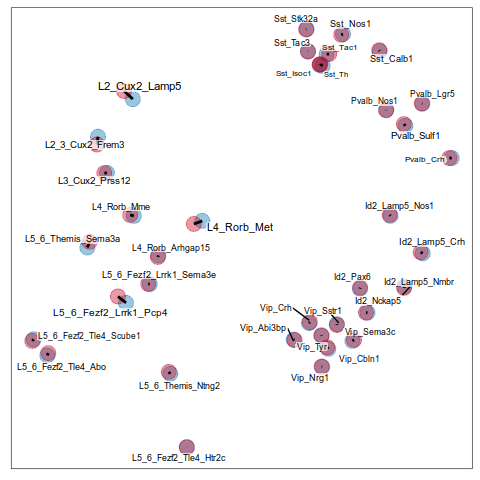
Step 2.2: expression similarity


Step 2.2: expression similarity
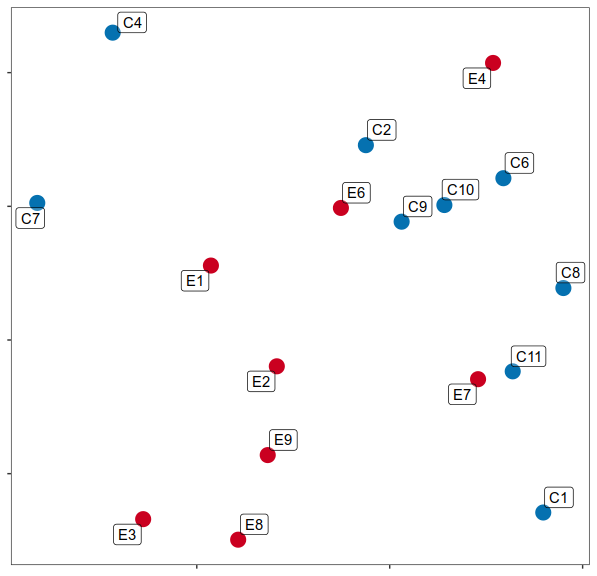
Step 2.2: expression similarity

Step 2.2: expression similarity
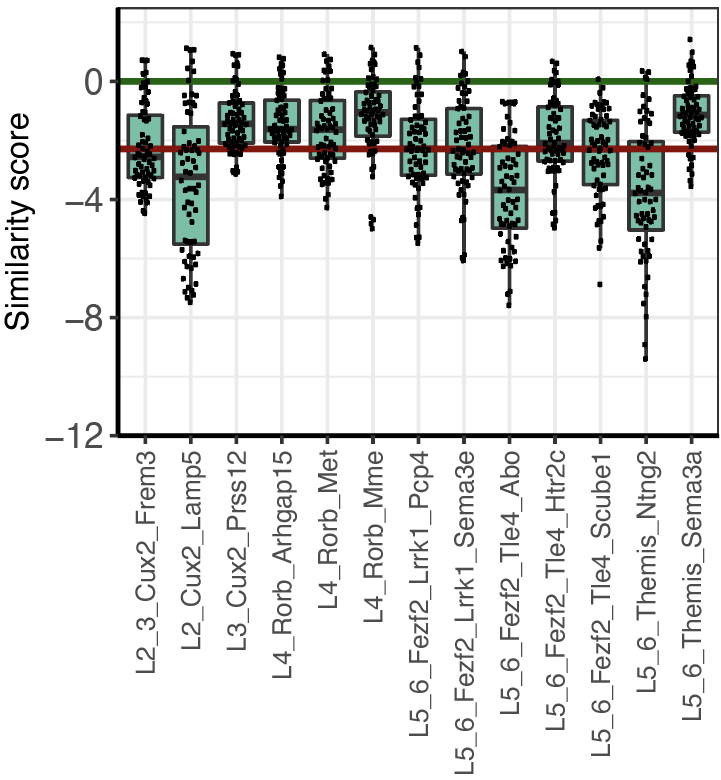
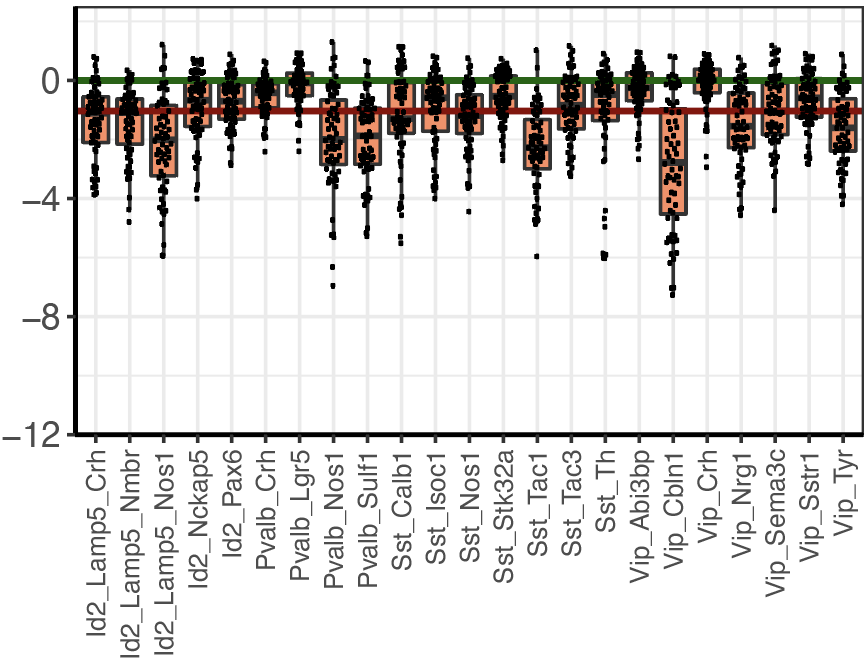
Step 2.3: cell type ranking
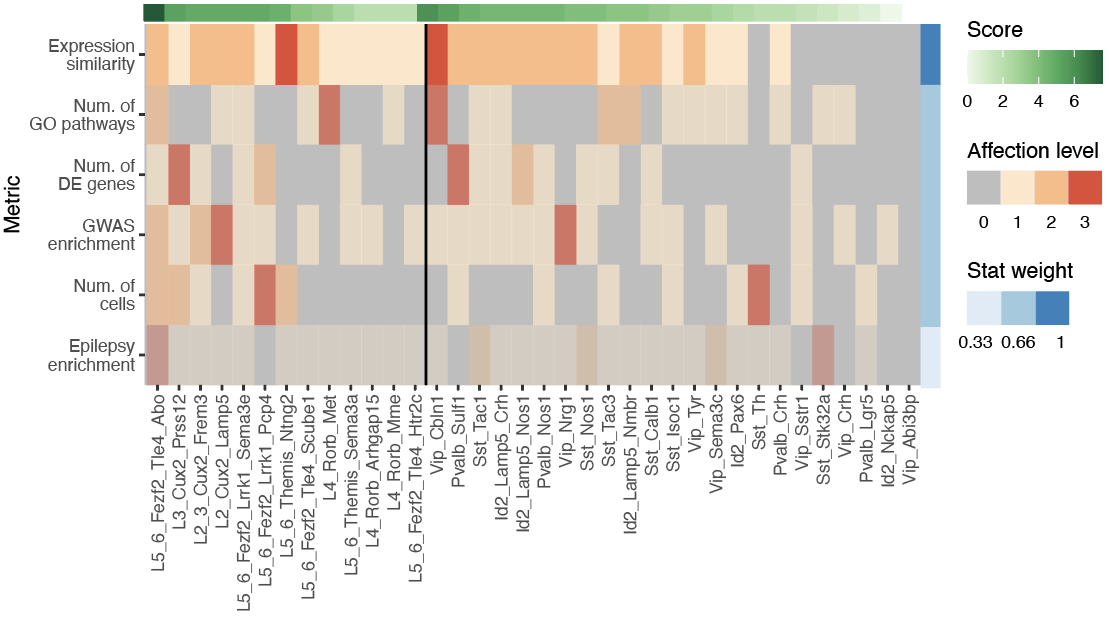
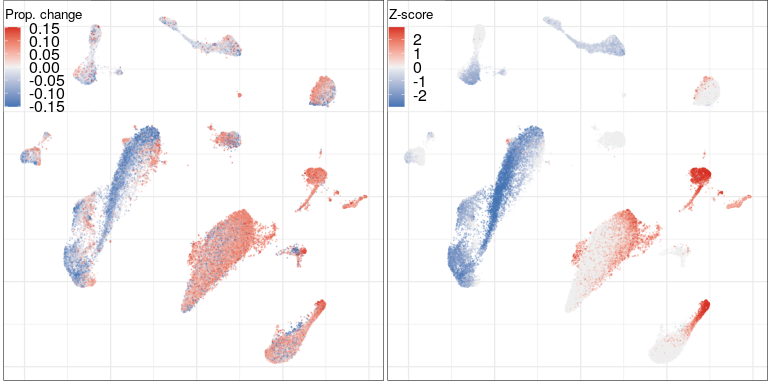
Step 3.1: global structure of changes
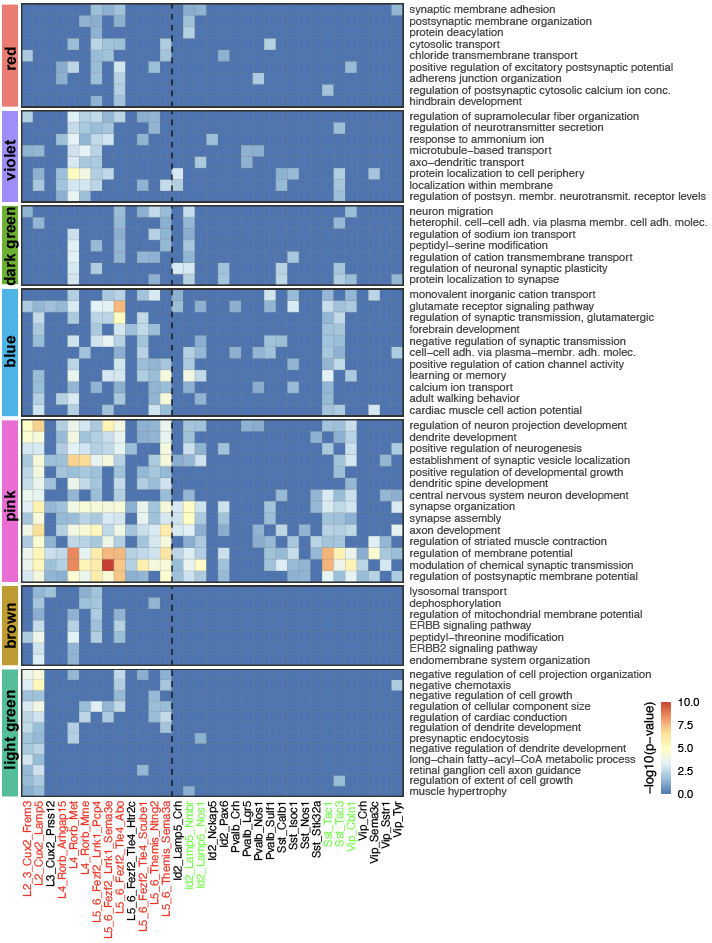


developmental processes, neural circuit re-organization and neurotransmission
ion transport and glutamate signaling
protein transport to axons/dendrites
cell adhesion, ion transport and synaptic plasticity
regulation of neuronal morphogenesis
Step 3.2: local structure of changes


Step 3.2: local structure of changes


Control

Epilepsy
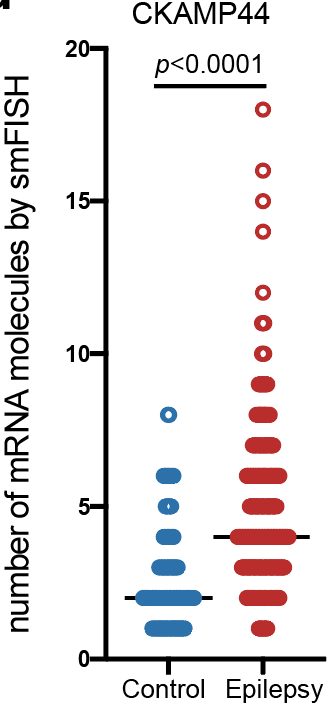
Step 3.2: local structure of changes

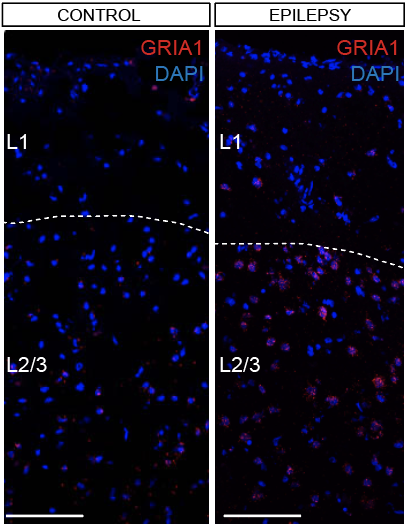
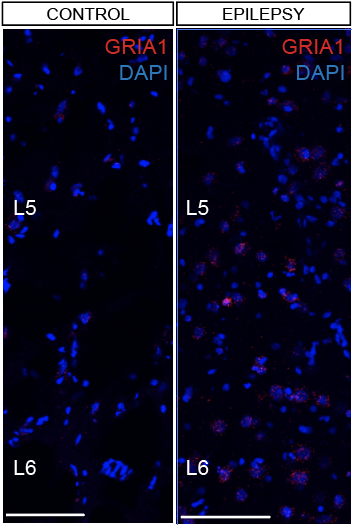
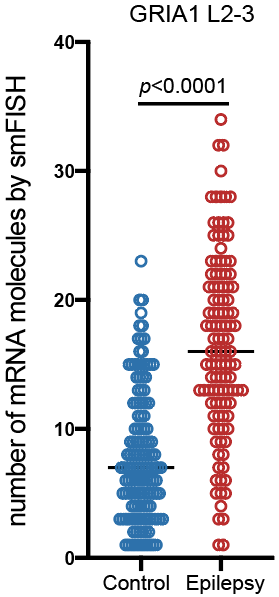
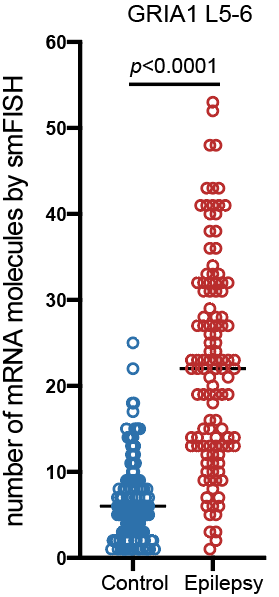
Step 3.2: local structure of changes

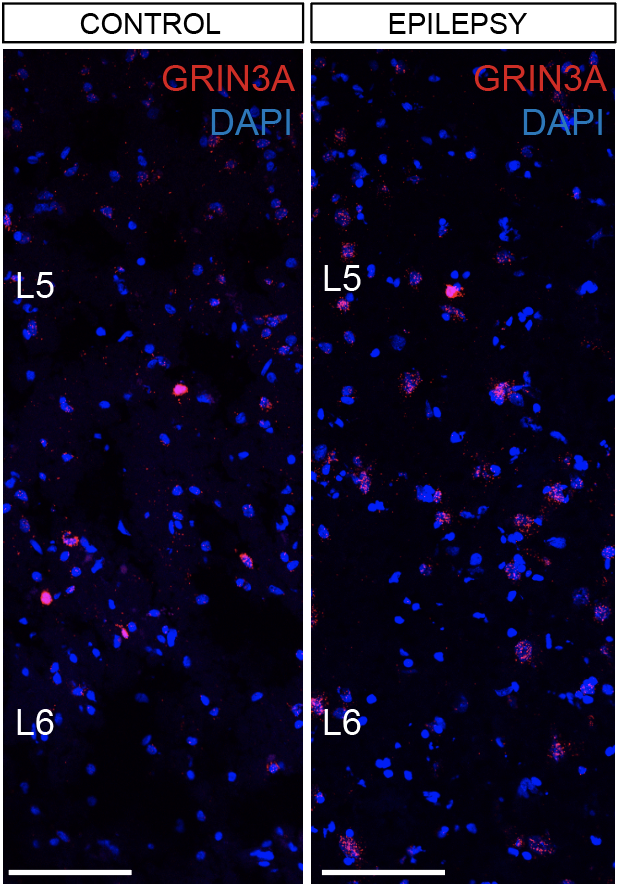
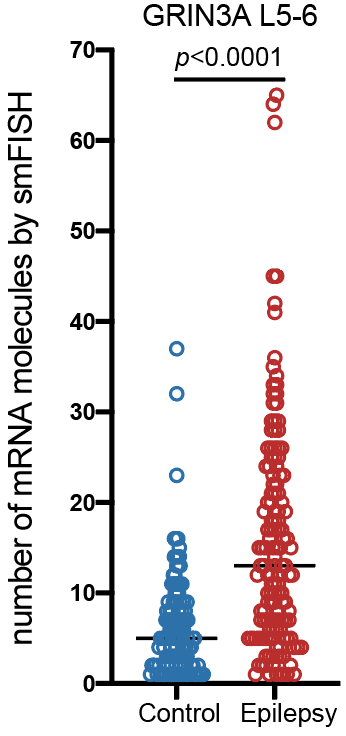

Projects



Conos
Epilepsy
Cacoa
Projects



Conos
Epilepsy
Cacoa
Future work
Cacoa
Epilepsy
Conos
Overview
Introduction
Goal: develop a comprehensive set of methods for analysis of scRNA-seq case-control experiments
State of the art: no methods were published when we started, several competitors exist now
Problem: how to measure changes between conditions?
Case-control analysis of single-cell RNA-seq studies
Case-control analysis of single-cell RNA-seq studies
Compositional analysis


Gene expression analysis

Case-control analysis of single-cell RNA-seq studies



Case-control analysis of single-cell RNA-seq studies
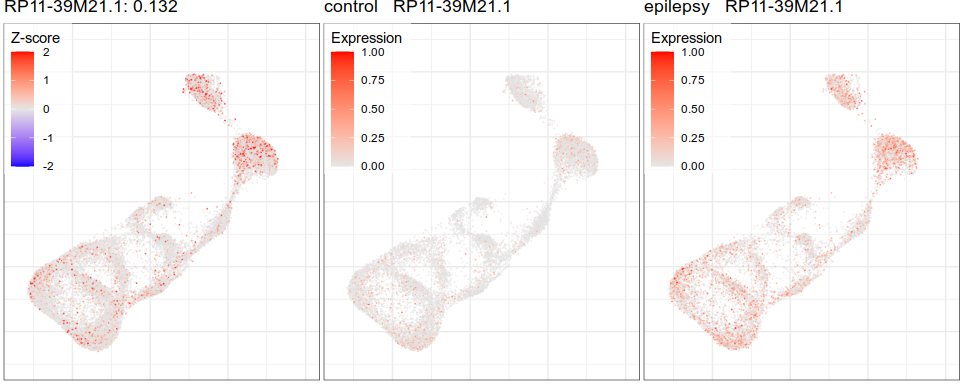
Gene expression analysis

Compositional analysis


Cluster-based
Cluster-free
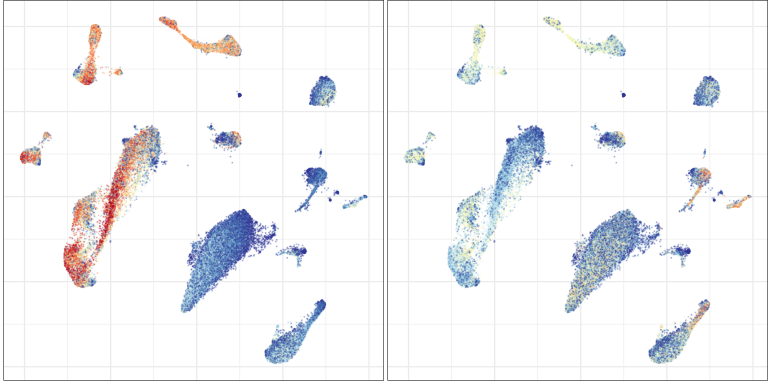
Control
Multiple sclerosis
Compositional analysis: cluster-based


Compositional analysis: cluster-based
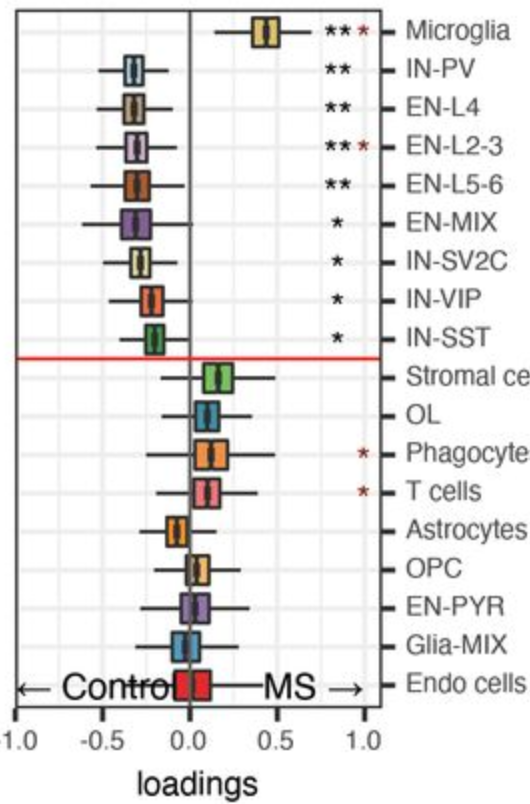
*: CoDA significance
*: proportion significance
Compositional analysis: cluster-based
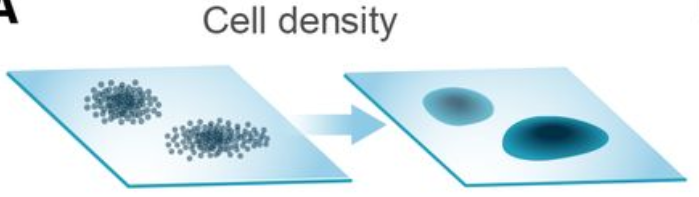
Compositional analysis: cluster-based


Control
Multiple sclerosis
Compositional analysis: cluster-based
Effect size
Significance
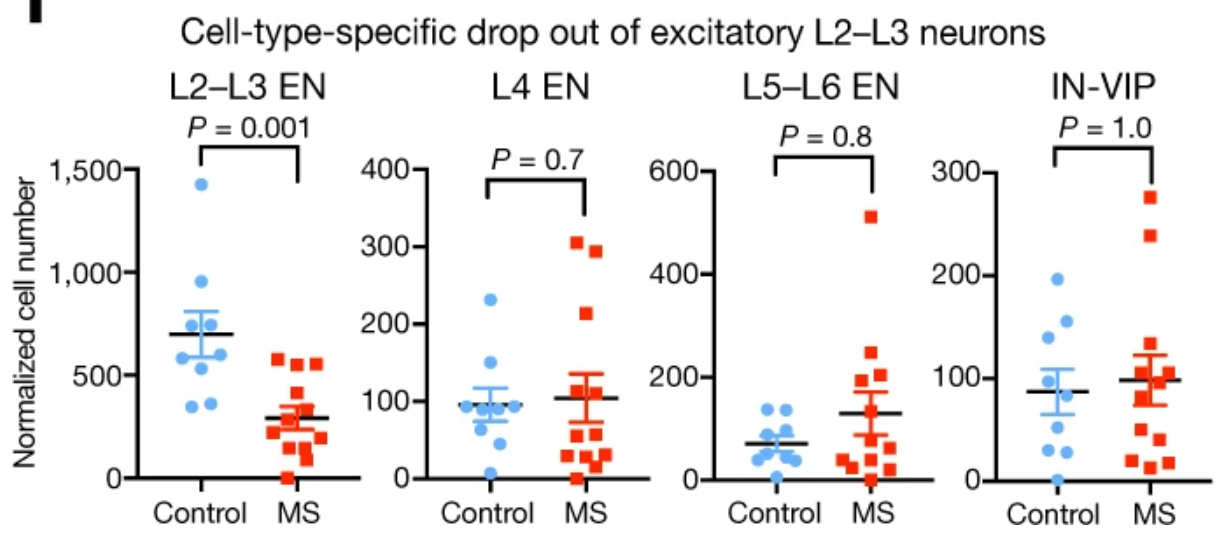
Expression analysis: cluster-based

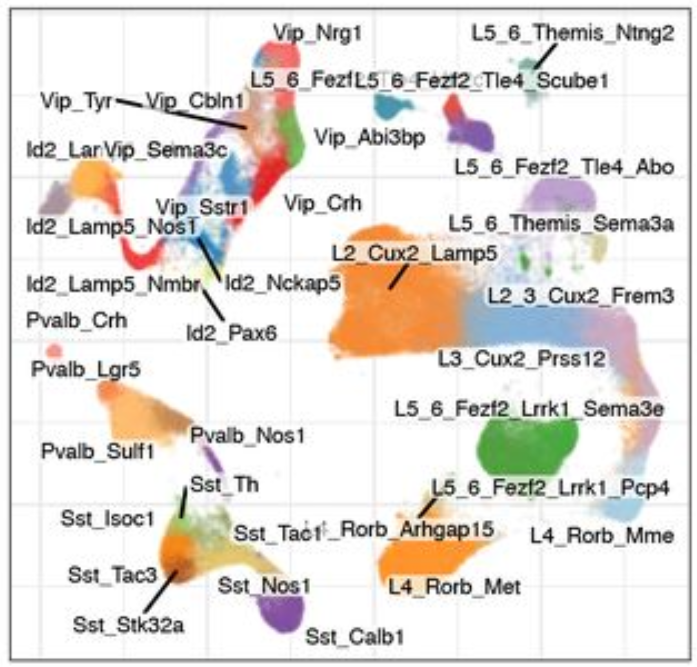
EN L2-L3
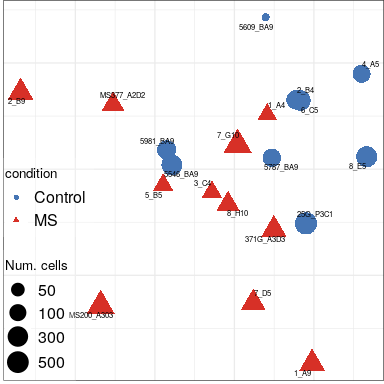
Expression analysis: sample structure

EN L2-L3
EN L2-L3
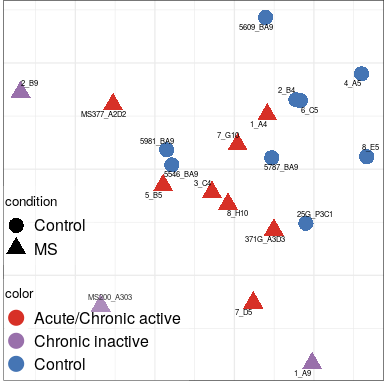
Expression analysis: sample structure
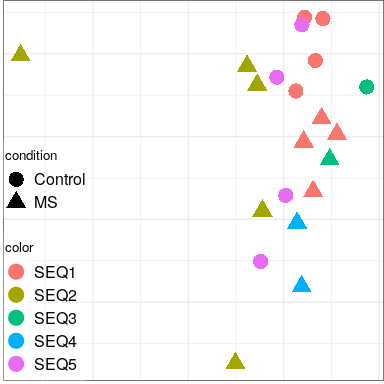
Color by batch
Aggregated across all cell types
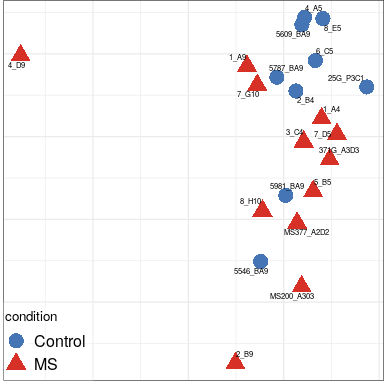
Expression analysis: sample structure
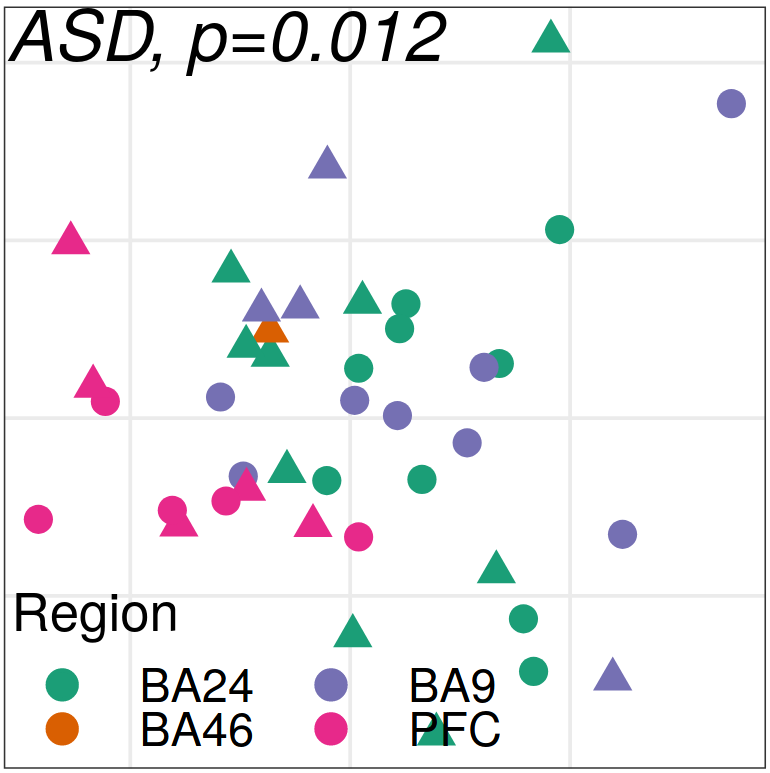
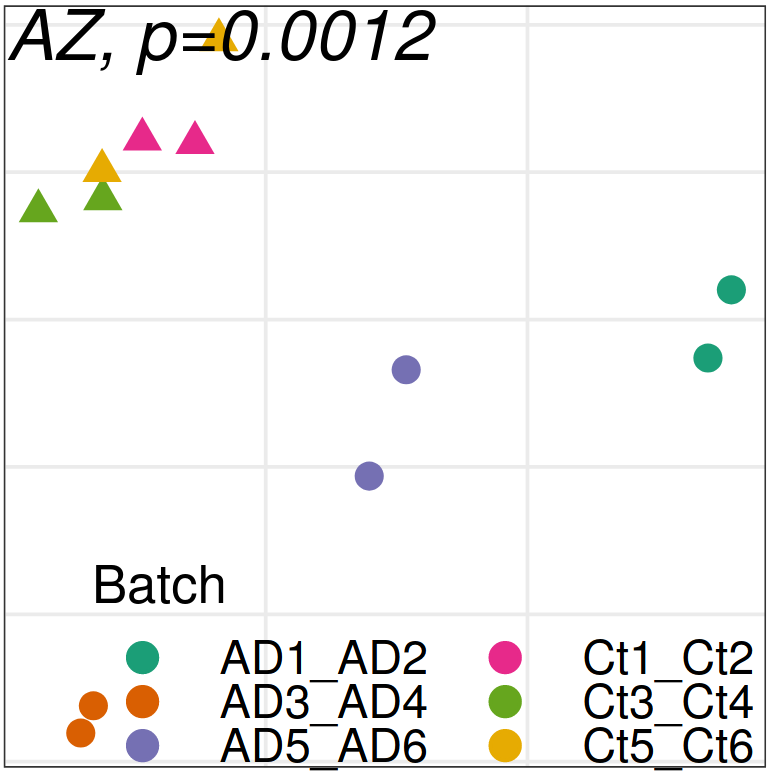
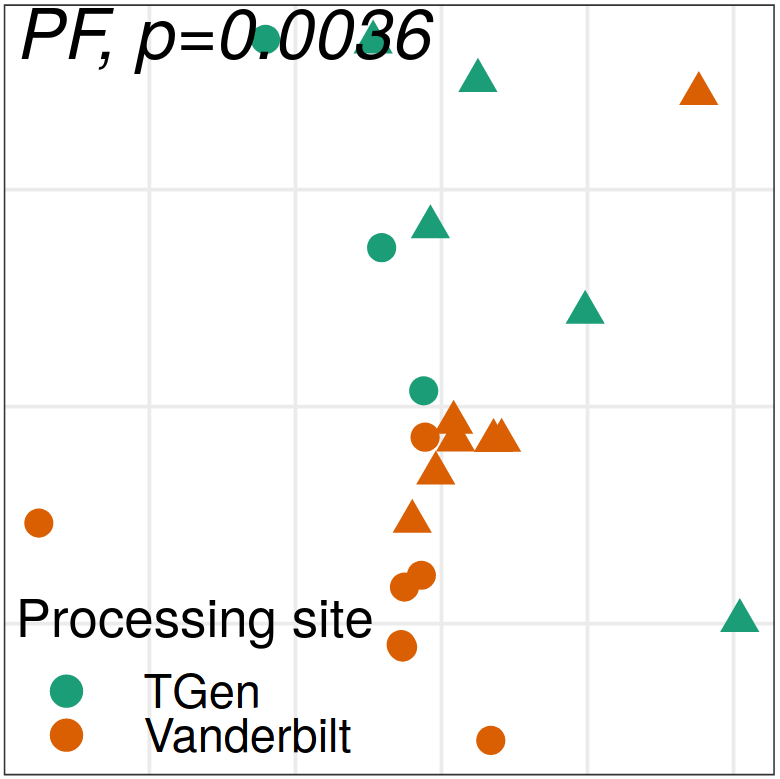
Expression analysis: expression distance
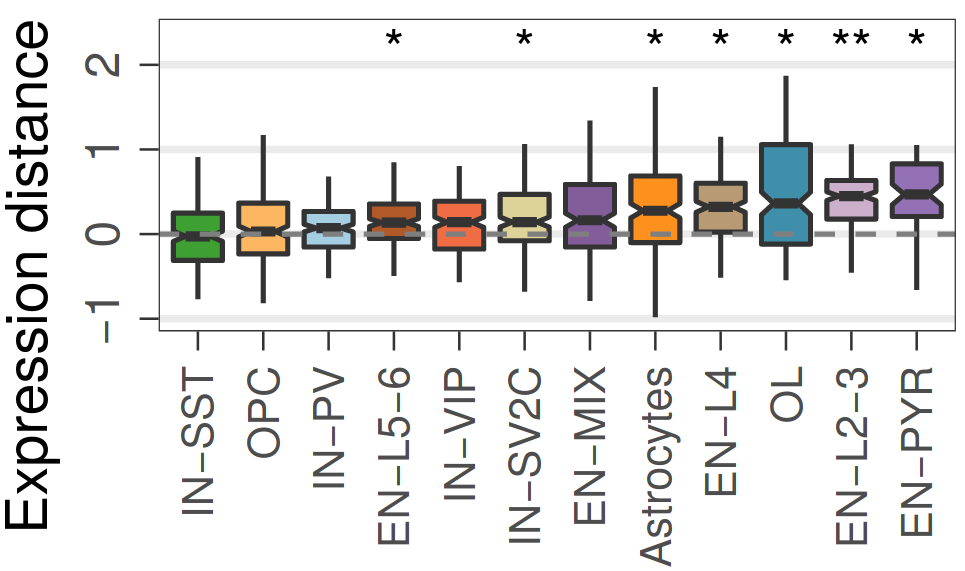
separation
EN L2-L3

Expression analysis: cluster-free

Expression analysis: cluster-free


Expression analysis: cluster-free
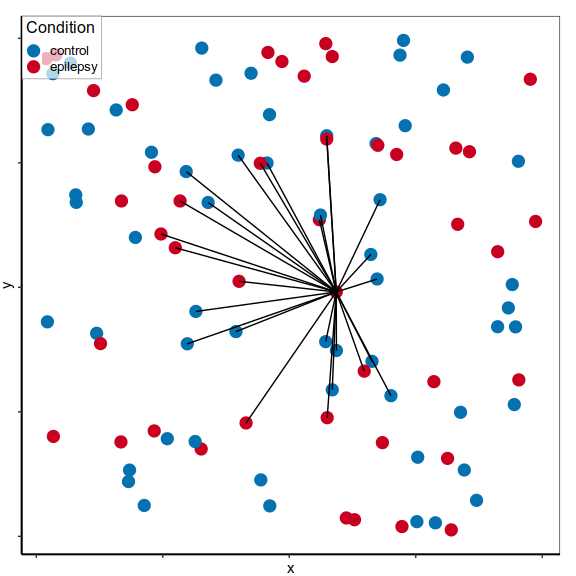
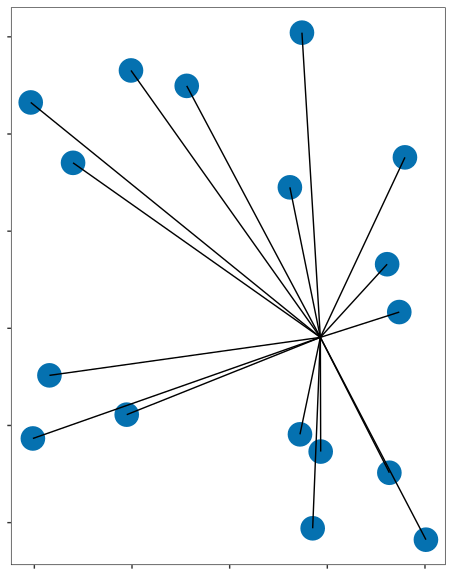
control
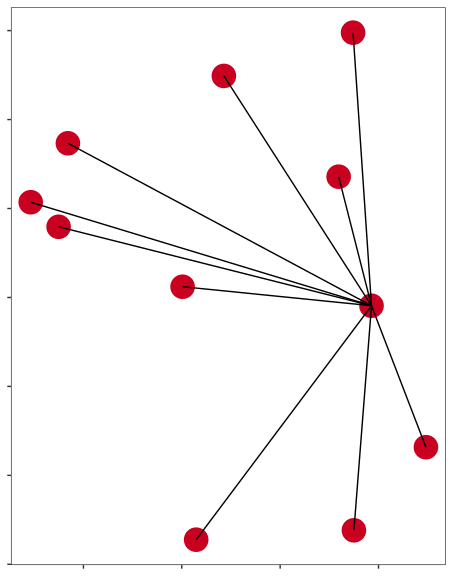
epilepsy
Expression analysis: cluster-free




control

epilepsy
Compare
Expression analysis: cluster-free shifts
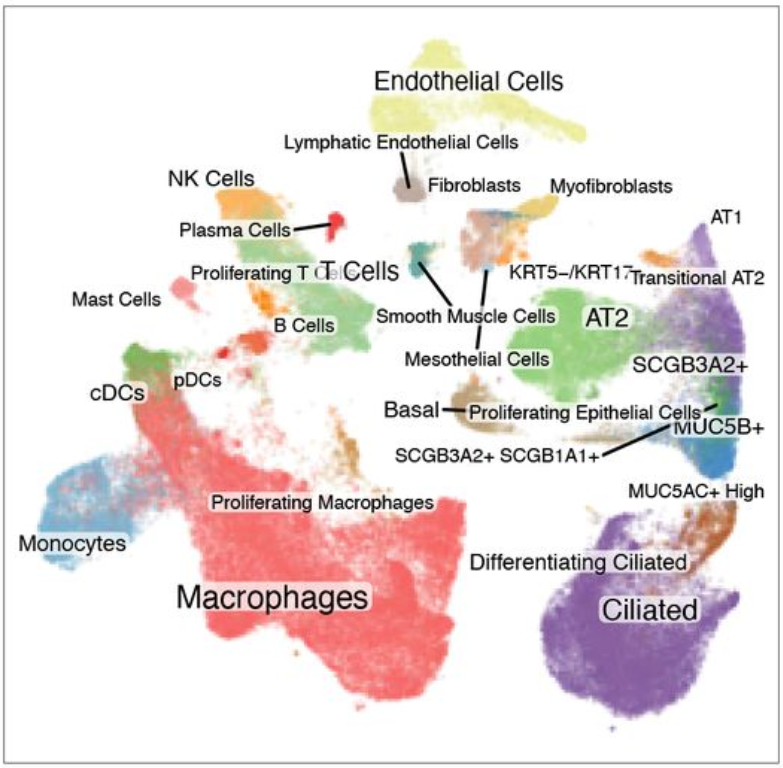
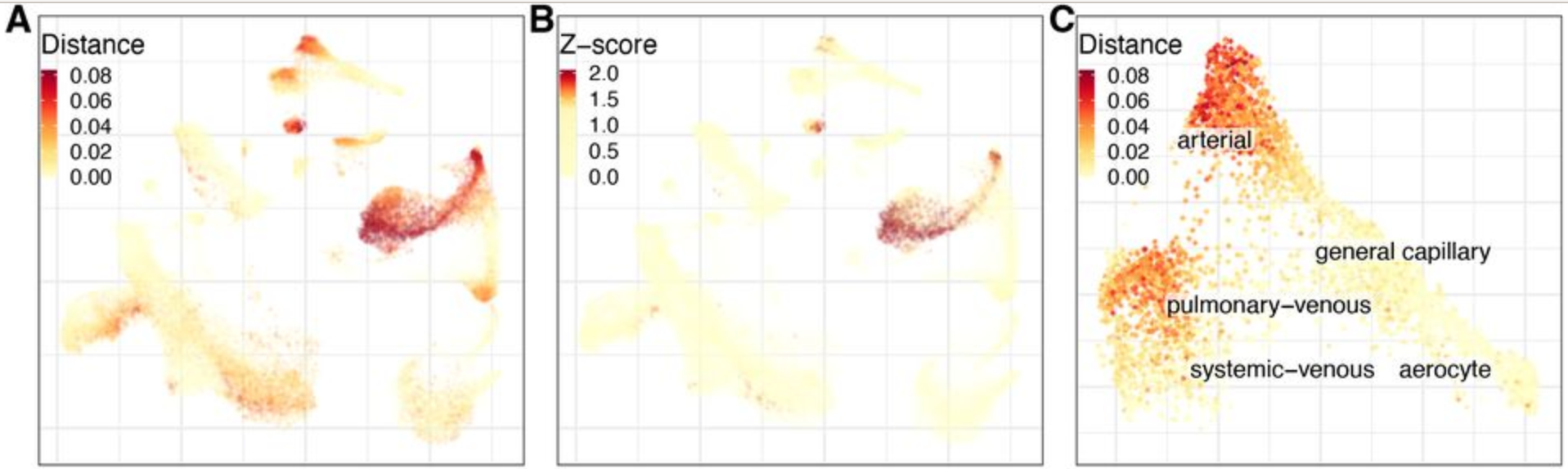


Expression analysis: cluster-free DE
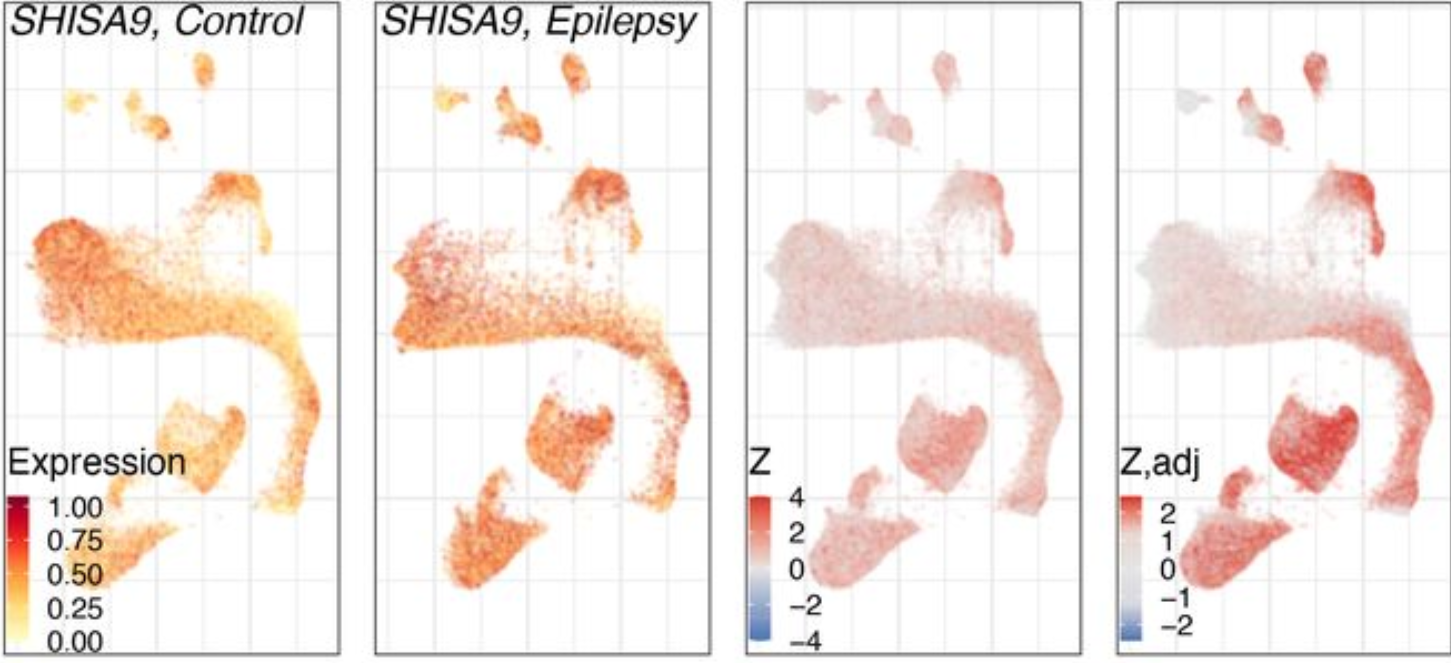
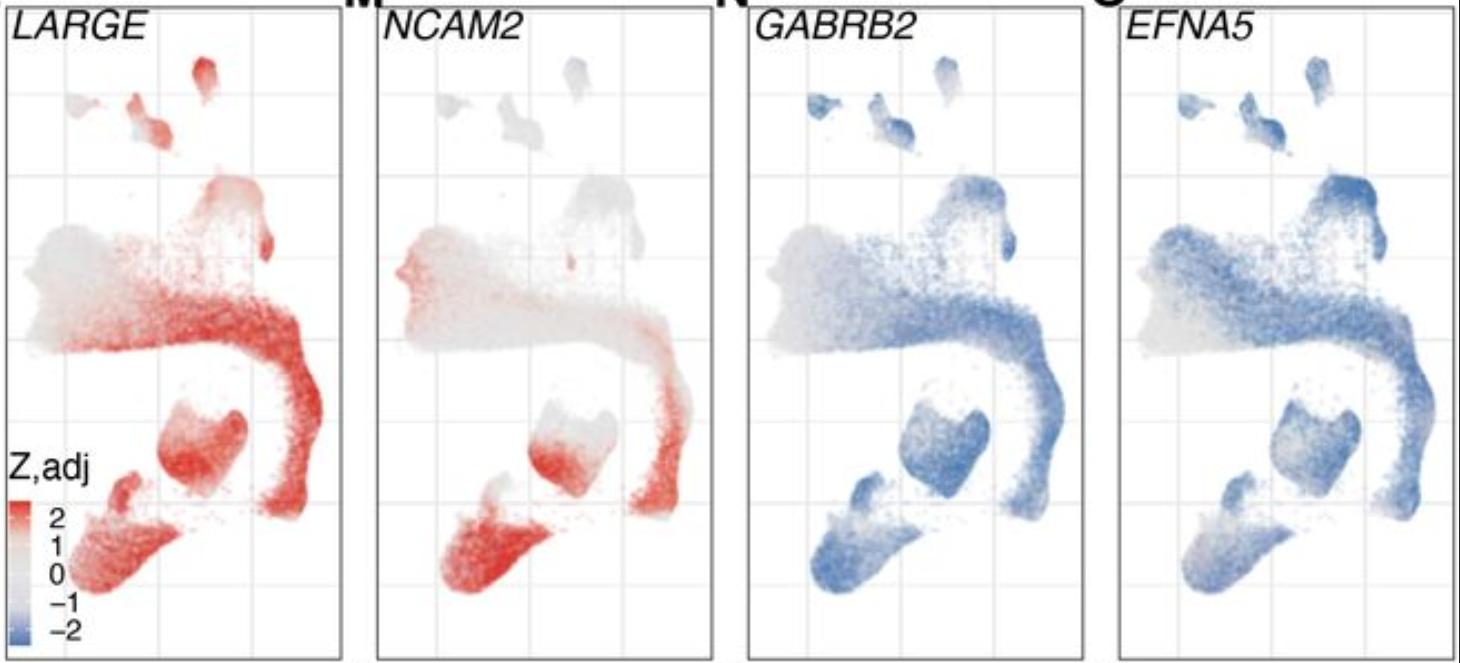


Expression analysis: cluster-free DE
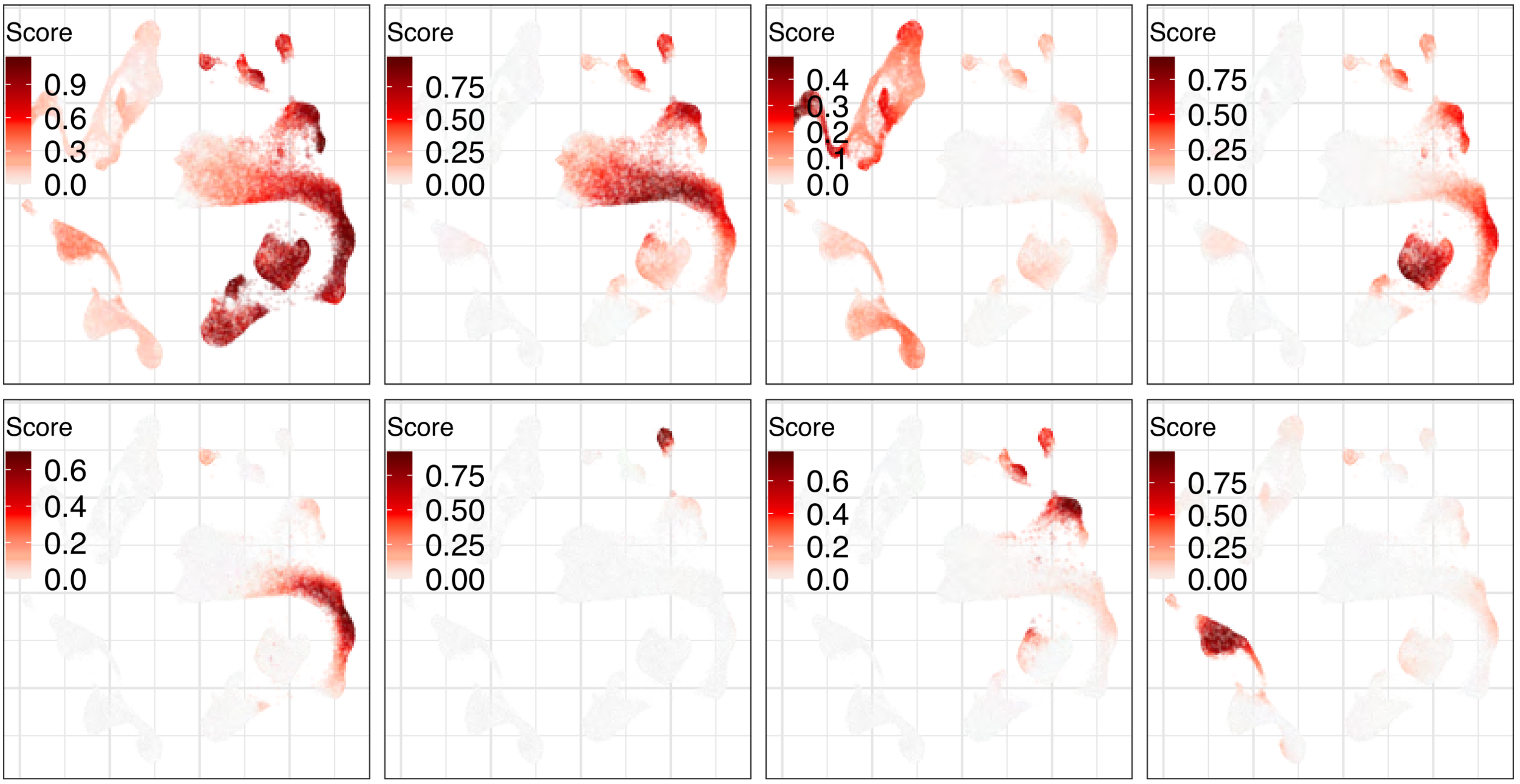


Gene programs:
Expression analysis: cluster-free DE

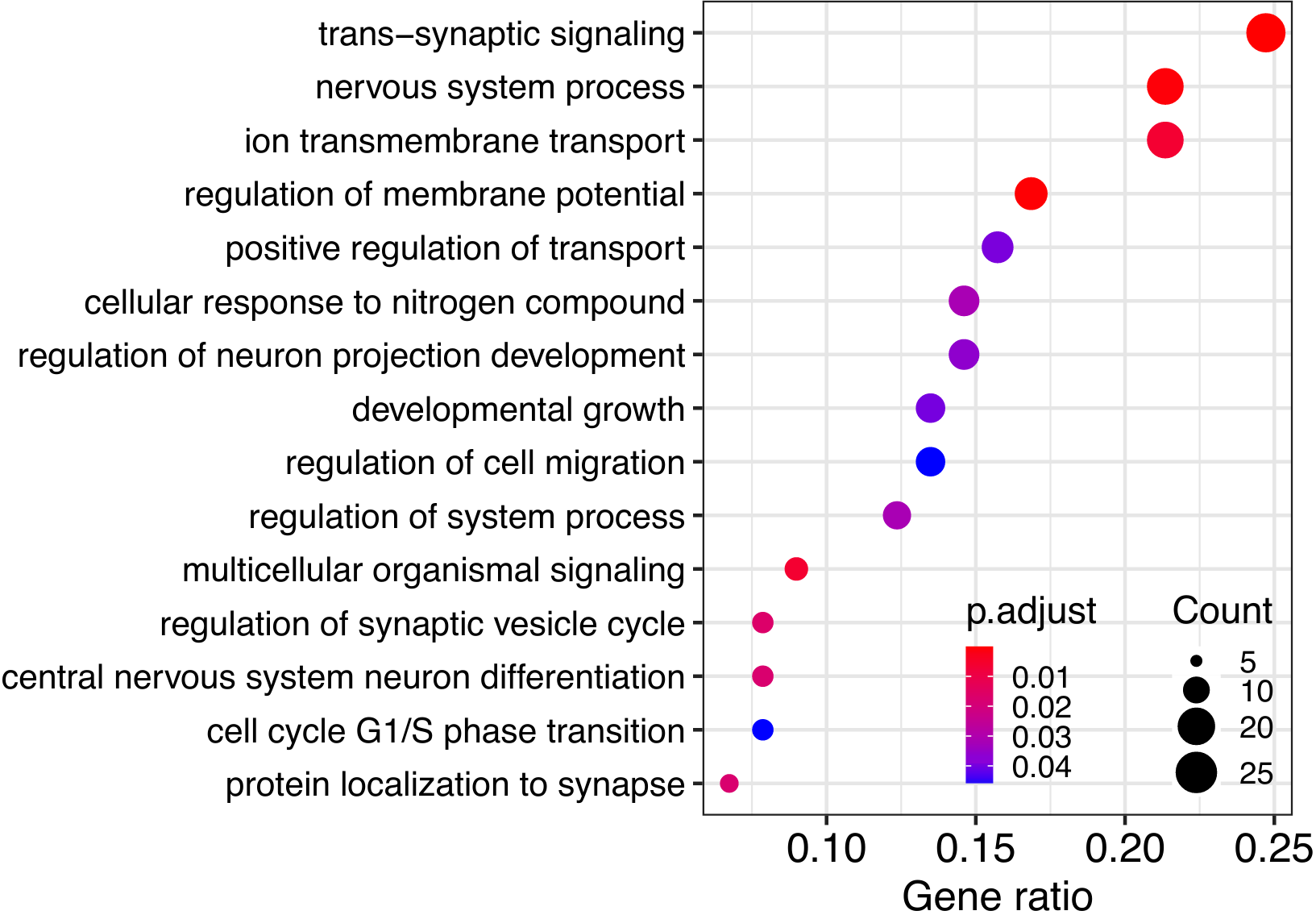

Science reproducibility
>70% of publications had
major flows, compromising main results
Science reproducibility


>70% of publications had
major flows, compromising main results
Science reproducibility
- Publish your code
- Double-check your results
- Don't get too excited

Future directions
- Improving precision and power of methods
- Including additional modalities
- Including multiple conditions
- Better handling covariates
Future work
Cacoa
Epilepsy
Conos
Overview
Introduction
Acknowledgements

Khodosevich Lab

Jonathan Mitchel

Ruslan Soldatov

Shenglin Mei

Evan Biederstedt
Navneet Vasistha
Konstantin Khodosevich
Rasmus Rydbirk
Irina Korshunova

Diego González

Katarina Dragicevic

Mykhailo Batiuk

Anna Igolkina

Peter Kharchenko
Acknowledgements









Thank you for your attention!
Petukhov Viktor
Khodosevich Lab, BRIC