The challenges and opportunities in imaging-based spatially resolved transcriptomics data analysis
Weize Xu
2023.12
The cover was created using DALLE3
Table of contents
- Spatially Resolved Transcriptomics
- Imaging-based SRT
- Up-stream analysis
- Down-stream analysis
- Prospect
SRT: Spatially Resolved Transcriptomics



Bulk cell
RNA-Seq
Single cell
RNA-Seq
Spatially Resolved
Transcriptomics
What and Why
Category


NGS-based
Imaging-based
- smFISH, seqFISH, MERFISH
- Super high spatial resolution.
- Geo-seq, Visium, DBiT-seq
- unbiased capture
History and tendency

- Higher spatial resolution.
- Capture more genes.
- Larger field of view.
Why imaging based?
Seeing is believing!
眼见为实!
smFISH

seqFISH and seqFISH+


seqFISH
seqFISH+
MERFISH

ExSeq

MiP-Seq

Commercial solutions





Upstream analysis
- Overview
- Bioimage processing
- Image reconstruction
- Image registration
- Cell segmentation
- Tool and frameworks for upstream analysis
- Starfish
- PipeFISH
Overview
Preprocessing: Image reconstruction
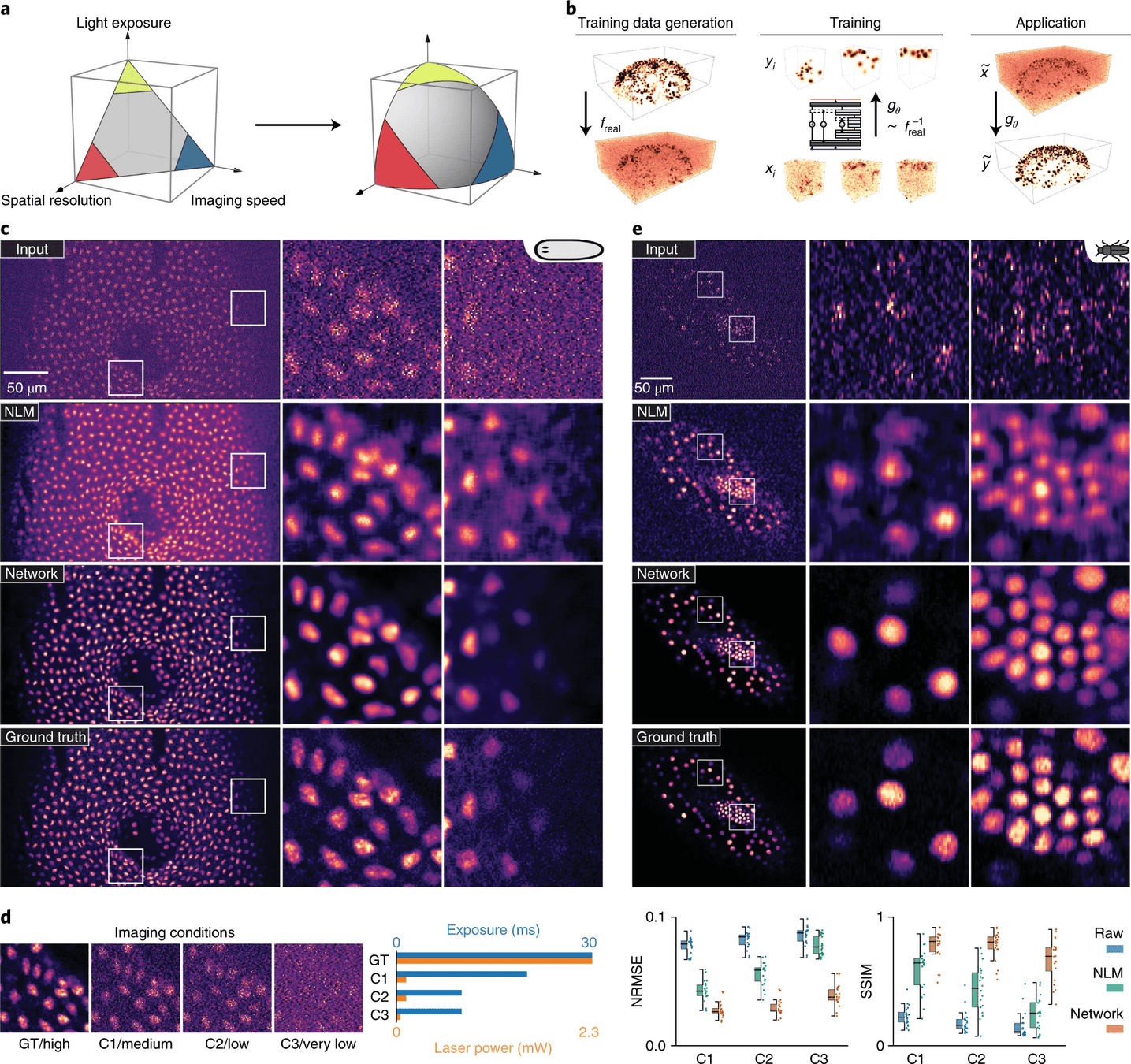
Content-aware image restoration(CARE) based on deep learning extends the range of biological phenomena observable by microscopy.

Use case: Thick tissue MERFISH


Preprocessing: Image registration
Challenge:
- Large scale registration
- Nonlinear deformation between rounds.


Spots Calling
Challenge:
- Accuracy!
- Workload of parameter adjustment.
- Optical crowding in high density region.


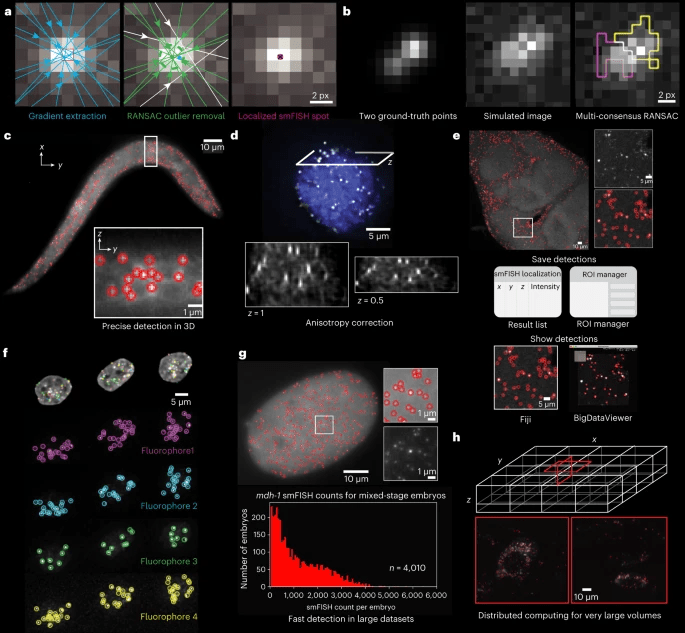
Spots Calling: U-FISH
Un-published work


Cell segmentation
Challenges
- 3d segmentation
- DAPI staining does not represent cell boundaries.

cellpose
Decode
Challenges
- Crowded signal
-
The balance issue between channels.
GraphISS

gene-to-cell
Challenges
- DAPI staining does not represent cell boundaries.
- The ambiguous area between two cells.

Framework and tools for upstream analysis
- Starfish
- PIPEFISH
- ImageJ ecosystem
- RS-FISH
- ...
- Python ecosystem
Chanllenges
- Scalability
- Universality
- User-friendly
Starfish

PIPEFISH

ImageJ ecosystem
- Spots calling: RS-FISH
- Image stiching: fiji/Stitching
- Visualization: Bigdata viewer
- Image registration: BigWrap
- ...
Python ecosystem
- Image processing: numpy, scipy, skimage
- Visualization: Napari viewer
- Image registration: itk-elastix
- Deep learning: Pytorch, Tensorflow
- ...
ImgFlow
- Node editor based image processing
- Based on Python ecosystem
- Composability

Un-published work
Downstream analysis
-
Some common spatial analysis
-
Integration with single-cell sequencing data
-
Subcellular level analysis
-
Alignment between different slides/experiments
-
Multi-modality integration
Common spatial analysis
Add spatial information to the analysis of single-cell RNA-Seq
-
Spatial Variable Genes
-
Spatial domain
-
Spatial trajectory
-
Spatial cell-cell interaction
-
...

Integration with single-cell sequencing data
Overcoming limitations in gene throughput.


Subcellular analysis
Find the pattern in subcellular level.



Alignment between different slides/experiments


Multi-modality integration
SRT + ?


Prospect
- Scalability: Large scale image processing
- AI (Deep learning)
- Analysis method
- Software infrastructure
- Multimodal integration
Thanks for your attention!
The cover was created using DALLE3