Clustering with graphs:
Spectral clustering and Louvain's algorithm
France ROSE
Machine Learning Journal Club
February 28th, 2018
Outline
-
From data to graph
-
Spectral theory and clustering
-
When your graph is too large: Louvain's algorithm
-
Retrieving cell categories with graph clustering
Ressources
-
A Tutorial on Spectral Clustering, by U. Luxburg (2007)
-
Fast unfolding of communities in large networks, by Blondel et al (2008)
-
Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis, by Levine et al (2015)
-
C++ and Matlab implementations of Louvain algorithm
-
phenoGraph python package
From data to graph
Data are already a graph
Data are not a graph yet
Ex: protein interactions
Similarity graph
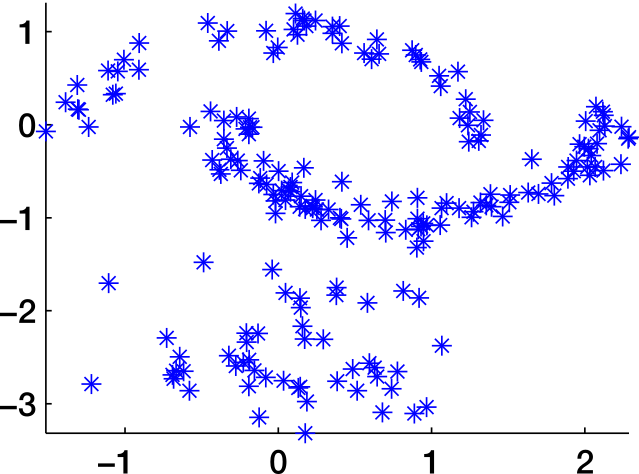
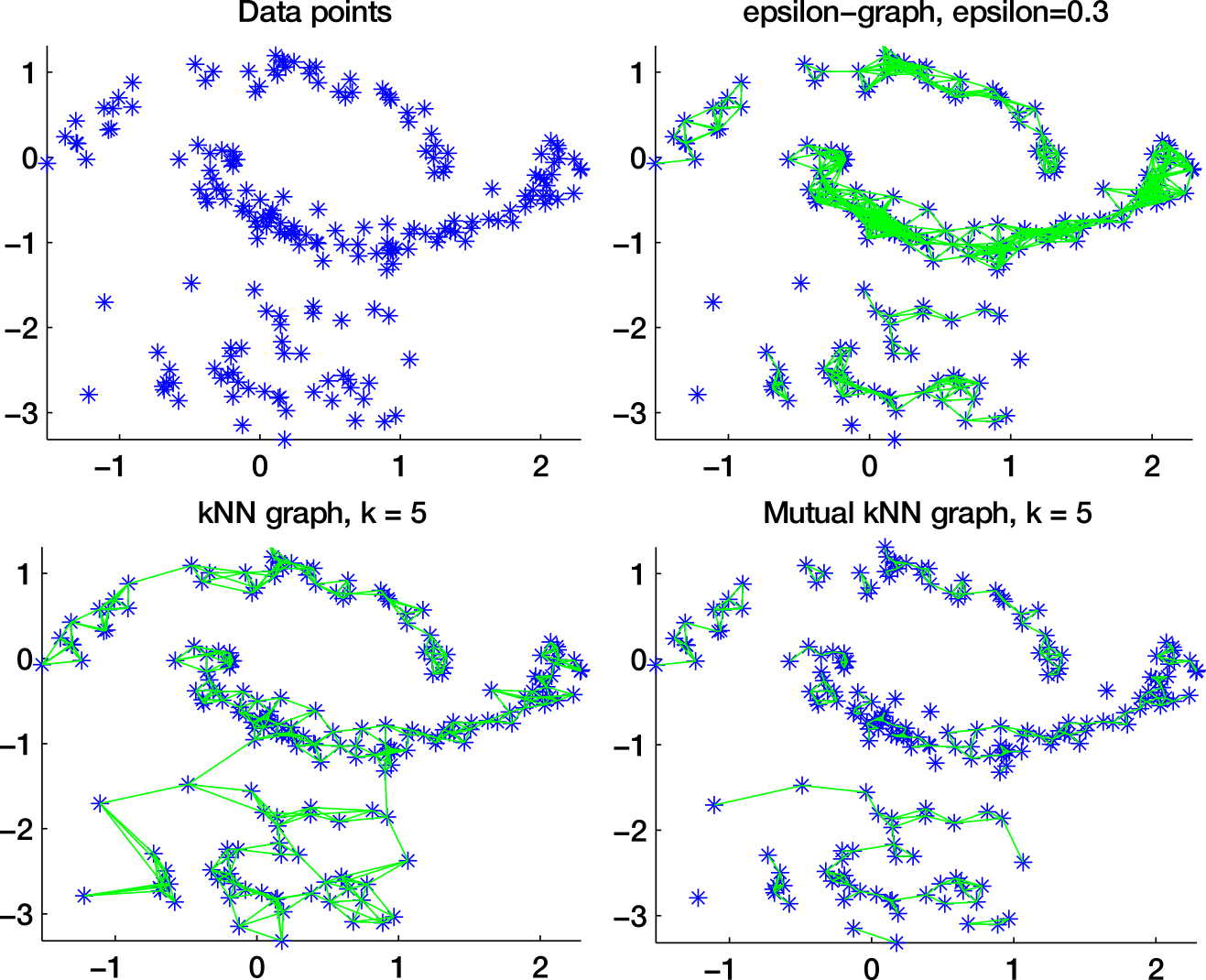
Similarity graphs
Distance function
-neighborhood graph
k-nearest neighbors graph

Compute
distances
RBF kernel
Euclidean, Mahalanobis, Manhattan...
Spectral Theory
Spectrum: eigenvalues and eigenvectors
Laplacian matrix = Degree matrix - Adjacency matrix
Laplacian matrix

- L is symmetric and positive semi-definite
- L has n non-negative, real-valued eigenvalues
- The smallest eigenvalue of L is 0, with the corresponding eigenvector 1
Laplacian matrix
- The smallest eigenvalue of L is 0, with the corresponding eigenvector 1
Intuition about spectral clustering

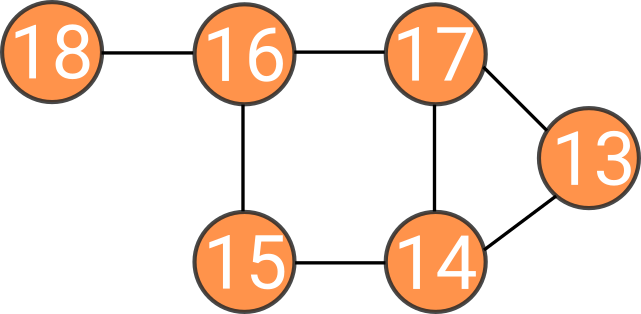
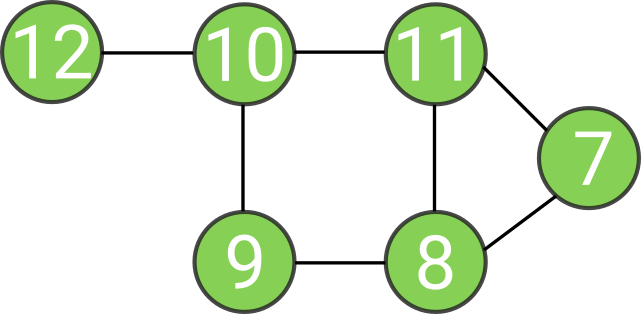






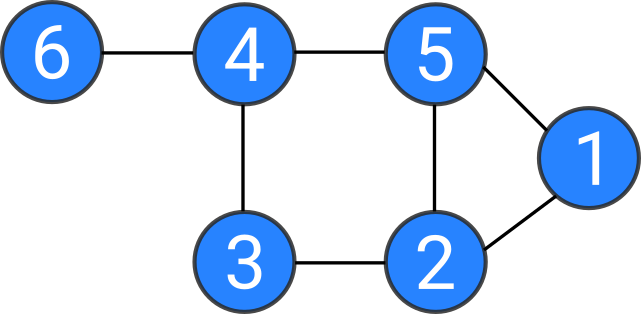
0 is now eigenvalue with 3 orthogonal vectors:
Intuition about spectral clustering



Count how many times 0 is eigenvalue
Matrix diagonalisation



Matrix diagonalization
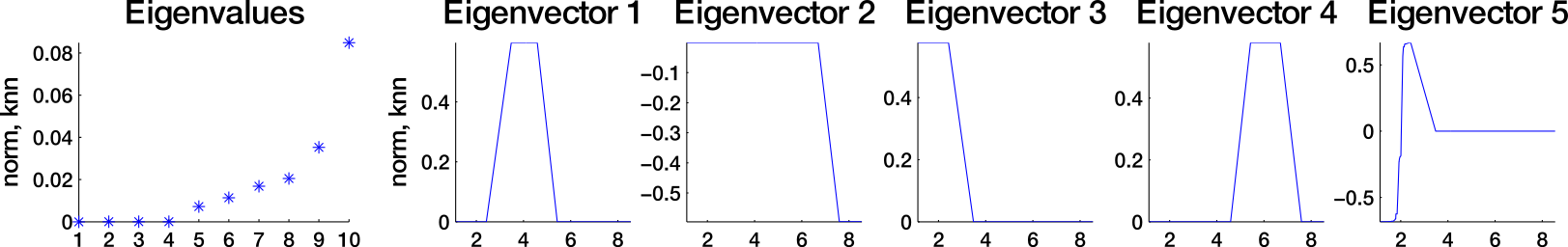

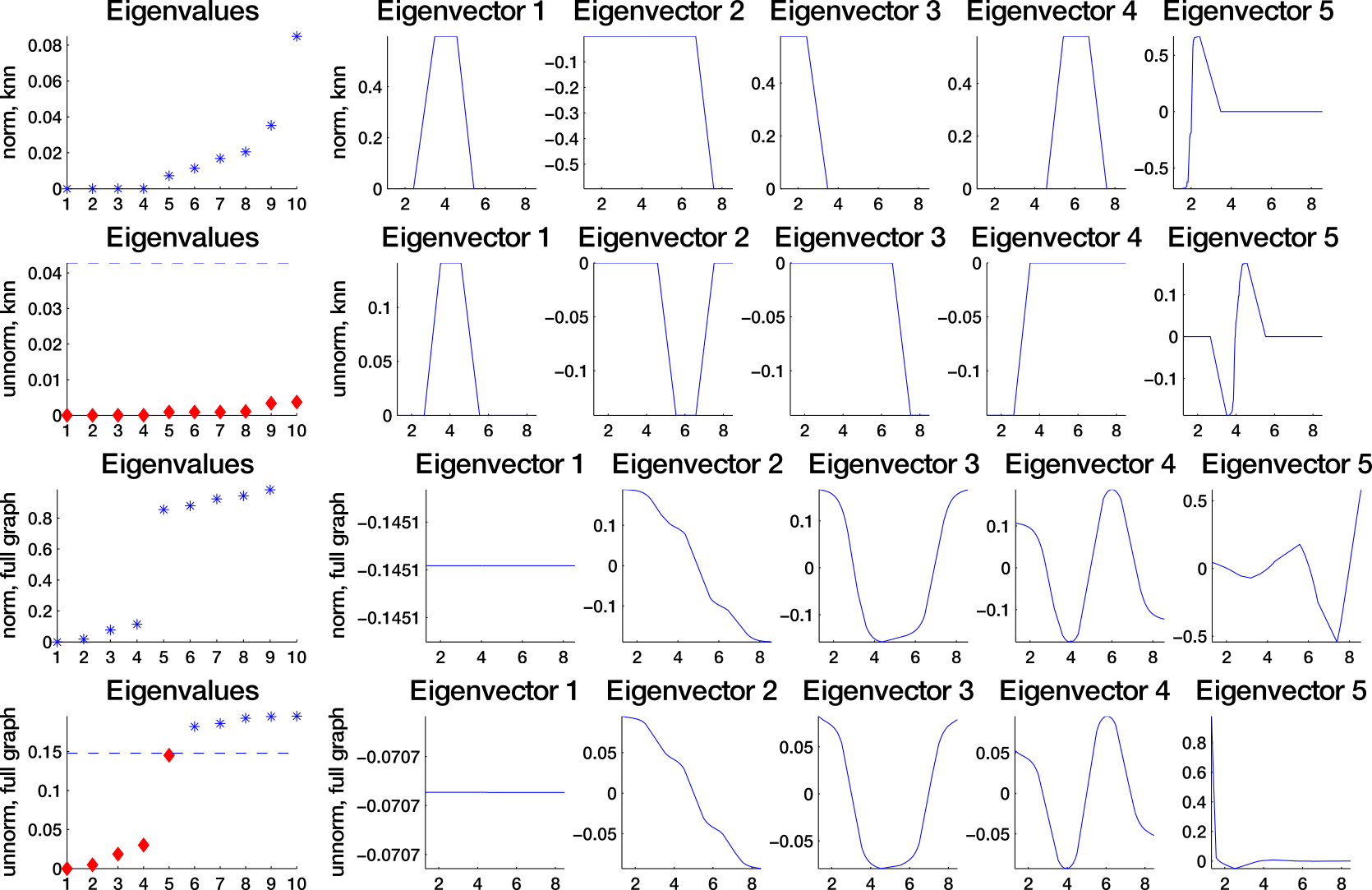
Which similarity graph?

fully connected graph (weighted)
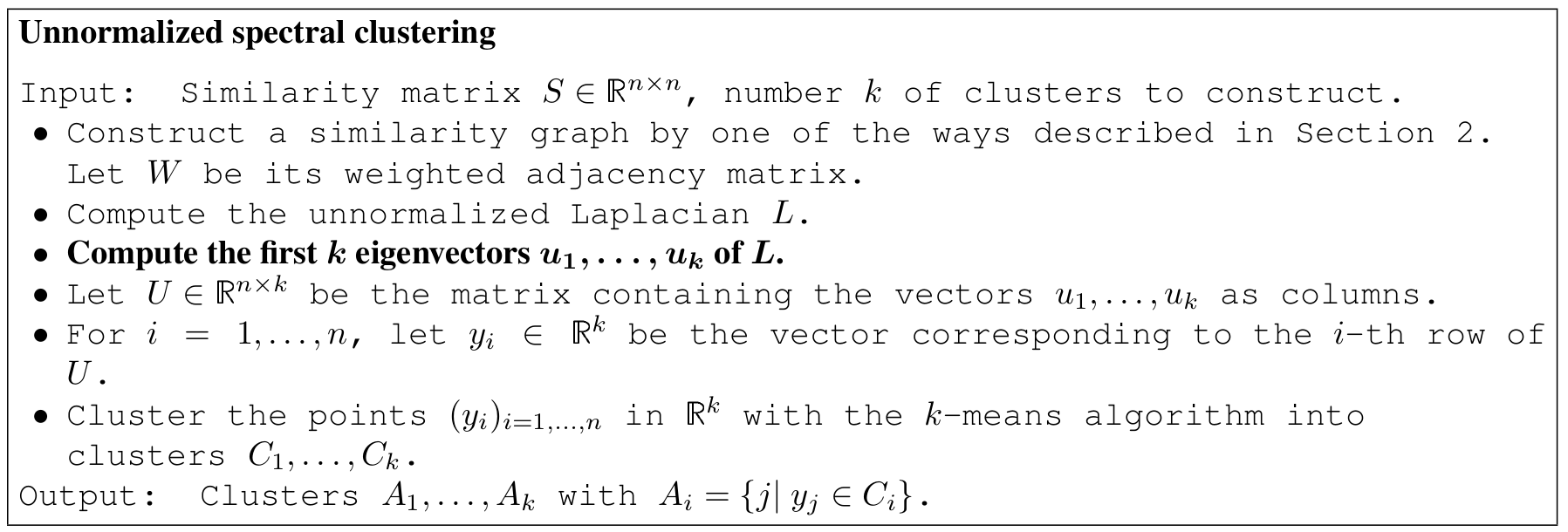
Spectral clustering algorithm
How to choose k?
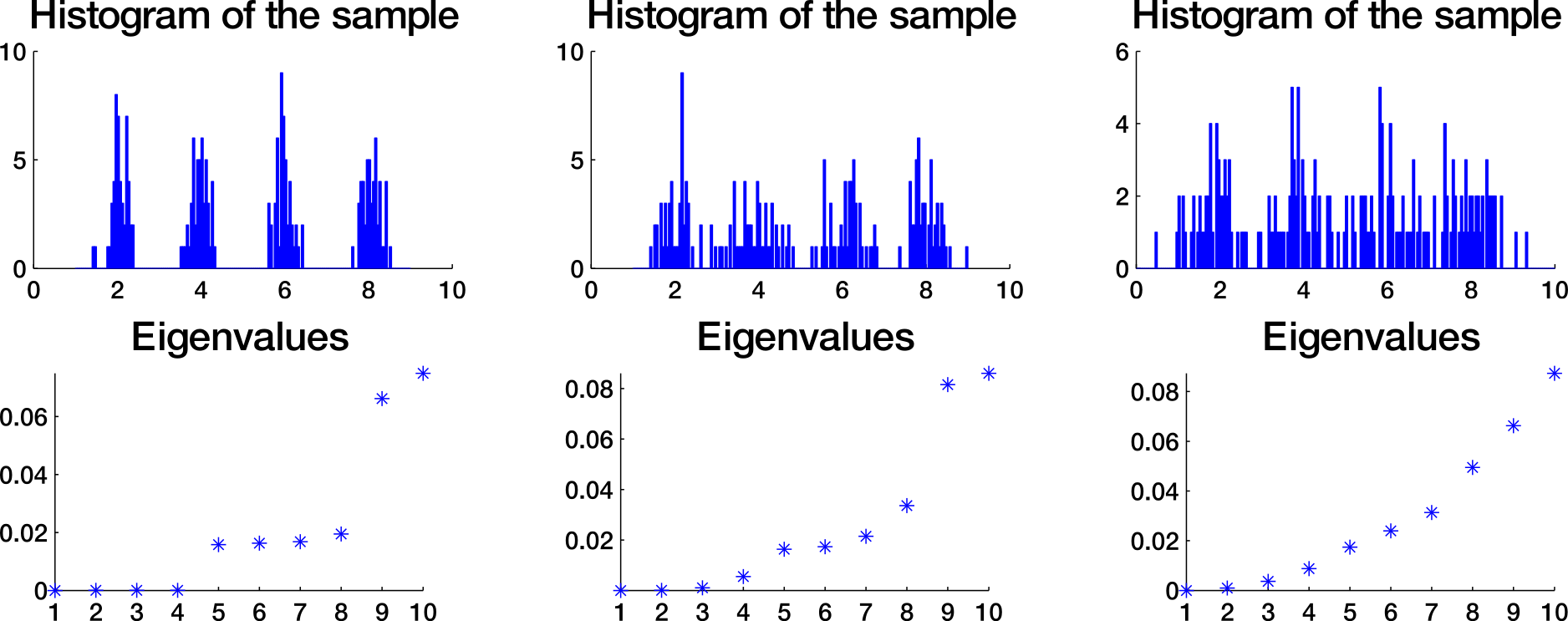
Eigengap heuristic
Spectral clustering algorithm
- Method to choose the number of clusters (eigengap)
- Based on strong theory
- Doesn't assume a certain shape for the clusters
- Problems when big matrices
- Tricky to define your similarity graph
Louvain's algorithm
Looking for communities/groups:
- many links inside a group
- few links between groups
Modularity
m: total number of edges
A: adjacency matrix
d: node degree
c: community membership
Optimize the modularity
m: total number of edges
A: adjacency matrix
d: node degree
c: community membership
Actual edge presence between v and w
Expected edge presence knowing the degrees and the total number of edges
Only count if v and w are classified in the same community
Sum over all pairs of nodes
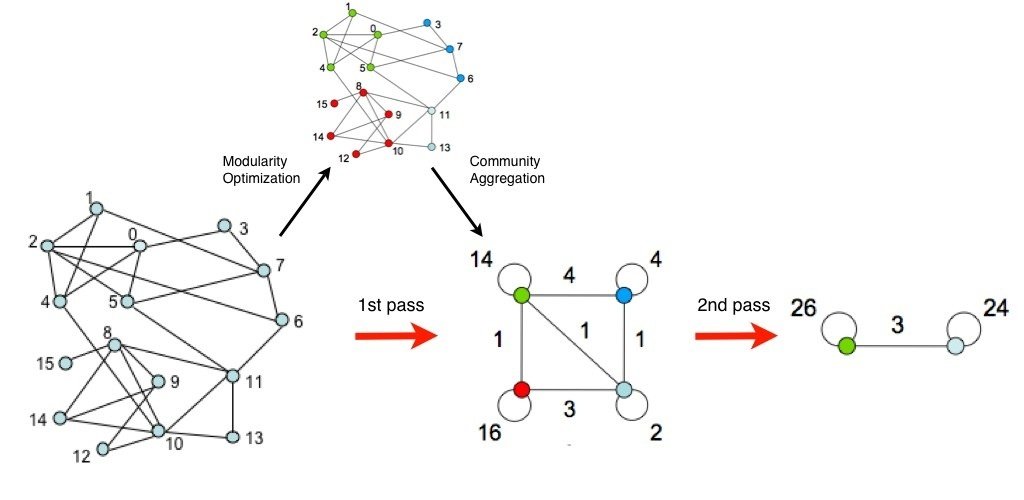
Louvain algorithm
Louvain algorithm
- Good because you don't choose the number of clusters you want
- Adapted to large graphs (www)
- Heuristics: depends on the order you aggregate nodes
- Tricky to define your similarity graph
- Can have problems of reproducibility
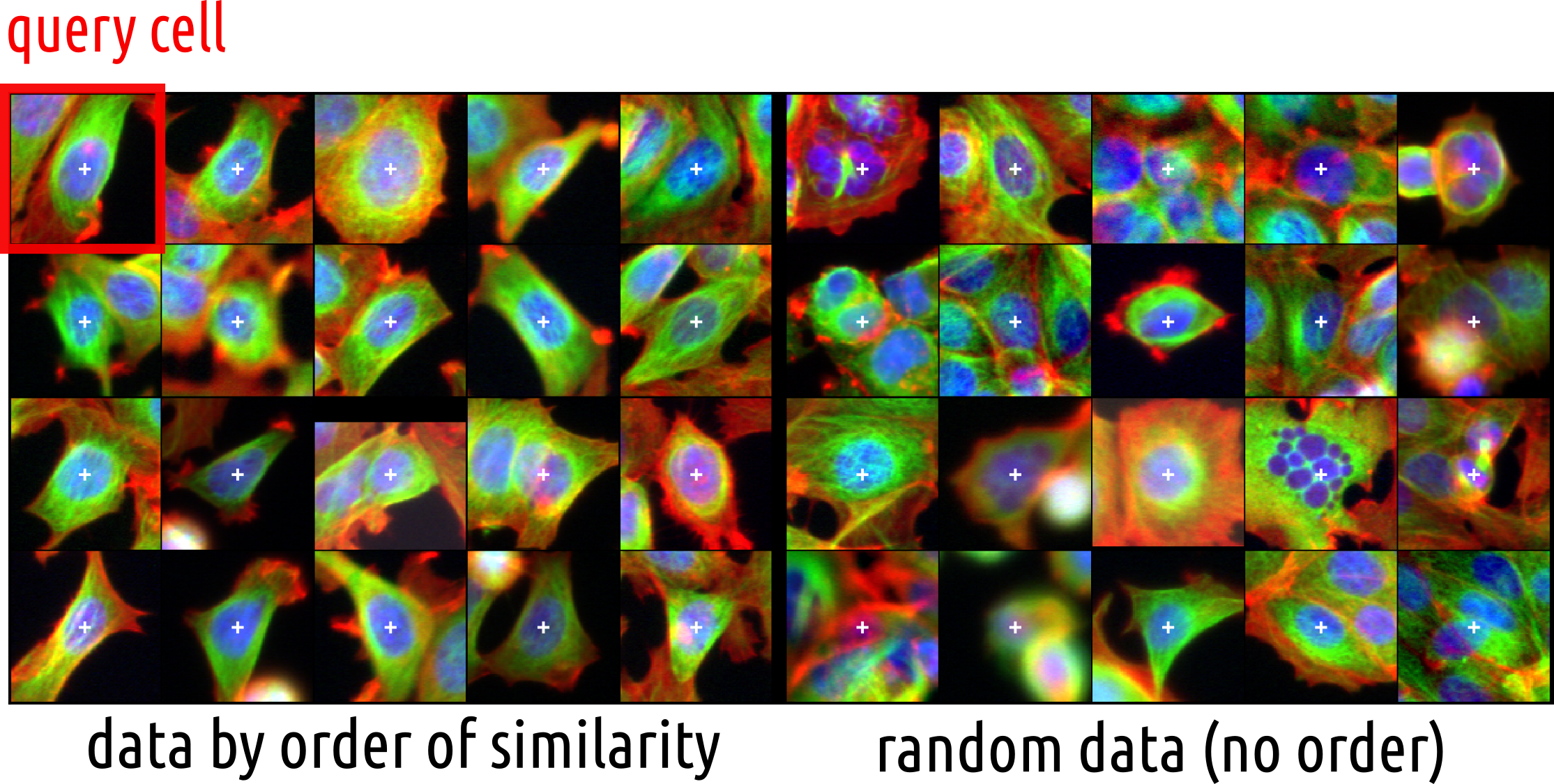
Using similarities between cells to cluster them
Clustering?
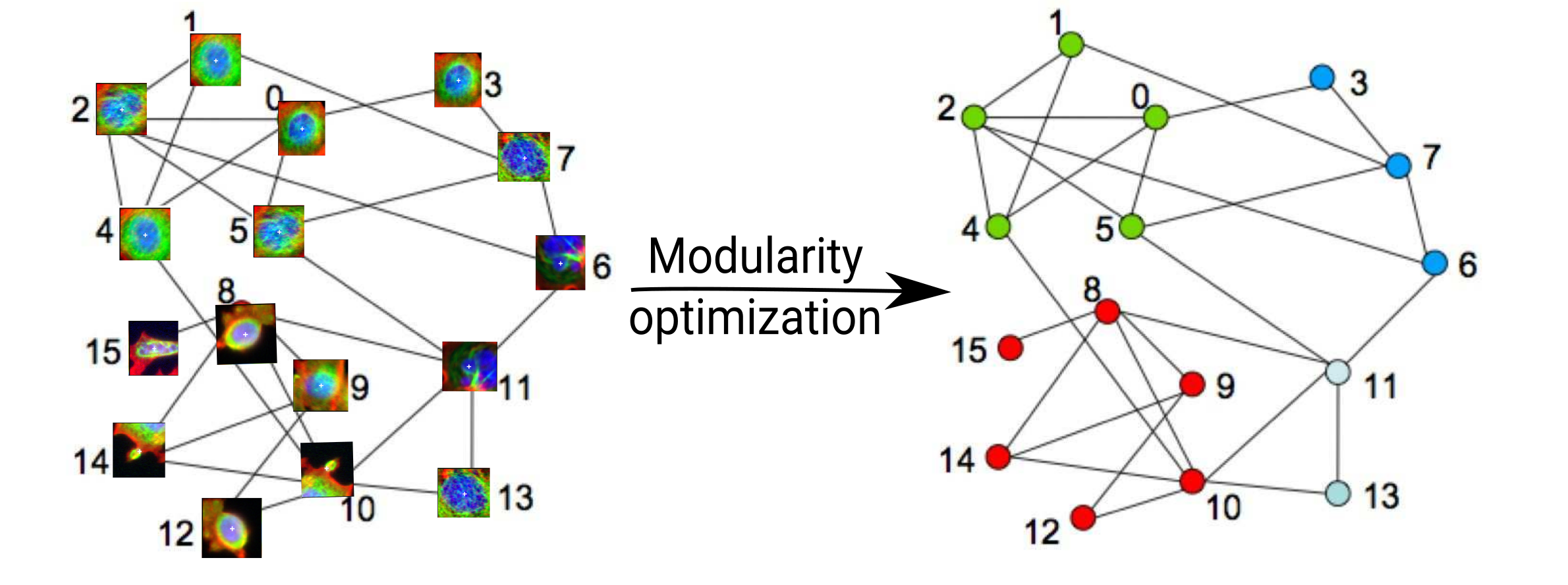
Applying Louvain algorithm
Cell categories
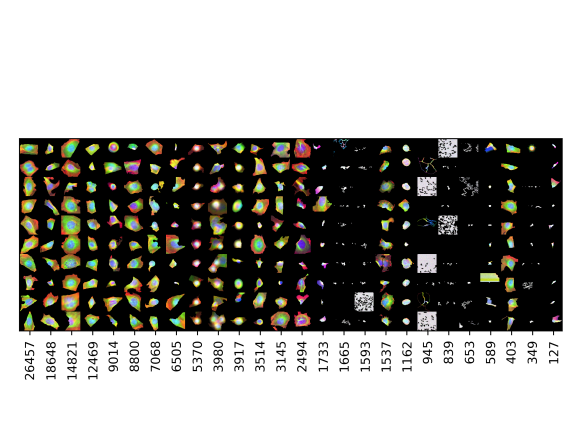
Cell categories
Number of cells
Examples of cells
Louvain algorithm
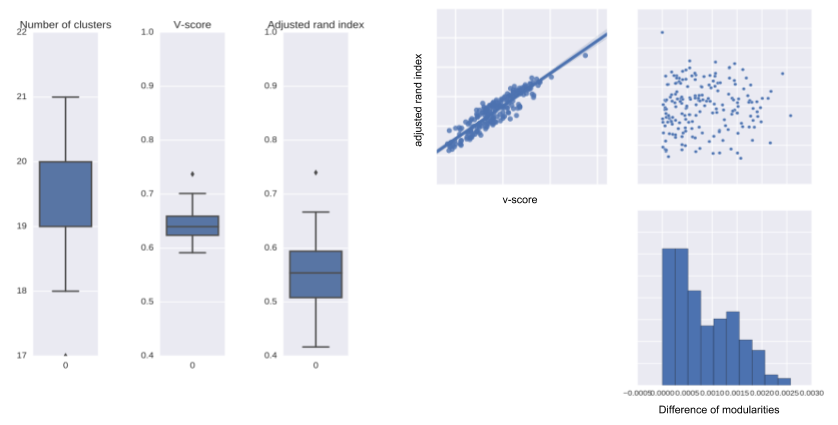
Take home message
- Similarity graphs describe relationships between data points
- Choosing a similarity measure can be tricky (high-dimension)
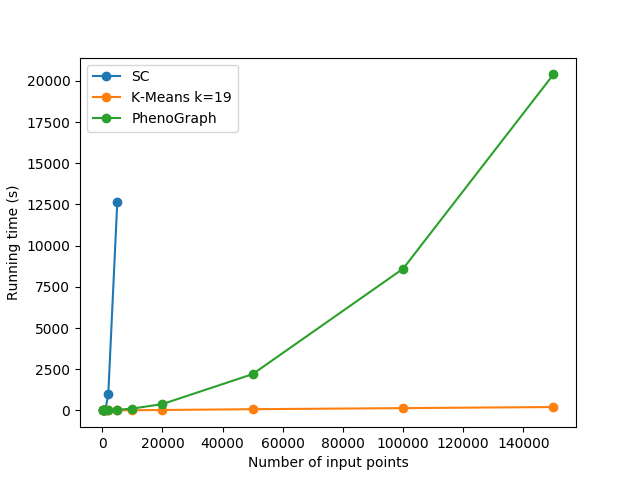
- Spectral Clustering: good if not too large dataset (memory)
- Louvain algorithm: heuristic to find communities in large data ! reproductibility !
Clustering with graphs:
By biocompibens



