Tools for CMIP6 data processing and standardization
exemplified by EMAC and for AerChemMIP
DReq Web-GUI
cdo cmor
DReq Web-GUI
cdo cmor
climate model setup
Data Request
model
output
CMIP6 conform data
Postprocessing
- A simple tool to provide a list of a customized DReq?
- How much data will be produced by my set up?
- A simple tool to convert data to CMIP standard?
- Can I automize parts of the postprocessing?
run
Simplified data workflow in CMIP6
DReq Web-GUI
- A simple tool to provide a list of a customized DReq?

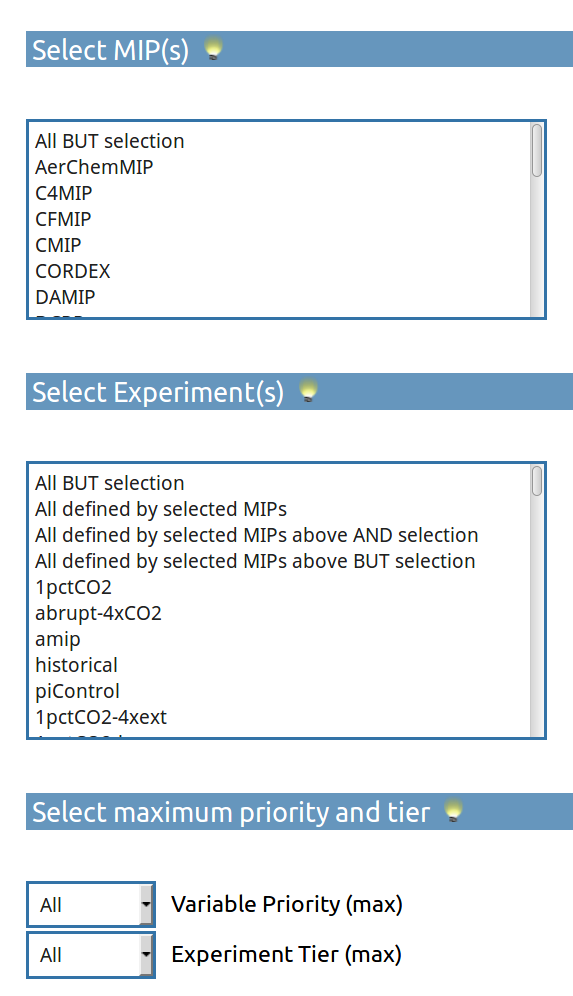

uses DreqPy API by Martin Juckes, NCAS BADC.
DReq Web-GUI
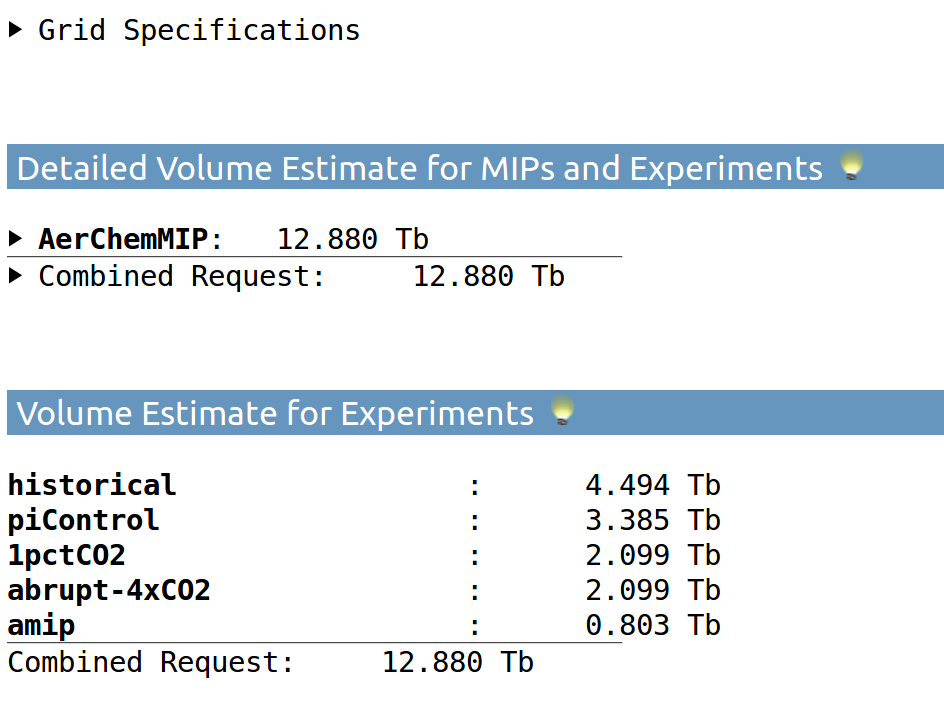
2. How much data will be produced by my set up?

uses DreqPy API by Martin Juckes, NCAS BADC.
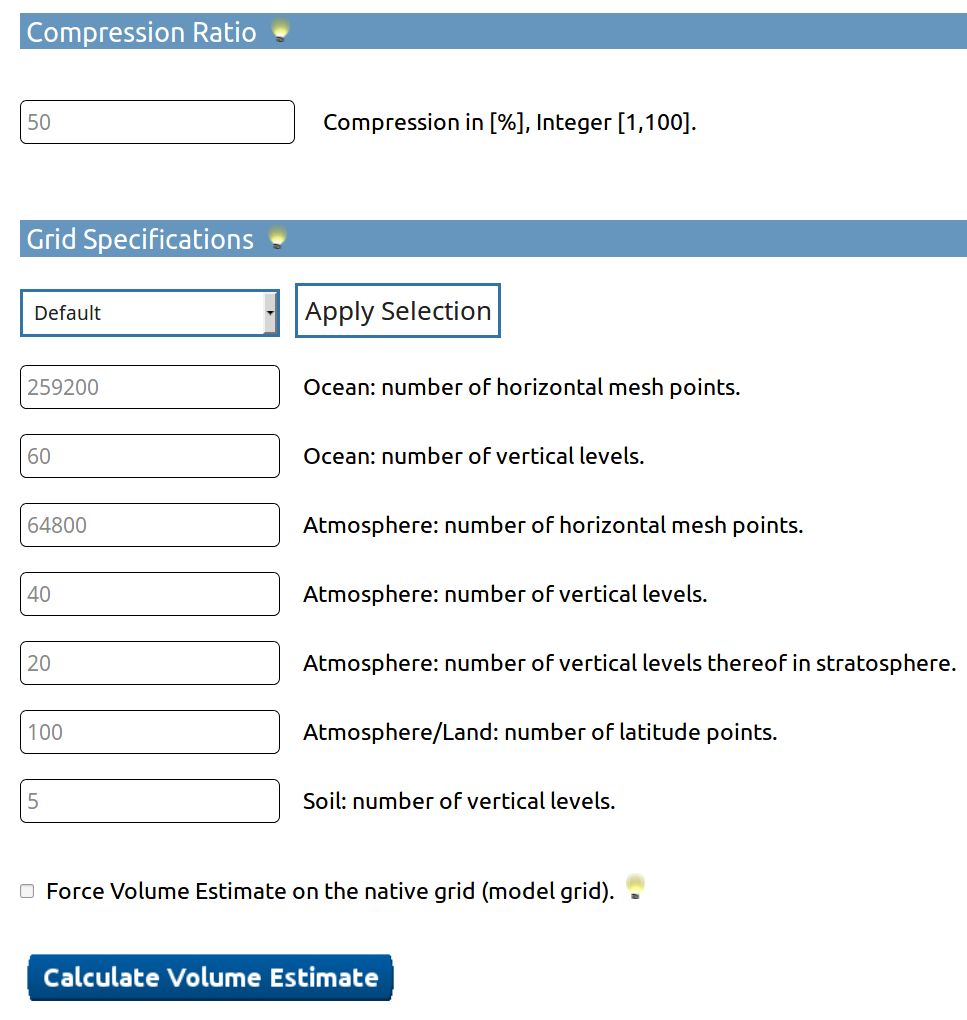
DReq Web-GUI
cdo cmor
climate model setup
Data Request
model
output
CMIP6 conform data
Postprocessing
The DReq Web-GUI
- can create a customized DReq!
-
estimates the volume of the custom DReq!
- A simple tool to convert data to CMIP standard?
run
Simplified data workflow in CMIP6
CMOR
Convention (1 and 2) about climate data accepted in Coupled Model Inter-comparison Project
Collection of operators to process climate data
Climate Model Output Rewrite to ensure correct conversion
introduction
cdo cmor
3. A simple tool to convert data to CMIP standard?
Aside from the specific data structure, extensive and diverse meta data from coordinate information to global attributes must be integrated into the file
Many functions needs to be called with specific arguments, for each variable differently
Was not able to create and maintain the CMIP standard.
-> cdo cmor
?
- CMOR ensures that ouput is CMIP compliant.
- Different CMIP standards can be produced
- Use synergies, avoid repeating work
- Why using CMOR to create CMIP standard?
- Why integrate CDO with CMOR?
No user side preparation of CMIP format description
cdo cmor
3. A simple tool to convert data to CMIP standard?
- CDO is a well known tool with an active support
- The CDO's interface allows
- Different infile formats
- enables access to all infile information no matter how structured
Use the power of CDOs...
?
- Why using CMOR to create CMIP standard?
- Why integrate CDO with CMOR?
cdo cmor
3. A simple tool to convert data to CMIP standard?
... to simplify CMOR usage:
?
- Why using CMOR to create CMIP standard?
- Why integrate CDO with CMOR?
cmor_setup();
cmor_dataset_json();
cmor_load_table();
cmor_set_table();
cmor_axis();
cmor_grid();
cmor_set_grid_mapping();
cmor_time_varying_grid_coordinate();
cmor_zfactor();
cmor_variable();
cmor_set_deflate();
cmor_set_variable_attribute();
cmor_create_output_path();
cmor_write();
cmor_close();are included by one cdo cmor operator
3. A simple tool to convert data to CMIP standard?
cdo cmor
cdo cmor,Amon,\
i=info_table.txt,\
gi=grid_info.nc,\
mt=mapping_table.txt\
infilecdo cmor,Amon,\
i=info_table.txt,\
gi=grid_info.nc,\
mt=mapping_table.txt\
infile"variable_entry":{
"ccb": {
"frequency": "mon",
"modeling_realm":"atmos"
}
}CMIP6_Amon.json
grid_
info.nc
mapping_
table.txt
ncdump -h mrsofc_fx_MPIESM-1-2-HR_historical_r1i1p1f1_gn.nc
#stdout:
netcdf test {
dimensions:
lat = 192 ;
lon = 384 ;
bnds = 2 ;
variables:
double lat(lat);
double lat_bnds(lat,bnds);
double lon(lon);
double lon_bnds(lon,bnds);¶meter cmor_name=tasmax code=201 units="K" cell_methods="m" project_mip_table=Amon /
¶meter cmor_name=tasmax code=201 units="K" cell_methods="m" project_mip_table=day /
¶meter cmor_name=tasmin code=202 units="K" cell_methods="m" project_mip_table=day /
¶meter cmor_name=tasmin code=202 units="K" cell_methods="m" project_mip_table=Amon /
¶meter cmor_name=rsds units="W m-2" cell_methods="m" positive="d" /
¶meter cmor_name=rlds units="W m-2" cell_methods="m" positive="d" /info_
table.txt
activity_id="CMIP"
experiment="AMIP"
experiment_id="amip"
forcing_index="1"
grid="All grid attributes are set for the native grid and based on information from attribute source."
grid_label="gn"
initialization_index="1"
institution="Max Planck Institute for Meteorology, Hamburg 20146, Germany"
institution_id="MPI-M"
license="CMIP6 model data produced by REQUIRED is licensed under a Creative Commons Attribution ShareAlike 4.0 International License (https://creativecommons.org/licenses). Consult https://pcmdi.llnl.gov/CMIP6/TermsOfUse for terms of use governing CMIP6 output, including citation requirements and proper acknowledgment. Further information about this data, including some limitations, can be found via the further_info_url (recorded as a global attribute in this file) and. The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law."
mip_era="CMIP6"
nominal_resolution="100 km"
physics_index="1"
realization_index="1"
source="MPIESM1.2-HR (2017): \naerosol: none, prescribed MACv2-SP\natmos: ECHAM6.3 (spectral T127; 384 x 192 longitude/latitude; 95 levels; top level 0.01 hPa)\natmosChem: none\nland: JSBACH3.2\nlandIce: none/prescribed\nocean: MPIOM1.63 (tripolar TP04, approximately 0.4deg; 900 x 450 longitude/latitude; 40 levels; top grid cell 0-12 m)\nocnBgchem: HAMOCC\nseaIce: unnamed (thermodynamic (Semtner zero-layer) dynamic (Hibler 79) sea ice model)"
source_id="MPIESM-1-2-HR"
source_type="AGCM"
sub_experiment="none"
sub_experiment_id="none"
parent_activity_id="no parent"
parent_experiment_id="no parent"
calendar="proleptic_gregorian"
required_time_units="days since REQUIRED"cdo cmor
4. Can I automize parts of the postprocessing?
cdo cmor
model
output
CMIP6 conform data
Postprocessing
1. Aggregation
2. Diagnostics
- needs a recipe
3. standardization
- requires attributes
- Provide recipes and attributes in a data base
- Based on 'cdo expr' and 'cdo cmor', script fragments can be created
cdo expr can evaluate expressions
cdo cmor can convert to CMIP6 standard
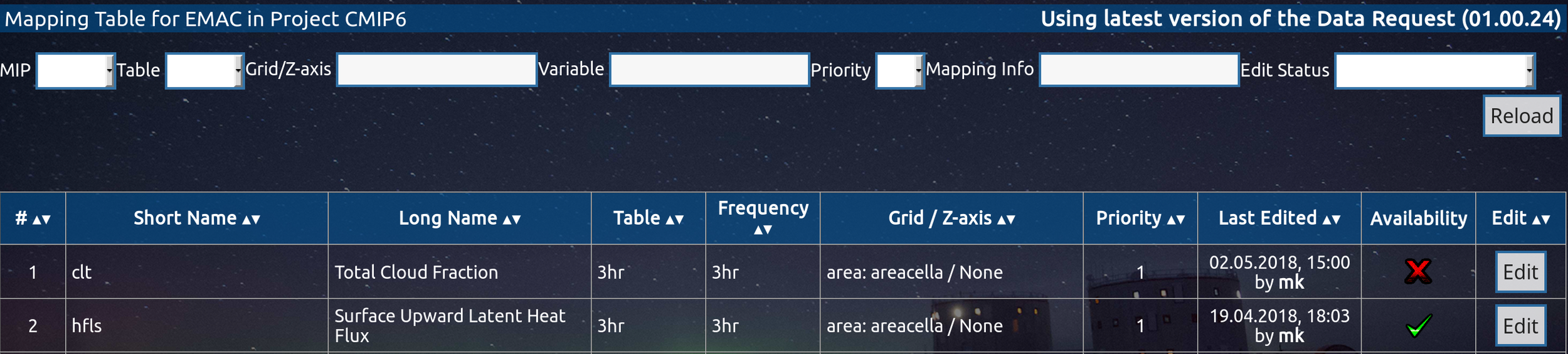
DReq Web-GUI
4. Can I automize parts of the postprocessing?
cdo cmor
Advantages of a webbased application:
- One data base always available
- Shared work
- Logs of every change with account info

model
output
CMIP6 conform data
Postprocessing
2. Diagnostics
- needs a recipe
3. standardization
- requires attributes
DReq Web-GUI
4. Can I automize parts of the postprocessing?
cdo cmor

DReq Web-GUI
4. Can I automize parts of the postprocessing?
cdo cmor


DReq Web-GUI
4. Can I automize parts of the postprocessing?
cdo cmor
cat EMAC_CMIP6_mapping.txt
¶meter cmor_name="ch4" name="CH4_ave" project_mip_table="AERmon" units="mol mol-1" cell_methods="m" /
¶meter cmor_name="co" name="CO_ave" project_mip_table="AERmon" units="mol mol-1" cell_methods="m" /
¶meter cmor_name="co2" name="CO2_ave" project_mip_table="AERmon" units="mol mol-1" cell_methods="m" /
¶meter cmor_name="dms" name="DMS_ave" project_mip_table="AERmon" units="mol mol-1" cell_methods="m" /
¶meter cmor_name="emibc" name="emibc" project_mip_table="AERmon" units="kg m-2 s-1" cell_methods="m" /
¶meter cmor_name="emico" name="emico" project_mip_table="AERmon" units="kg m-2 s-1" cell_methods="m" /
¶meter cmor_name="eminox" name="eminox" project_mip_table="AERmon" units="kg m-2 s-1" cell_methods="m" /
¶meter cmor_name="emiso2" name="emiso2" project_mip_table="AERmon" units="kg m-2 s-1" cell_methods="m" /
¶meter cmor_name="hcho" name="HCHO_ave" project_mip_table="AERmon" units="mol mol-1" cell_methods="m" /
¶meter cmor_name="hcl" name="HCl_ave" project_mip_table="AERmon" units="mol mol-1" cell_methods="m" /
¶meter cmor_name="hno3" name="HNO3_ave" project_mip_table="AERmon" units="mol mol-1" cell_methods="m" /
¶meter cmor_name="isop" name="C5H8_ave" project_mip_table="AERmon" units="mol mol-1" cell_methods="m" /
¶meter cmor_name="jno2" name="J_NO2_ave" project_mip_table="AERmon" units="s-1" cell_methods="m" /
¶meter cmor_name="lwp" name="xlvi_ave" project_mip_table="AERmon" units="kg m-2" cell_methods="m" /
¶meter cmor_name="n2o" name="N2O_ave" project_mip_table="AERmon" units="mol mol-1" cell_methods="m" /
¶meter cmor_name="no" name="NO_ave" project_mip_table="AERmon" units="mol mol-1" cell_methods="m"DReq Web-GUI
4. Can I automize parts of the postprocessing?
cdo cmor
model
output
CMIP6 conform data
Postprocessing
2. Diagnostics
- needs a receipe
3. standardization
- requires attributes
#-- Diagnostic for emac (ESM: EMAC-2-53-AerChem) variable pr / table 3hr
(if_requested $member $esmod 3hr pr $chunk >$err.pr.3hr 2>&1 && {
find_file -e -p $period "$sdir" "*3hr*" $err.find_file.3hr ifile
$cdo -f nc \
expr,"pr=aprl_ave+aprc_ave+aprs_ave" \
$ifile in_cmor/3hr_pr_${period}.nc >>$err.pr.3hr 2>&1 || { echo ERROR >>$err.pr.3hr; }; }; )& #-- CMOR-rewrite for emac (ESM: EMAC-2-53-AerChem) 3hr
cn='clt pr rlds rsds rsdscs rsuscs tos'
for var in $cn; do
(if_requested $member $esmod 3hr $var $chunk 1>>$err.$var.3hr 2>&1 || continue
ifile=in_cmor/3hr_${var}_${period}.nc
$cdo cmor,3hr,mt=$mt,dr=$dr,cn=$var $ifile \
1>>$err.$var.3hr 2>&1 || { echo ERROR >>$err.$var.3hr;};)&
doneDReq Web-GUI
DReq Web-GUI
cdo cmor
climate model setup
Data Request
model
output
CMIP6 conform data
Postprocessing
The DReq Web-GUI
- can create a customized DReq!
-
estimates the volume of the custom DReq!
cdo - cmor can convert data to CMIP standard
- expr can evaluate expressions
run