visbio
Metabolic maps for the masses
Zak King, Ali Ebrahim, Niko Sonnenschein
Winter retreat
Dec. 19, 2013
What is visbio?
A collection of web visualizations for systems biology.
Heat maps,
interactive scatter plots,
dendrograms, ...
But, for now,
it's all about the
maps.
Why I am doing this?
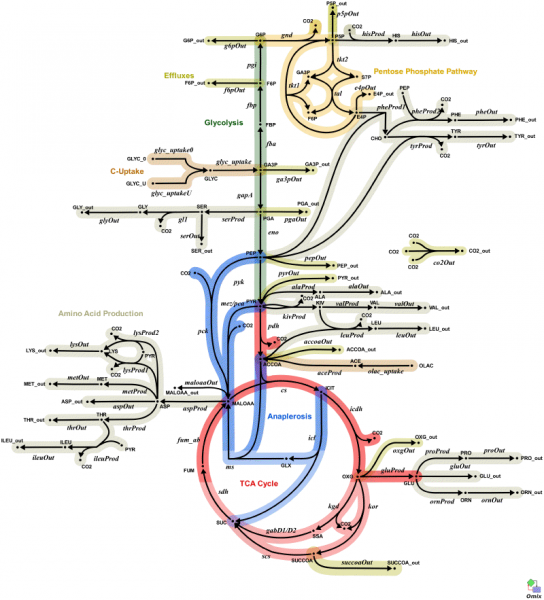
Visualizing metabolism is hard

Human metabolism:
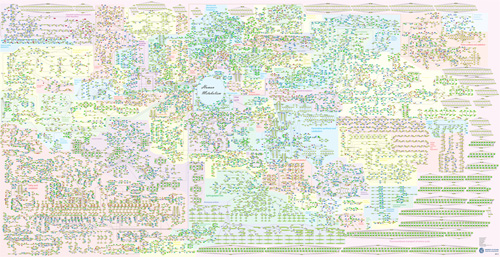
Is this better?
KEGG
Specialized figures are the most informative:
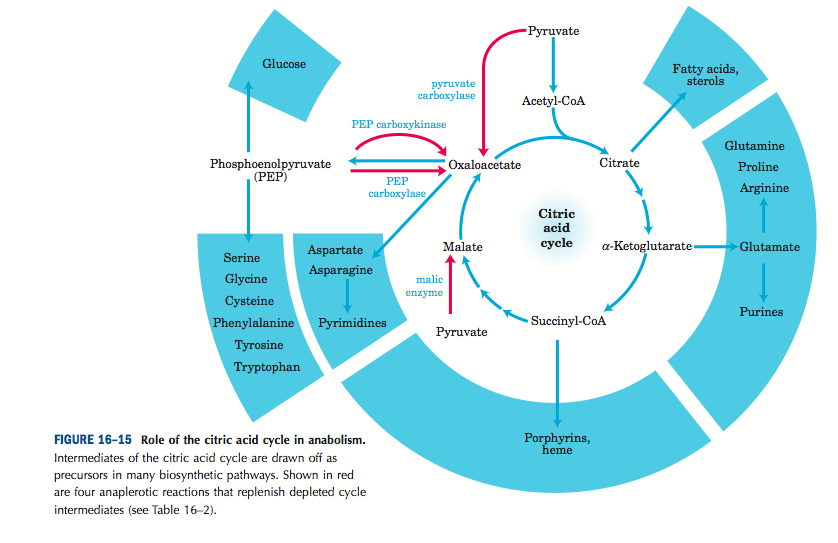
Lehninger, Principles of Biochemistry, 4th Ed.
And some nice tools exist
to build them:
Omix
SimPheny is one
of the best!
But what about data integration?
Customized figures for publication?
Sharing?

-
Load SimPheny maps
- Apply any dataset
- Build features on the fly
- Share in any browser, including mobile browsers
- Run from Python and IPython notebooks (& NBViewer)
How does import work?
Demo time!
-
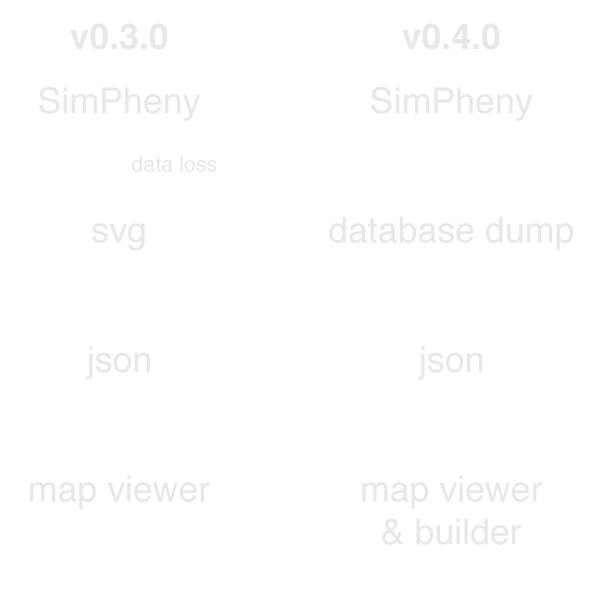
v0.3.0
-
Metabolic maps in Python, IPython, & web
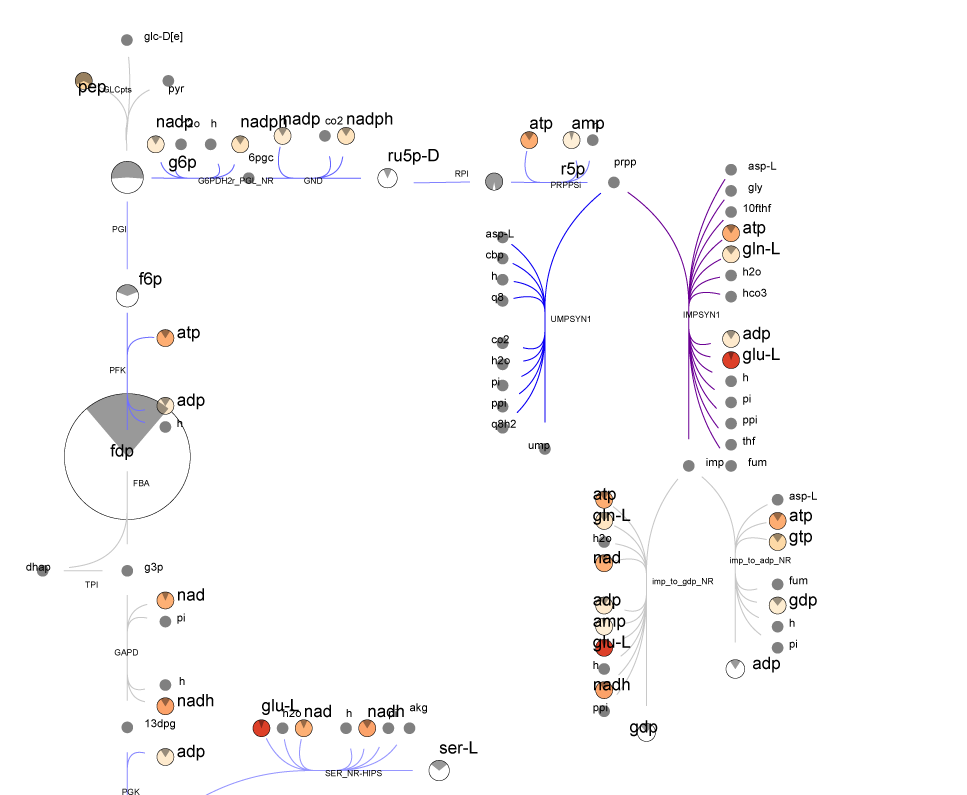
McCloskey, D. et al. Biotechnology and Bioengineering. 1–31 (2013).
Coming Soon!
-
v0.4.0
-
View and edit maps
-
Direct import from BIGG database
Let's share the maps
- We have 828 maps from SimPheny
- We could curate and share the collection online
-
Community development = free features

 What do you think?
What do you think?Getting started
↓
a) Download
Or,
> git clone git@github.com:zakandrewking/visbio.git
b) Install Python package
> python setup.py install
In Python and IPython, you can:
import visbio
map = visbio.Map()
map.create_standalone_html()
map.view_browser()
More instructions at github.com/zakandrewking/visbio
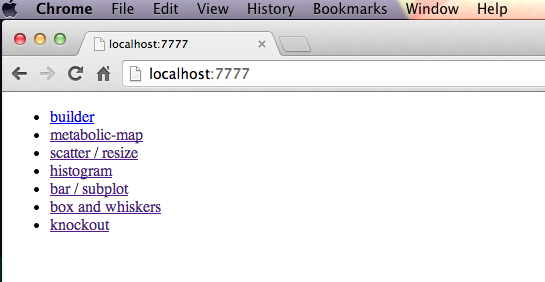
c) Run local server
The only dependencies are Python, tornado, and cobrapy.
> python server/tornado_main.pyserving directory /Users/zaking/www/visbio-public on port 7777
What's next:
visbio: Metabolic maps for the masses
By zakandrewking
visbio: Metabolic maps for the masses
- 2,787