Daniel Himmelstein, PhD
Data-Driven Drug Repurposing Workshop
2023-10-03
Integrating All Biomedical Knowledge to Systematically Find the Best Opportunities:
From Hetionet to Related Sciences

slides.com/dhimmel/related-sciences
Workshop Information
Data-Driven Drug Repurposing Workshop: Unlocking disease biology and advancing systematic approaches
- CZI Headquarters
- Hosted by CZI & Every Cure
- Redwood City, CA
- 2023-10-03 at 1:30 PM
Session 2: Curating and Representing Biomedical Knowledge in Network-Based Approaches
Session Abstract: Biomedical knowledge graphs (KGs) represent rich relationships and semantics between drugs, targets and diseases, which support novel methods for inferring biological pathways and predicting drug-target links across a network of interacting genes and proteins. In this session, panelists will share efforts to build and apply KGs for drug repurposing, including advances in optimizing predictions as well as challenges in extracting biomedical knowledge and overcoming inconsistencies, contradictions and other limitations and complexities.

Sandler Neurosciences Center

Sergio
Greene Lab
http://www.greenelab.com/
Postdoc at the University of Pennsylvania

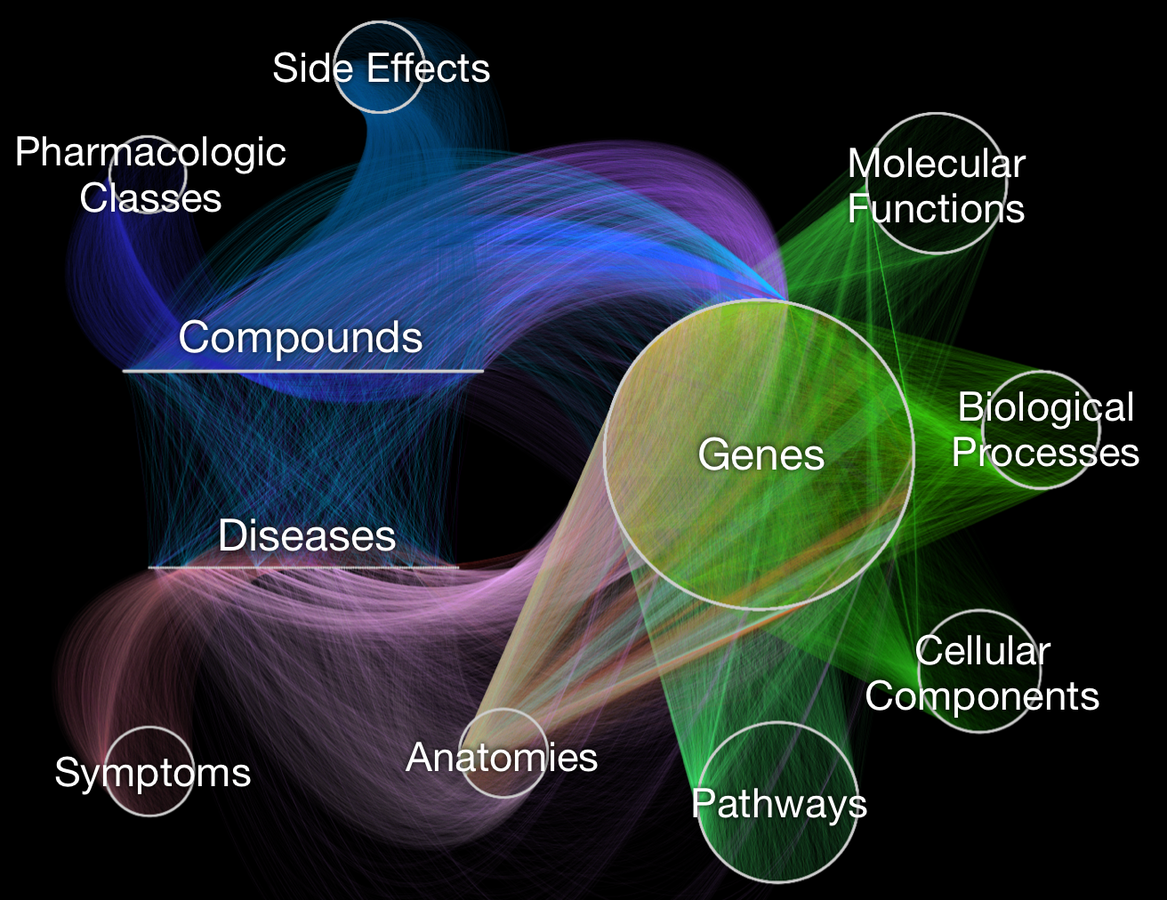
- knowledge graph for drug repurposing
- integrates 29 public resources
knowledge from millions of studies
- ~50 thousand nodes
11 types (labels)
- ~2.25 million relationships
24 types
Hetionet v1.0
Systematic integration of biomedical knowledge prioritizes drugs for repurposing
Daniel S Himmelstein, Antoine Lizee, Christine Hessler, Leo Brueggeman, Sabrina L Chen, Dexter Hadley, Ari Green, Pouya Khankhanian, Sergio E Baranzini
eLife (2017) https://doi.org/cdfk
Hetionet metagraph (schema)

https://neo4j.het.io
github.com/hetio/hetionet
observations =
compound–disease pairs
features = types of paths
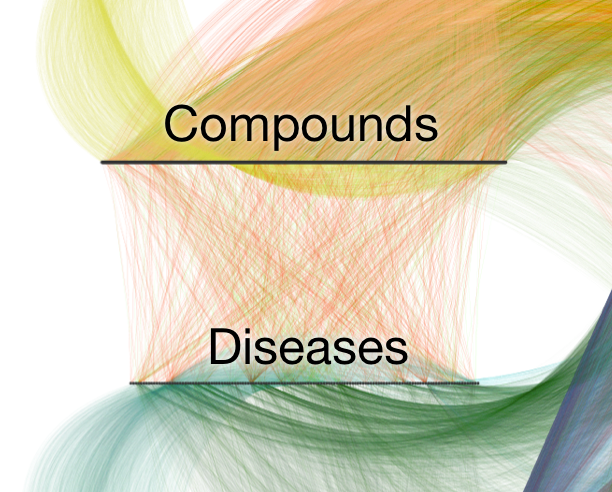
Project Rephetio
Systematic integration of biomedical knowledge prioritizes drugs for repurposing
Daniel S Himmelstein, Antoine Lizee, Christine Hessler, Leo Brueggeman, Sabrina L Chen, Dexter Hadley, Ari Green, Pouya Khankhanian, Sergio E Baranzini
eLife (2017) https://doi.org/cdfk
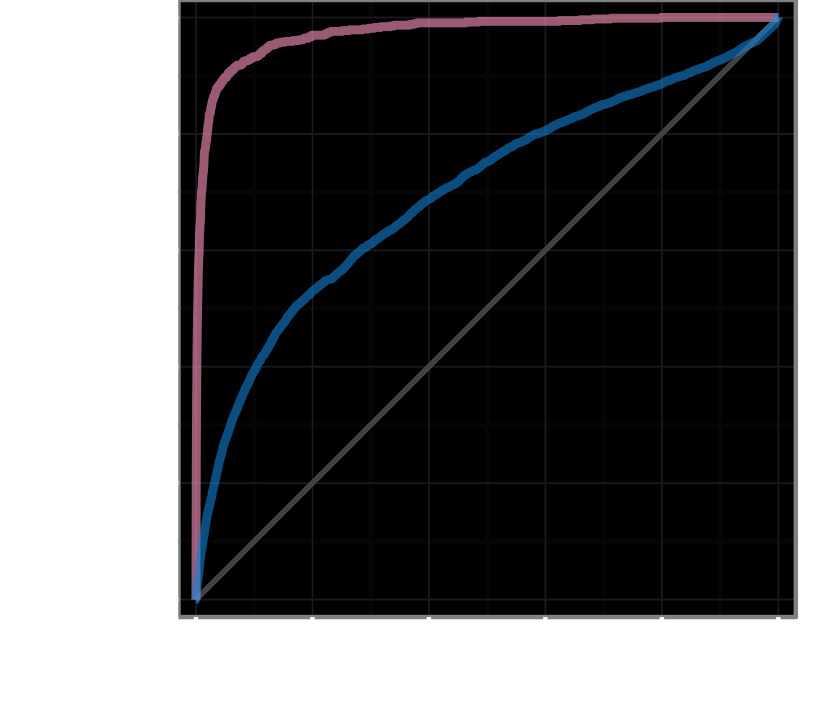
predicted probability of treatment for 209,168 compound–disease pairs
https://het.io/repurpose/
1,538 connected
138 connected



disease modifying treatments
+755, −208,413
AUROC = 97.4%
treatments with clinical trials
+5,594, −202,186
AUROC = 70.0%

Project Rephetio: Does bupropion treat nicotine dependence?
- Bupropion was first approved for depression in 1985
-
In 1997, bupropion was approved for smoking cessation
- Can we predict this repurposing from Hetionet? The prediction was:
- 99.5th percentile for nicotine dependence
- probability 2.50-fold greater than null

Compound–causes–Side Effect–causes–Compound–treats–Disease

Compound–binds–Gene–associates–Disease

Compound–binds–Gene–participates–Pathway–participates–Disease
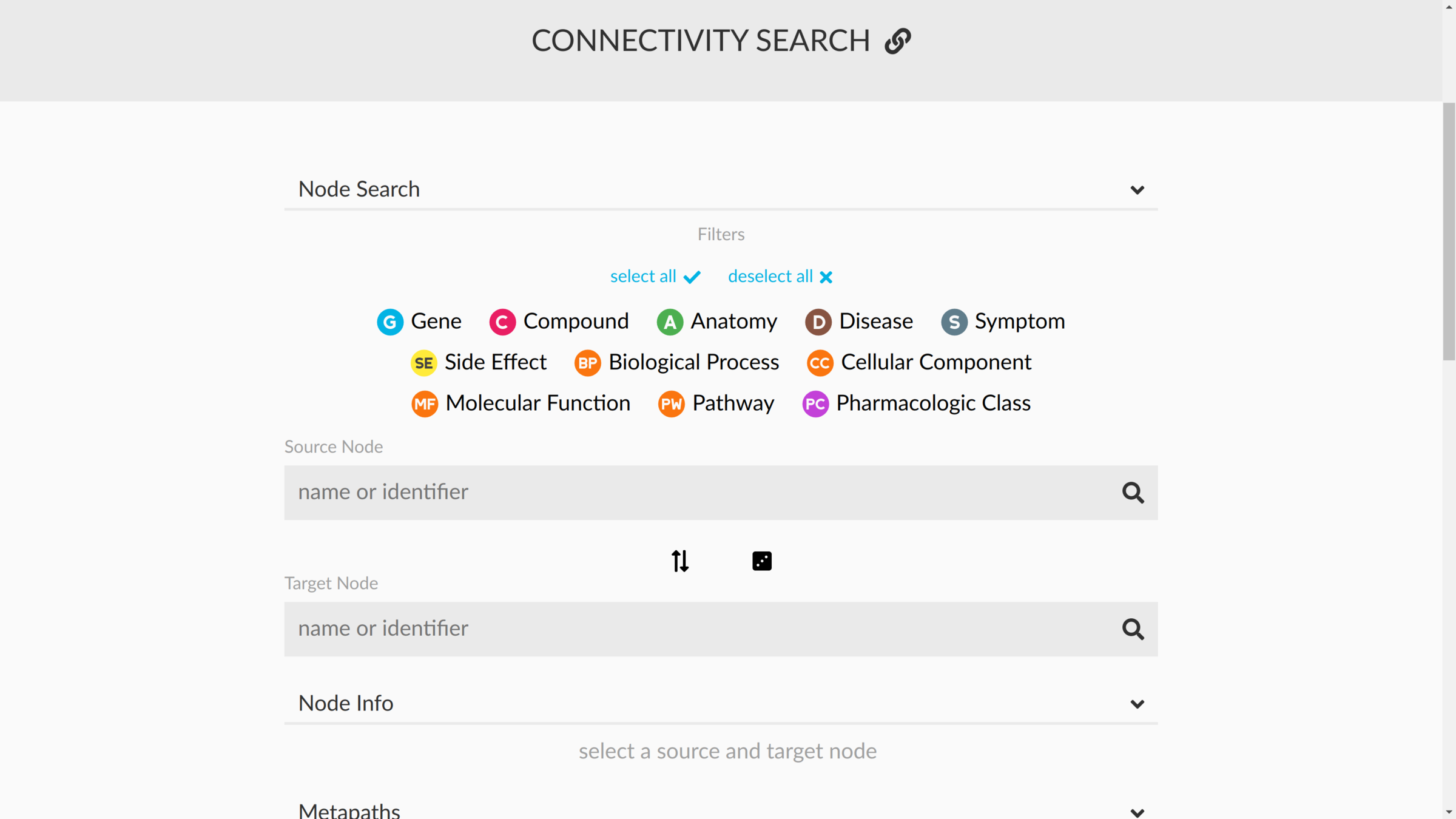
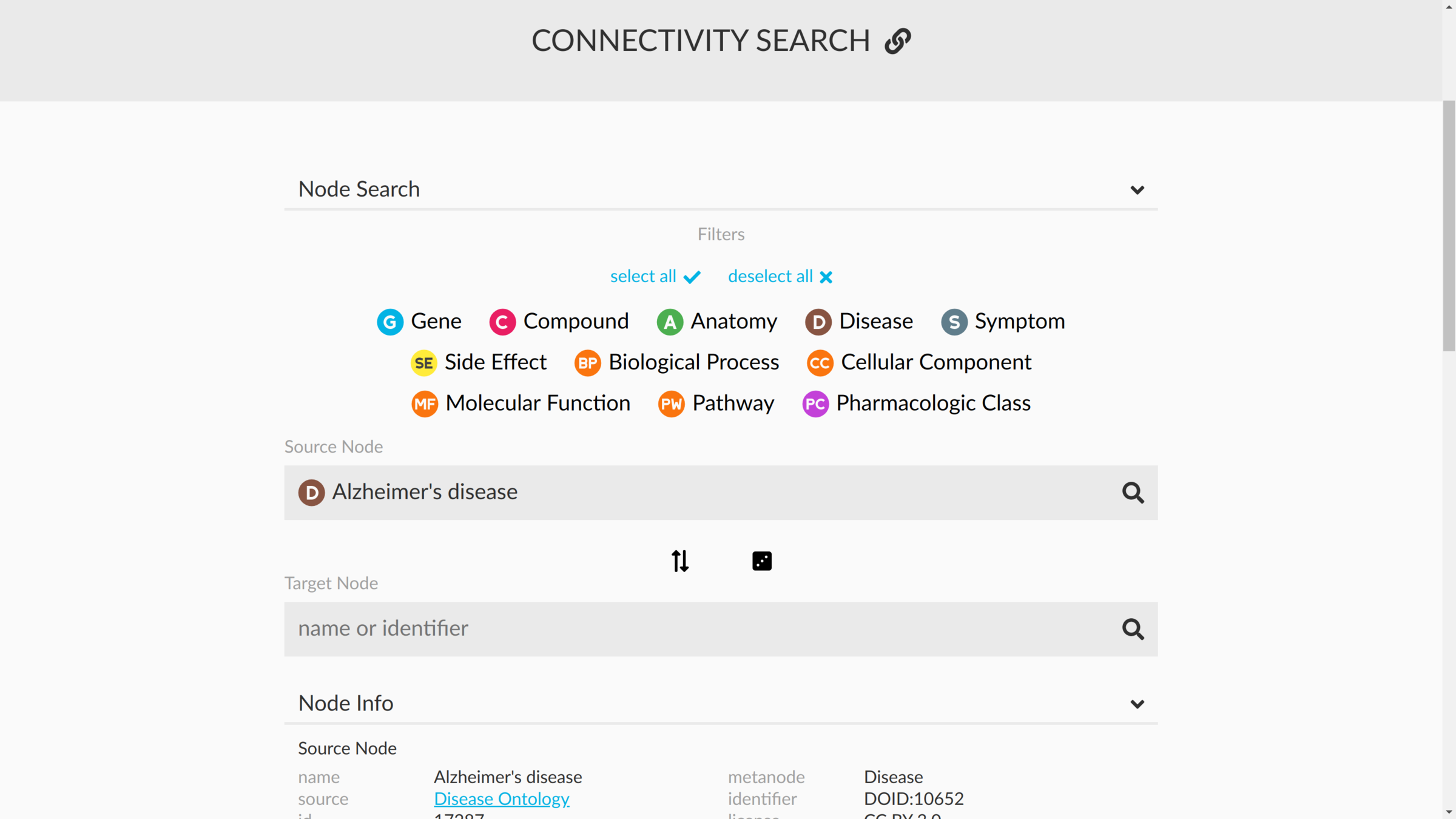
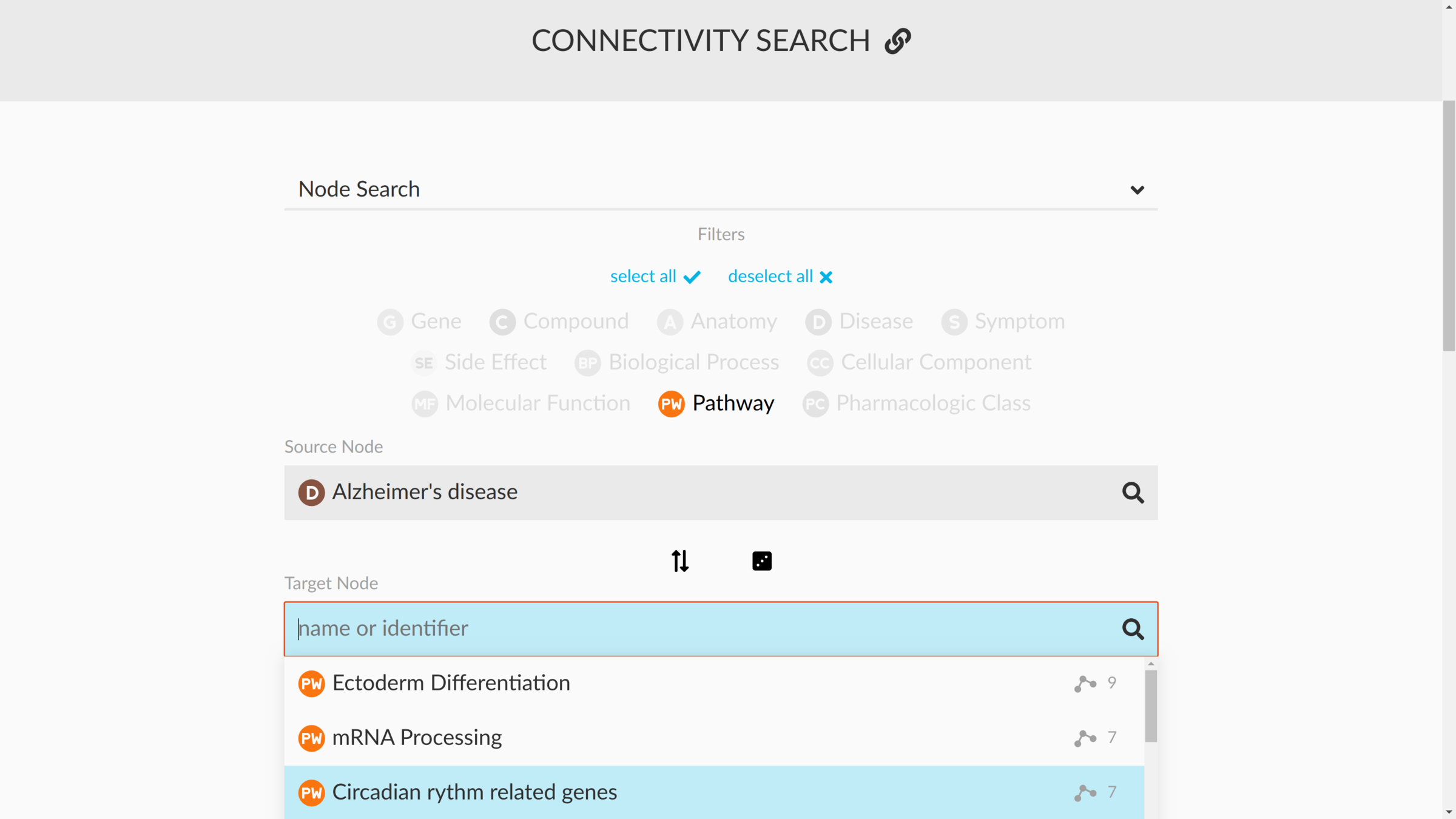
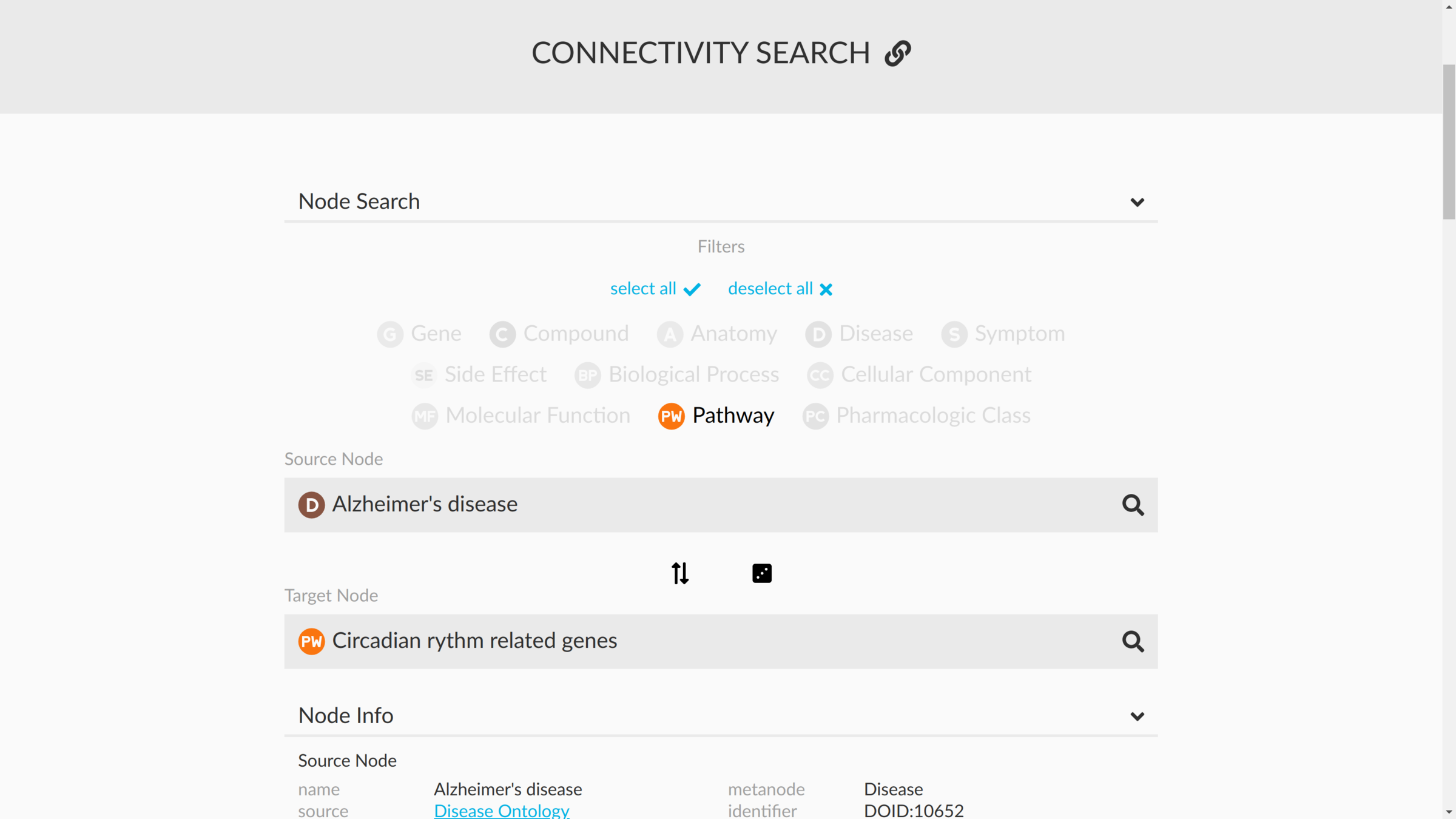
connectivity search
how are two nodes connected?
sans supervision
Hetnet connectivity search provides rapid insights into how biomedical entities are related
Daniel Himmelstein, Michael Zietz, Vincent Rubinetti, Kyle Kloster, Benjamin Heil, Faisal Alquaddoomi, Dongbo Hu, David Nicholson, Yun Hao, Blair Sullivan, Michael Nagle, Casey Greene
GigaScience (2023) https://doi.org/gsd85n
https://het.io/search/
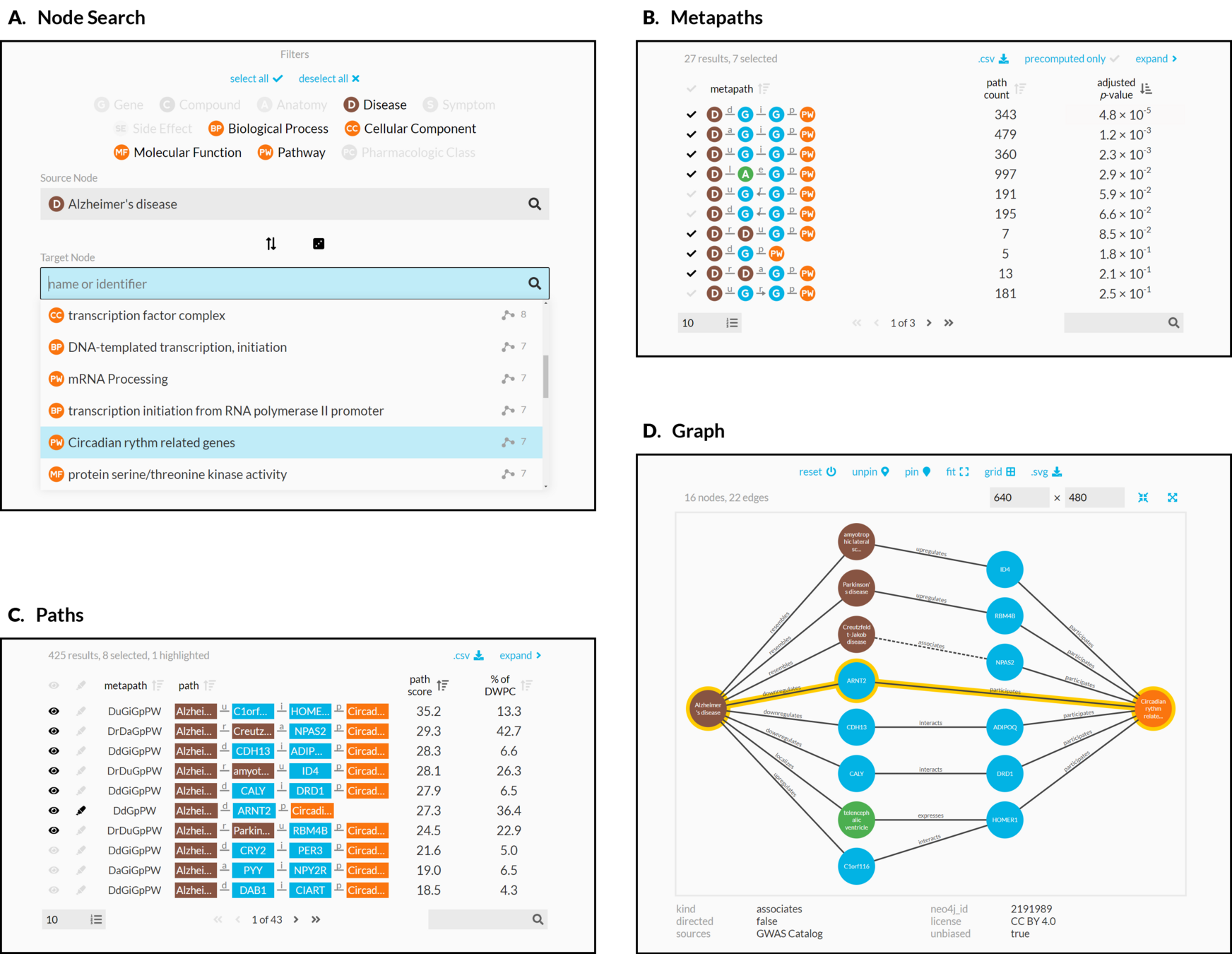
enriched metapaths

enriched paths

visualizing subgraphs
- What we learned?
- unsupervised is hard
- unsupervised is hard
- Hetionet wishlist
- time resolution
- automated updates
- greater disease coverage
- data on real world entities
https://related.vc
continuing the journey at
SUPPLY
of great new drug targets
evidence from 3 million global researchers
DEMAND
to acquire new drugs
350+ large
biopharma acquirers

https://related.vc/team






https://related.vc
how to build biotechs that fail less often?


an efficient R&D operating
model
a new data science platform
- new scientific staffing model
- hub-and-spoke R&D partnerships

- rank 250 million target-disease pairs
- leverage ML to predict outcomes over time
RS Facets

AI/ML Opportunity Ranking Platform

RS Facets
-
Time-Resolved Biomedical Atlas
70+ public and private data sets -
Feature Design
100s of metrics designed to quantitatively assess risk and reward for 250 million target-disease pairs -
Predictive ML Models
Outcomes like probability of clinical success, commercial interest, or economic value creation -
Validation and Back-testing
Enable objective comparative performance benchmarking and iterative improvement

RS Facets™ ingests all activities in global biomedicine to systematically predict the best new drug discovery opportunities.
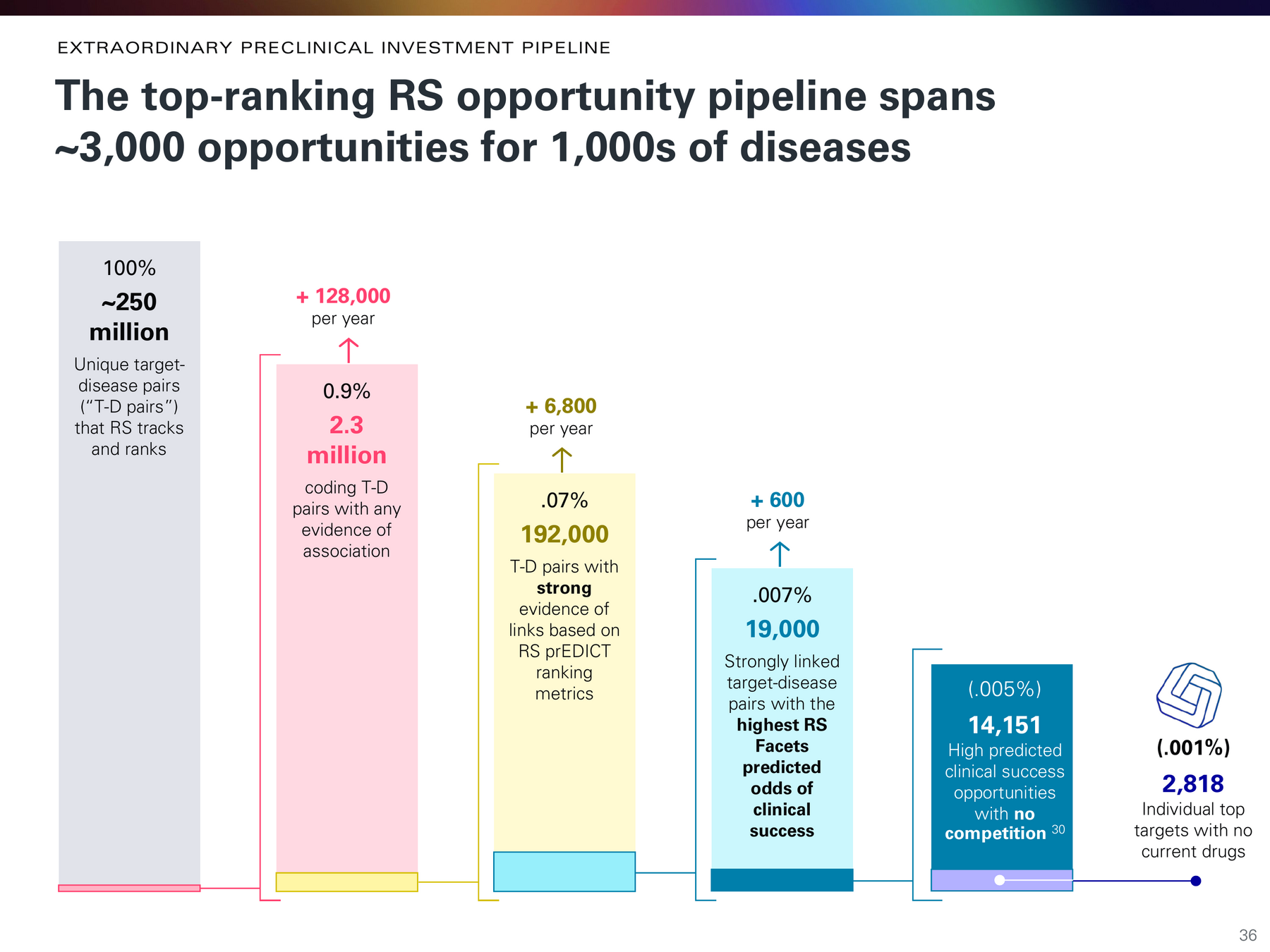






Identifying 1,000s of the very best opportunities from 250 million
Investing in things most likely to work.
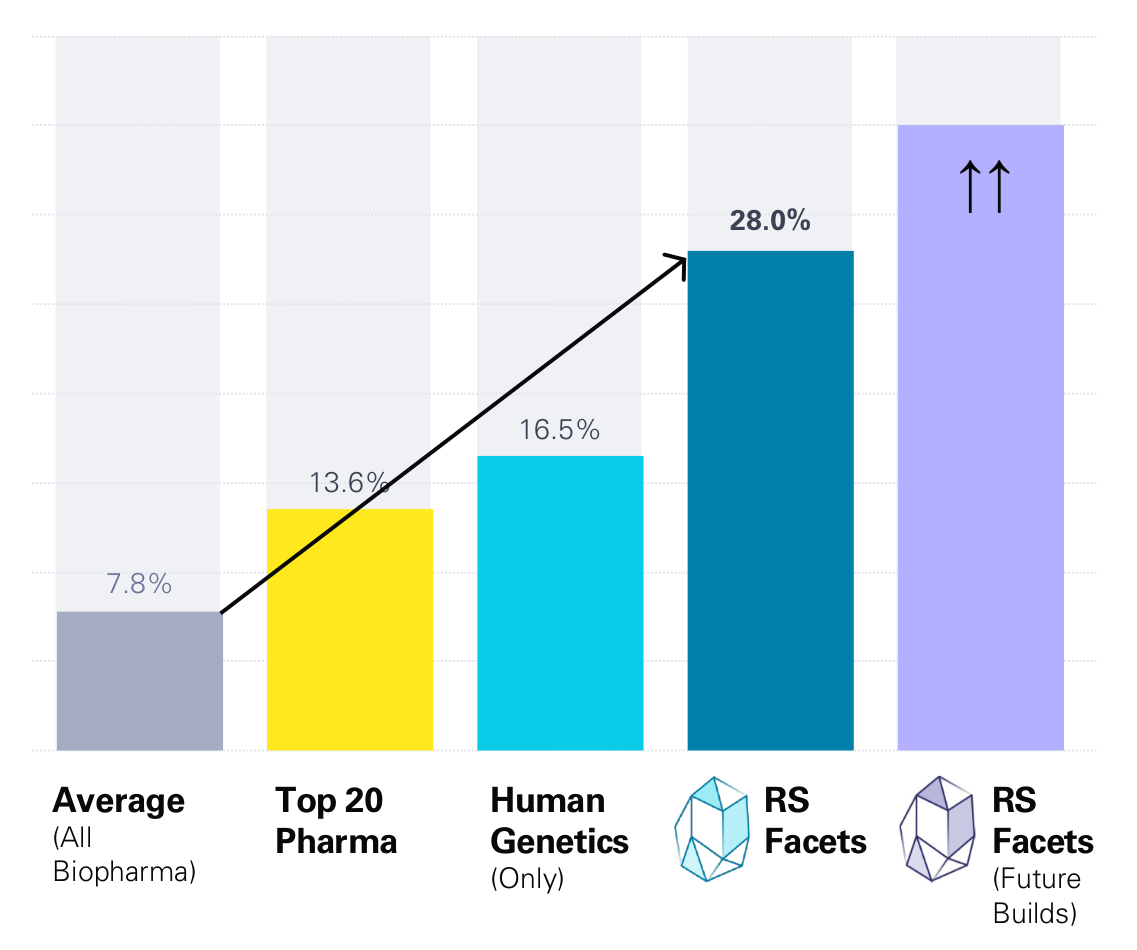




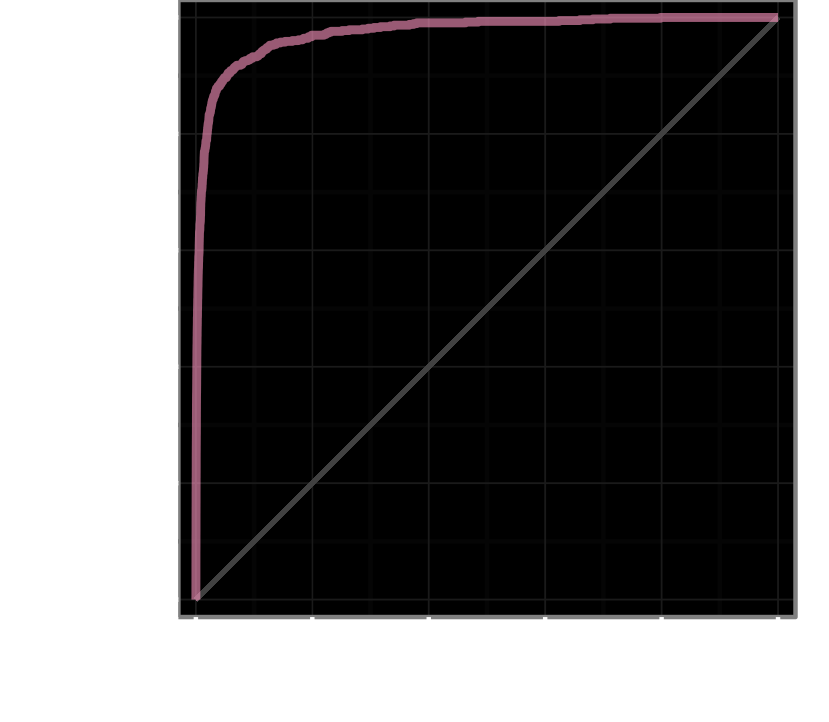
Back-testing performance covering all indications except infectious diseases and oncology
RS Facets back-testing and validation data for its Q1 2023 clinical prediction model build; comparisons reflect the performance the Facets system’s top predictions on a bet- and time-matched basis would have had in a given historical year, based only on what was known in that year.

Likelihood of FDA Approval from Phase I
- diving into one data team project at Related Sciences
- EFO OTAR Slim
- cause of the tangle
- diagnosable diseases
- grouping terms
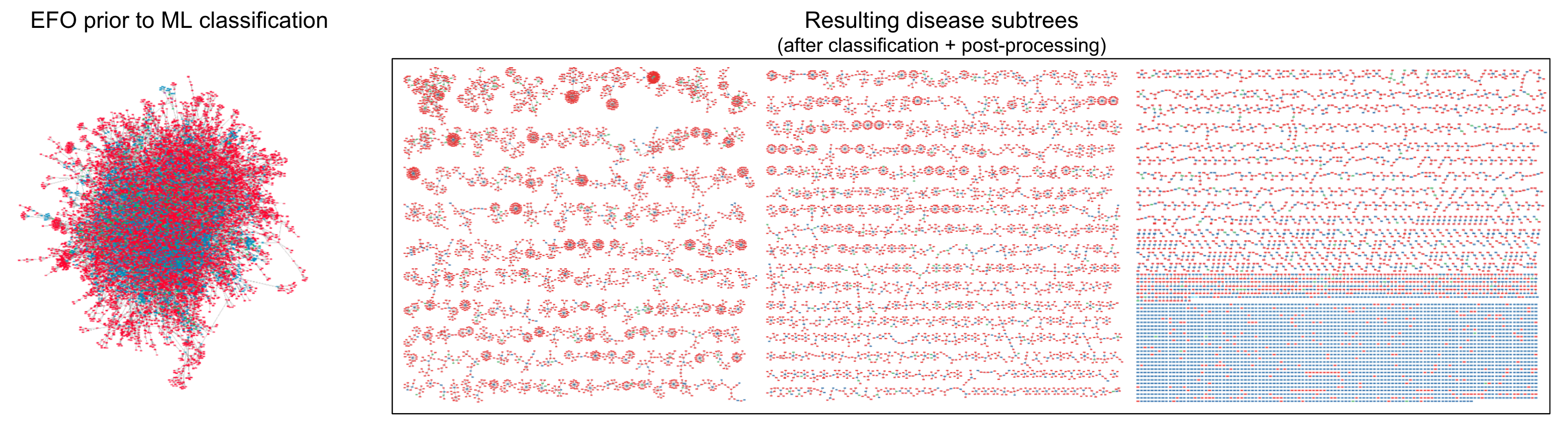
- How many diseases are there?
the hairball of disease

github.com/related-sciences

classifying EFO nodes
Audience poll from the MONDO Outreach Workshop
https://slides.com/dhimmel/efo-disease-precision


predicted EFO classifications
Node outline shows precision
- low = dotted
disease groupings - medium = dashed
diseases - high = solid
disease subtypes


nxontology software suite
-
nxontology
NetworkX-based Python library for representing ontologies.
-
nxontology-ml
Machine learning to classify ontology nodes. -
nxontology-data
Making ontologies accessible as simple JSON files.- EFO
- MeSH
- HGNC Gene Groups
- PubChem Classifications
Figure from the obo-community slack by Philip Strömert generated with Midjourney prompt:
We cannot interpret our research data anymore because we did not annotate it with ontologies

github.com/related-sciences
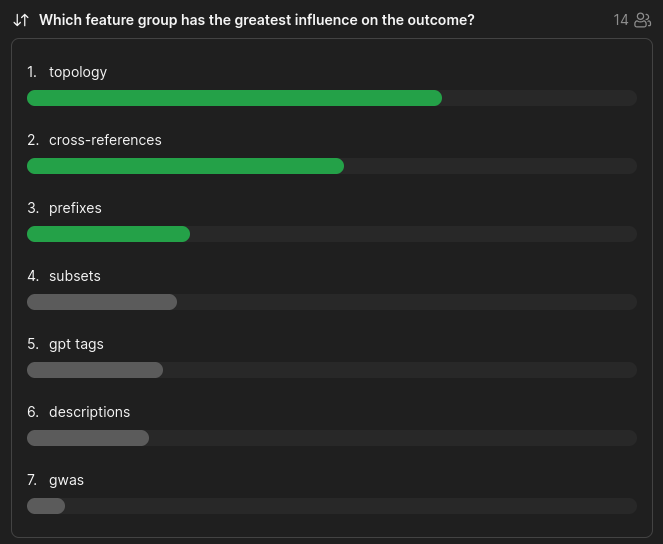
feature group importance
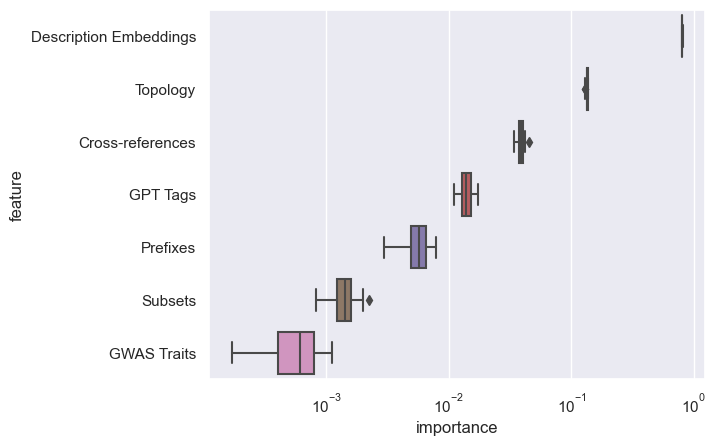
poll
Which feature group has the greatest influence on the outcome?
Slido: 1342 945
- topology
- cross-references
- prefixes
- subsets
- descriptions
- gpt tags
- gwas

Looking forward to discussing!
Say hi to Adam Kolom and me.