Collective Cell Migration &
Cell-Cell Communication
Julien Varennes and Prof. Andrew Mugler

Collective Cell Migration
- A variety of different cells use inter-cellular communication in order to coordinate behavior.
- Groups of cells may use inter-cellular communication in order to better sense their environment.
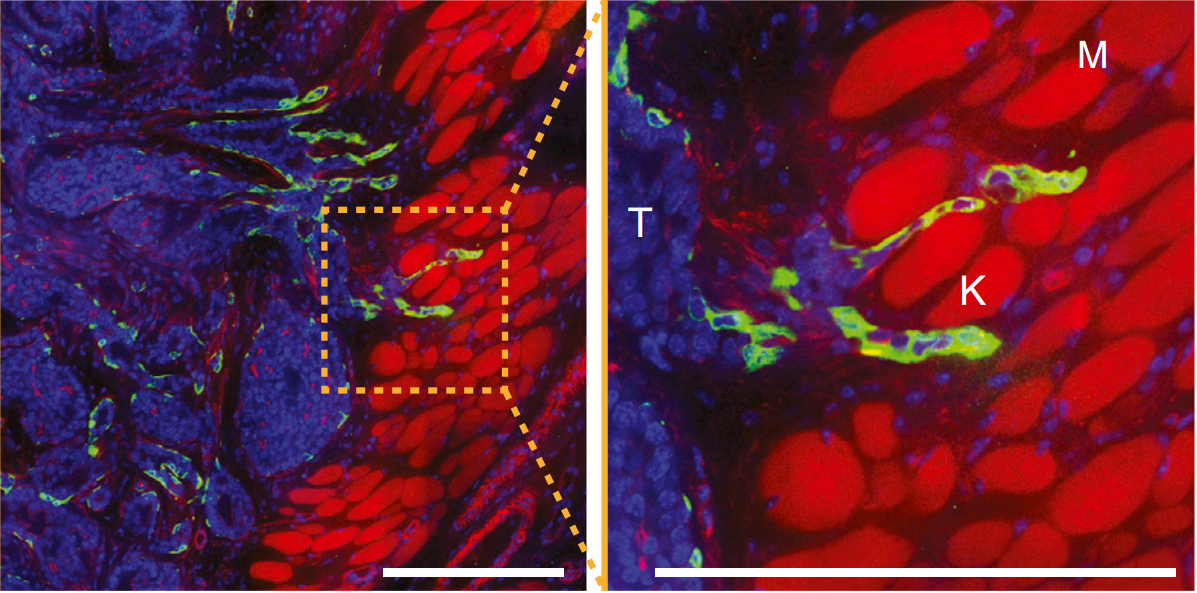
- Tumors exploit this trait during the invasion of tumor cells into the surrounding tissue.
200 \mu m
200μm
Tumor
Collective Cell Migration

- In the case of breast cancer, it has been observed that invasion occurs in the form of cell clusters.
- Recently, it has been shown that multicellular clusters can sense their environment with a higher precision than single cells.
Is there an optimal group size for most efficient collective cell migration?
Model Basics
- Cells migrate in groups.
- Cell's interact with their surroundings via adhesive energies.
- Cell-cell adhesion
- Cell-ECM adhesion
- Cell's interact with their surroundings via adhesive energies.
Model Basics
- Cells have a desired (resting) size
- Cell movement may be biased via cell polarization.
- Each cell has a polarization vector.
Cell-Cell Communication Model
- We use a minimal adaptive model based on local excitation and global inhibition (LEGI).
- S is the signaling molecule
- X is the local reporter, Y is the global reporter
- R is the downstream read-out
S \rightarrow S+X+Y
S→S+X+Y
X \rightarrow R
X→R
Y \dashv R
Y⊣R
Cell-Cell Communication Model
Reactions in each cell
s_k \rightarrow s_k + x_k: \ \kappa
sk→sk+xk: κ
s_k \rightarrow s_k + y_k: \ \kappa
sk→sk+yk: κ
x_k \rightarrow \emptyset: \ \mu
xk→∅: μ
y_k \rightarrow \emptyset: \ \mu
yk→∅: μ
y_k \rightleftharpoons y_{k\pm1}: \ \gamma_{k,k\pm1}
yk⇌yk±1: γk,k±1
R_k = x_k - y_k
Rk=xk−yk

S \rightarrow S+X+Y
S→S+X+Y
X \rightarrow R
X→R
Y \dashv R
Y⊣R

How does communication influence cell migration?
- We assume that cells that are in contact are repelled from one another.
- This affects the dynamics via the cell's polarization.
\vec{p}_k \equiv \text{cell polarization,} \ \ \vec{q}_k \equiv \text{cell repulsion}
p⃗k≡cell polarization, q⃗k≡cell repulsion
\frac{d\vec{p}_k}{dt} = -\frac{1}{\tau} \vec{p}_k + \epsilon R_k \ \vec{q}_k
dtdp⃗k=−τ1p⃗k+ϵRk q⃗k
polarization
repulsion
Implementation: Cellular Potts Model

Cell behavior is simulated using CPM.
u = \sum_{i,j} J_{i,j} + \lambda \sum_{i=1}^N (V_T-V_i)^2
u=∑i,jJi,j+λ∑i=1N(VT−Vi)2

\alpha
α
\beta
β
\sigma_i \in 1, 2, 3, ...
σi∈1,2,3,...
\sigma_i = 0
σi=0
For individual cells:
For ECM:
Implementation: Cellular Potts Model

A change in the spin value of a pixel is accpeted with the following probabiliy.
w = \sum_{k = \sigma_a, \sigma_b} \frac{\Delta\vec{x}_{k(a \to b)} \cdot \vec{p}_k}{ |\Delta\vec{x}_{k(a \to b)}|}
w=∑k=σa,σb∣Δx⃗k(a→b)∣Δx⃗k(a→b)⋅p⃗k

u = \sum_{i,j} J_{i,j} + \lambda \sum_{i=1}^N (V_T-V_i)^2
u=∑i,jJi,j+λ∑i=1N(VT−Vi)2
Chemical Concentration
Collective Cell Migration
By Julien Varennes
Collective Cell Migration
- 1,578

