Johannes Köster
2019
https://koesterlab.github.io

dataset
results
Data analysis
"Let me do that by hand..."

dataset
results
dataset
dataset
dataset
dataset
dataset


"Let me do that by hand..."
Data analysis
dataset
results
dataset
dataset
dataset
dataset
dataset
automation
From raw data to final figures:
- document parameters, tools, versions
- execute without manual intervention
Reproducible data analysis
dataset
results
dataset
dataset
dataset
dataset
dataset
scalability
Handle parallelization:
- execute for tens to thousands of datasets
- efficiently use any computing platform
automation
Reproducible data analysis
dataset
results
dataset
dataset
dataset
dataset
dataset
Handle deployment:
be able to easily execute analyses on a different system/platform/infrastructure
portability
scalability
automation
Reproducible data analysis
190k downloads since 2015
















Widely used

~3 new citations per week
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
shell:
"some-tool {input} > {output}"Concise and readable DSL
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
script:
"scripts/mytask.py"Python scripts
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
script:
"scripts/mytask.R"R scripts
import matplotlib.pyplot as plt
import pandas as pd
d = pd.read_table(snakemake.input[0])
d.hist(bins=snakemake.config["hist-bins"])
plt.savefig(snakemake.output[0])No boilerplate
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
script:
"scripts/mytask.py"rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
wrapper:
"0.24.0/bio/mytool"Reusable tool wrappers
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
cwl:
"https://github.com/some/cwl-tool"CWL integration

rule mytask:
input:
"path/to/{dataset}.txt"
output:
"result/{dataset}.txt"
script:
"scripts/myscript.R"
rule myfiltration:
input:
"result/{dataset}.txt"
output:
"result/{dataset}.filtered.txt"
shell:
"mycommand {input} > {output}"
rule aggregate:
input:
"results/dataset1.filtered.txt",
"results/dataset2.filtered.txt"
output:
"plots/myplot.pdf"
script:
"scripts/myplot.R"Implicit dependencies



workstation
compute server
cluster






grid computing
cloud computing



Scalability
rule mytask:
input:
"path/to/{dataset}.txt"
output:
"result/{dataset}.txt"
conda:
"envs/mycommand.yaml"
shell:
"mycommand {input} > {output}"channels:
- bioconda
- conda-forge
dependencies:
-mycommand =2.3.1Conda integration
rule mytask:
input:
"path/to/{dataset}.txt"
output:
"result/{dataset}.txt"
singularity:
"docker://some/container"
shell:
"mycommand {input} > {output}"Singularity integration
singularity: "docker://some/os"
rule mytask:
input:
"path/to/{dataset}.txt"
output:
"result/{dataset}.txt"
conda:
"envs/mycommand.yaml"
shell:
"mycommand {input} > {output}"Singularity + Conda
Interactive reports
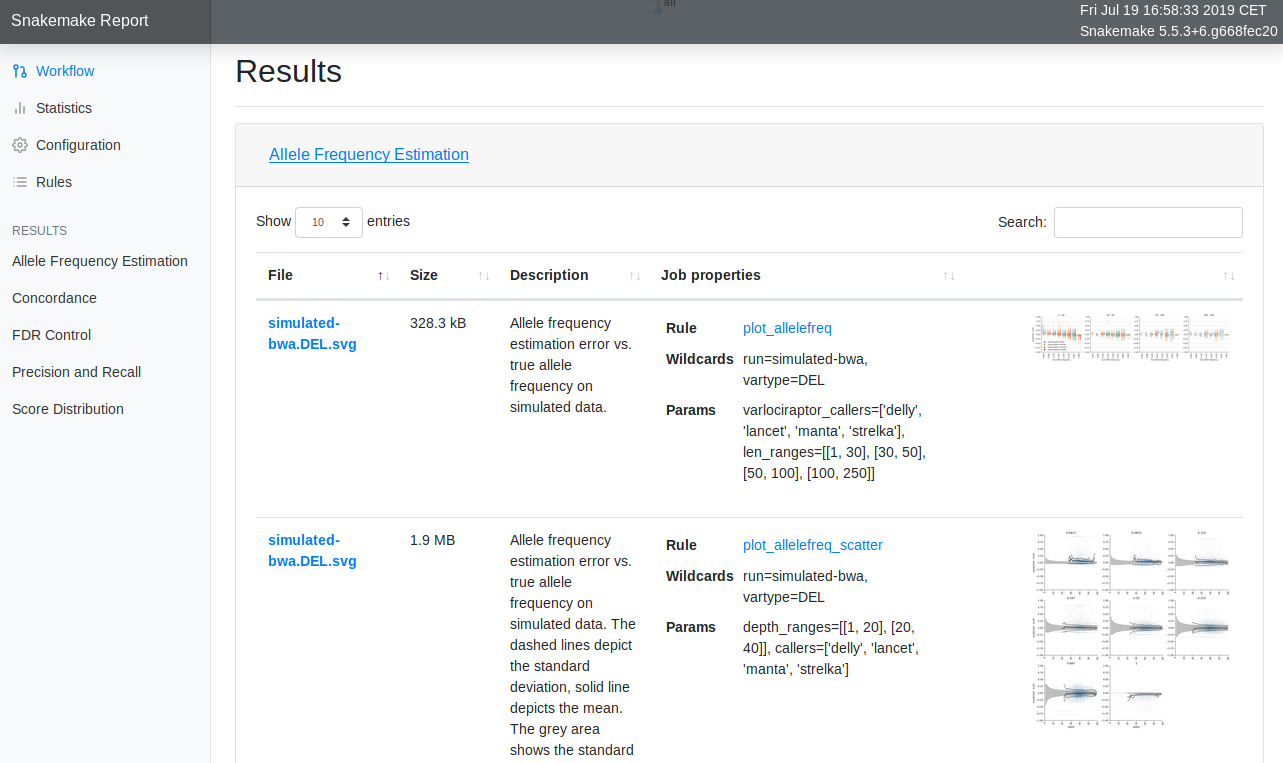
Interactive reports

Snakemake in short
By Johannes Köster
Snakemake in short
- 12,751



