Galaxy for Genomic Data Science

What is Galaxy?
- Galaxy is an open,web-based platform for data intensive bioinformatics research
- All tools required for Bioinformatics Data Analysis are accumulated in a single platform.
- It is maintained by John Hopkins and Penn State University with support from NSF.
- Galaxy provides the required cloud platform to run your calculations and data processing.
- One can choose from over 70+ Public Galaxy Servers for working on Bioinformatics.
- If not,You can run your Galaxy instance locally or by creating your own cloud server.
Galaxy 101
Working in Galaxy
- To use the resources of Galaxy,you need an account.Go to the Link to make a new account.
- One can import your own datasets into the Galaxy instance or one can obtain data from the Table Browser.
- We will look at a simple example of finding the number of coding exons with highest number of variations in them.
Storing Workflows
- The Galaxy also enables scientists and bioinformaticians to share their research in the form of workflows.
- Workflows, as a part of reproducible research,can be extracted easily from a history in Galaxy.
- Galaxy also allows for editing of workflows.
- You can also import workflows into your Galaxy account to work on and reproduce the results of a specific research.
Running Galaxy locally
- One can run a Galaxy instance locally and customize it according to their requirements.
- Requirements for running Galaxy locally-
- UNIX/Linux or Mac OS X
- Python 2.6 or 2.7
- Git(optional)
- GNU Make,gcc to compile and install tools and dependencies
To run Galaxy,Download the latest source from Github:
% git clone https://github.com/galaxyproject/galaxy/Change to the directory where Galaxy is downloaded
% cd galaxy
% git checkout -b master origin/masterStart up the project
% sh run.shConfigure your Port if the default port is occupied by editing the file galaxy/config/galaxy.ini
host = 0.0.0.0Add Root Users
# this should be a comma-separated list of valid Galaxy users
admin_users = user1@example.com,user2@example.comRunning Galaxy on Cloud
- To make your own Galaxy on Cloud,you will require an account in Amazon AWS.
- After getting an account in Amazon AWS,log into the Management console and set AWS region to US East.
- Make a Key Pair for your Galaxy Console and save it carefully for later use.Create Security Group and Inbound rules for the group
- After this,Create your own instance for Galaxy by supplying cluster name,password and putting your Access Key and Secret Key.
- Load up your Galaxy Instance by going to the link of your master instance.
Setting up AWS for your Galaxy Instance
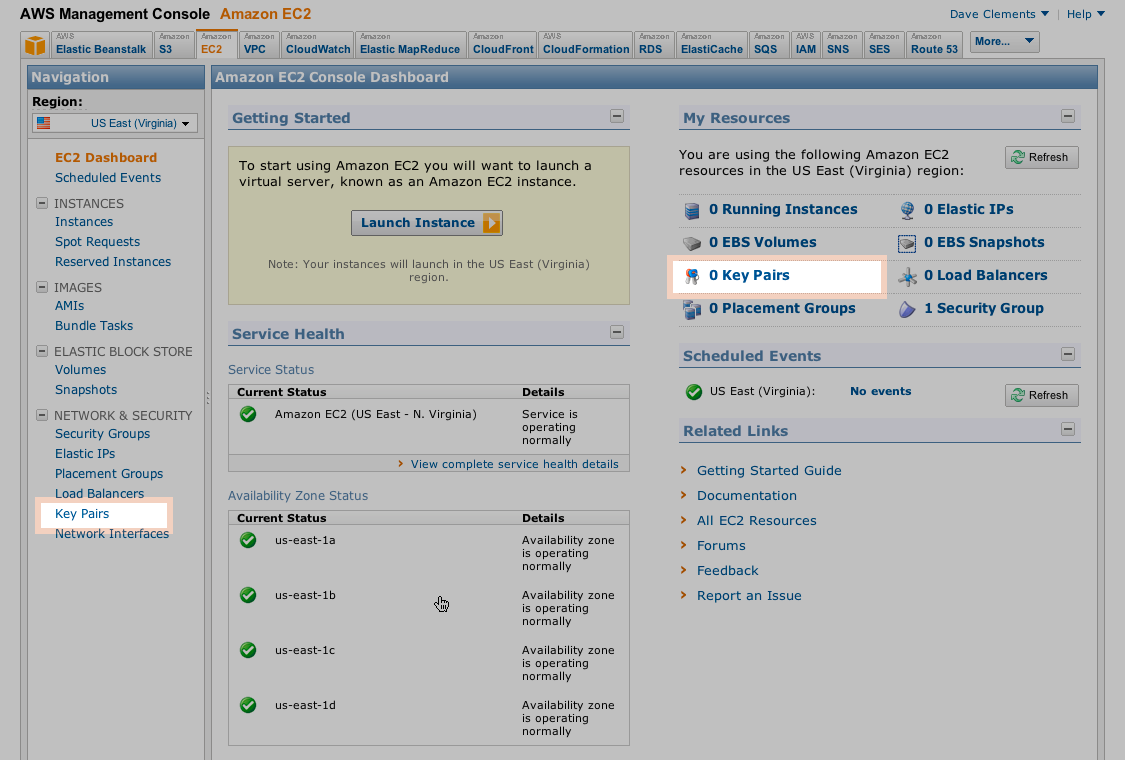
Adding Key Pairs
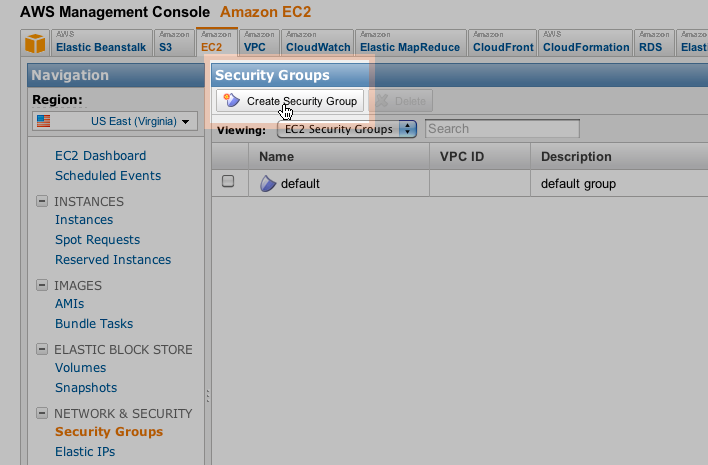
Create a Security Group
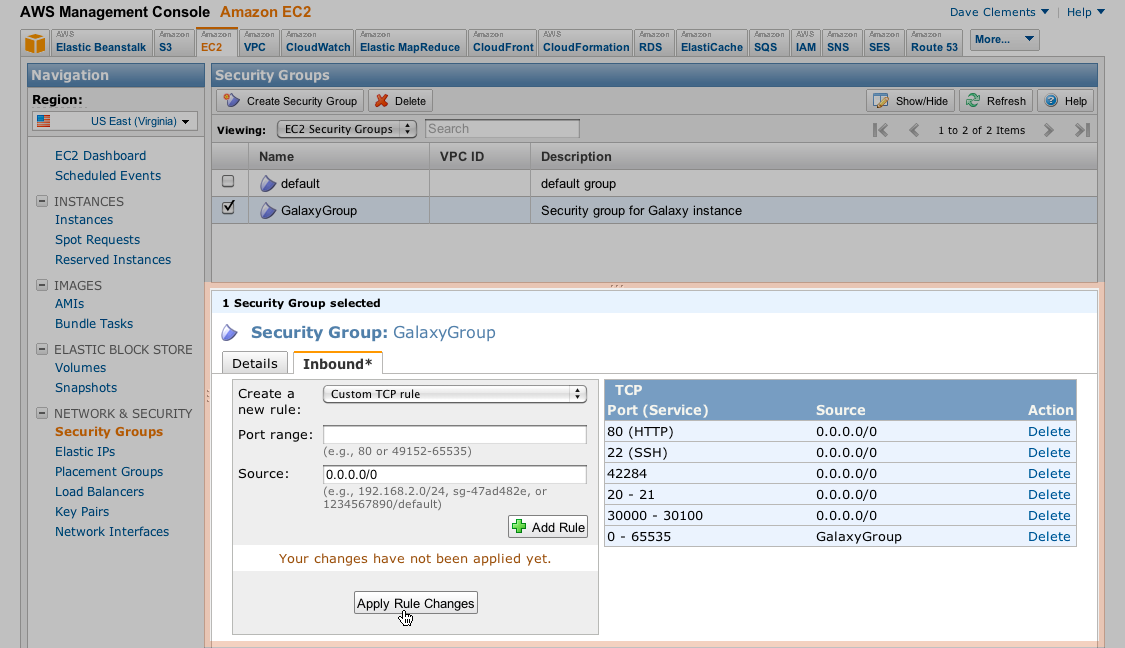
Apply Rules for your Security Group
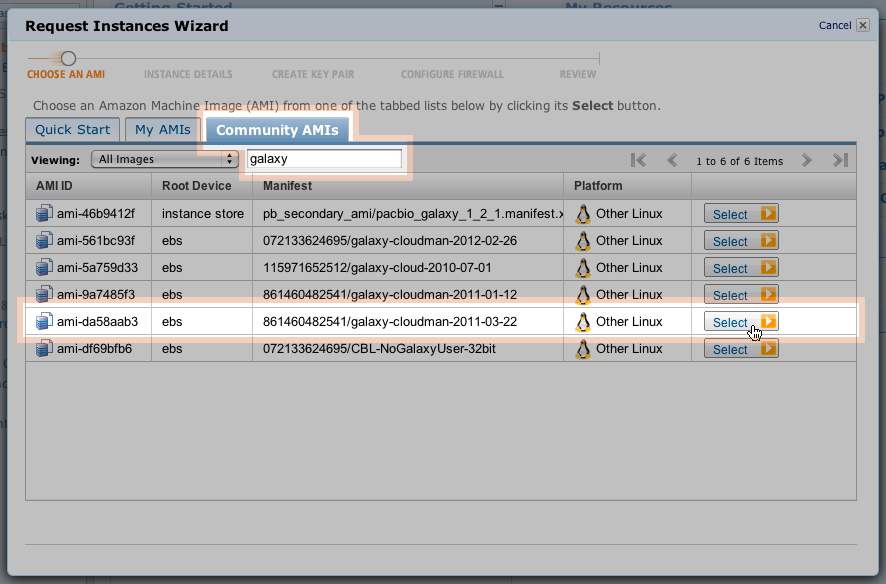
Select the AMI required
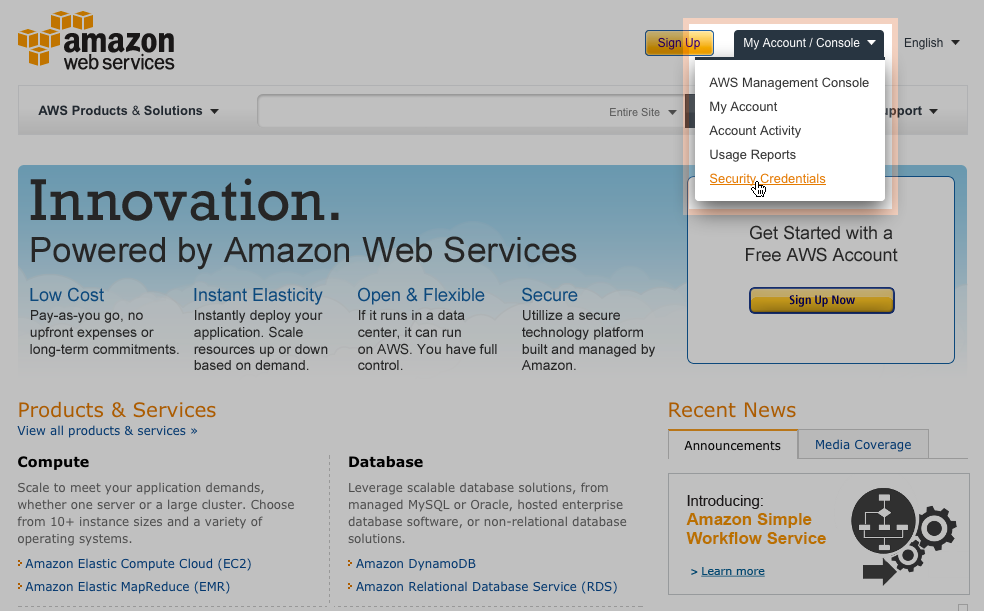
Login with Security Credentials
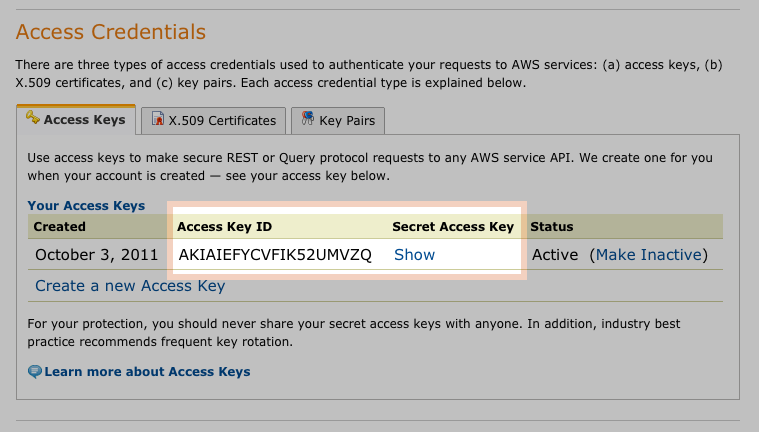
Give Access Credentials
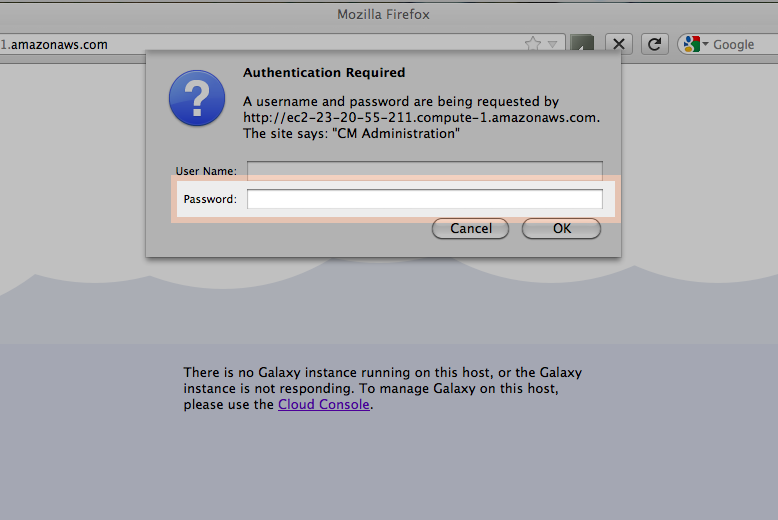
Login into your Amazon Instance
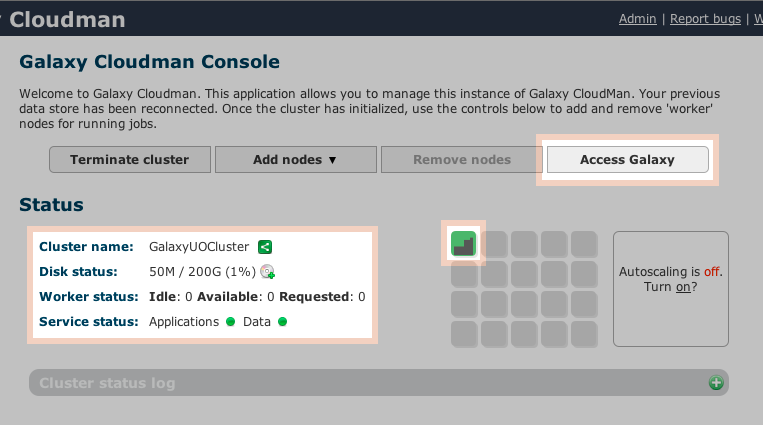
Add nodes for your Galaxy Cloudman Console
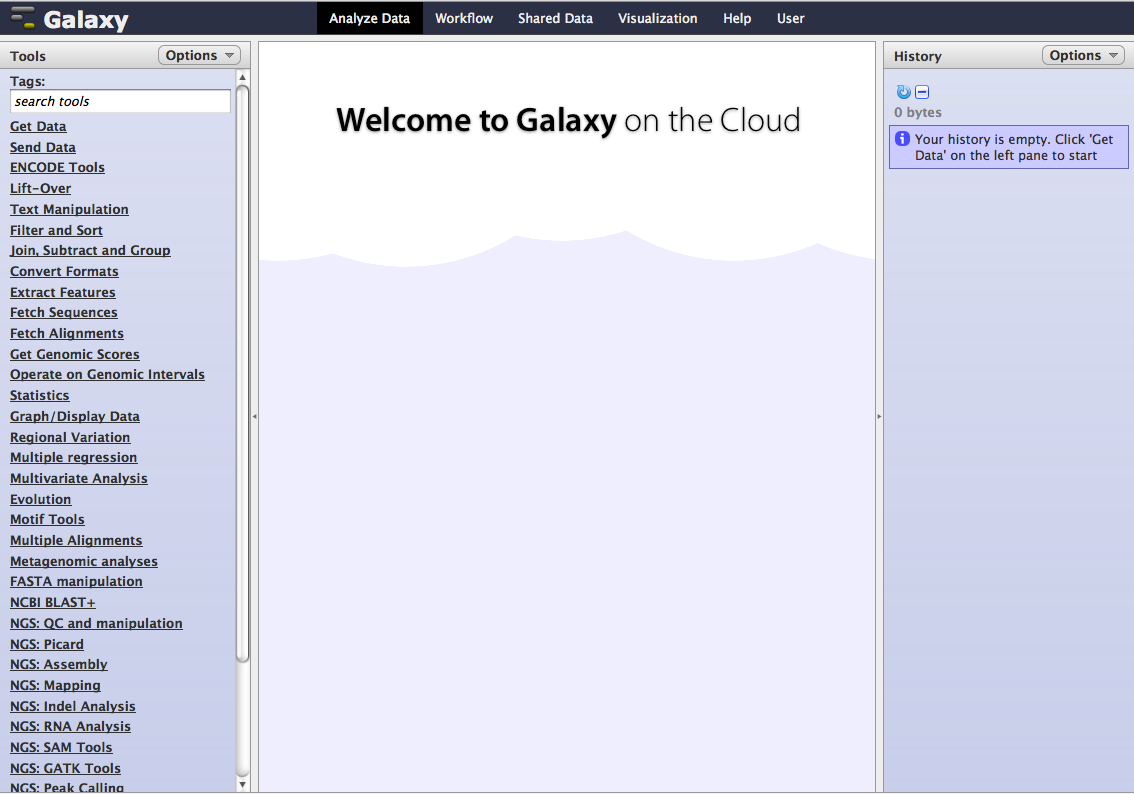
Welcome to Galaxy on Cloud!
For help and other FAQs
- To learn how to use Galaxy and more tutorials,Here is the Tutorial link
- For deploying Galaxy and deploying your own sets of tools for Galaxy,One can refer this link on Galaxy Toolshed.
- For Problems in Running a Galaxy functionality,Galaxy has a nice forum in Biostars
- For Development of Galaxy,Refer to the Project page for Galaxy.
Special Thanks
- Jennifer Hillman-Jackson
- Dannon Bakker
- Daniel Blankenberg
- Nicholas Stoler
- James Taylor
Questions?
Thank You
Galaxy for Genomic Data Science
By Sourav Singh
Galaxy for Genomic Data Science
- 2,034

