
Computational Biology Seminar
Class 03

September 12, 2023
(BIOSC 1630)
Feedback: Homework load has been too much

We have a minimum amount of pages to write

Reading literature—the main purpose of this course—takes lots of time

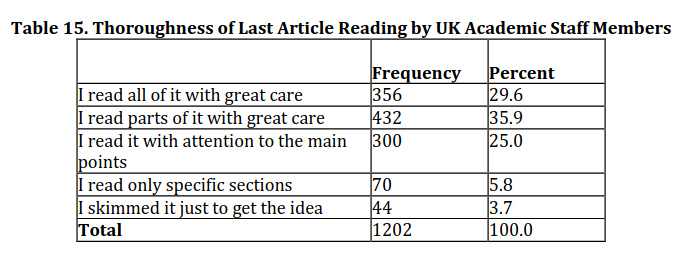
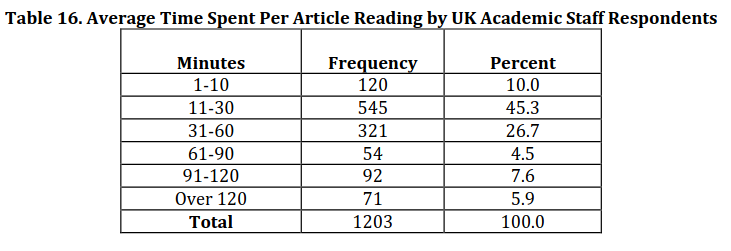
Academics in the UK spend, on average, 49 minutes reading one article
Tenopir, 2012, URL
Assignment 01 was more work than I intended
Submissions contained, on average, 900 words

Active reading asked for too many bullet points
Assignment 02 deadline was extended to Sep. 20
Reduced the

- number of proposed papers to two (instead of three)
- number of references
Feedback: Not getting much from class

I scaffold where you are reading

Read literature on your own outside of class
Read literature where I can immediately provide assistance
Semester start
Semester end
I design my activities to have realistic context

Read literature for homework and critically discuss during class
Journal club:
I do not find this engaging enough (for me or you)
I will ignore feedback that is not constructive

Today's learning objectives

-
Apply critical thinking skills to assess the suitability of research methods for addressing specific research questions.
-
Analyze and critique the limitations and potential biases of chosen research methods.
-
Explain the purpose of specific computational methods and algorithms in the context of computational biology.
-
Evaluate the performance of computational algorithms by comparing their results to experimental data or gold standard benchmarks.
We will be reading and evaluating two method papers for protein function annotation

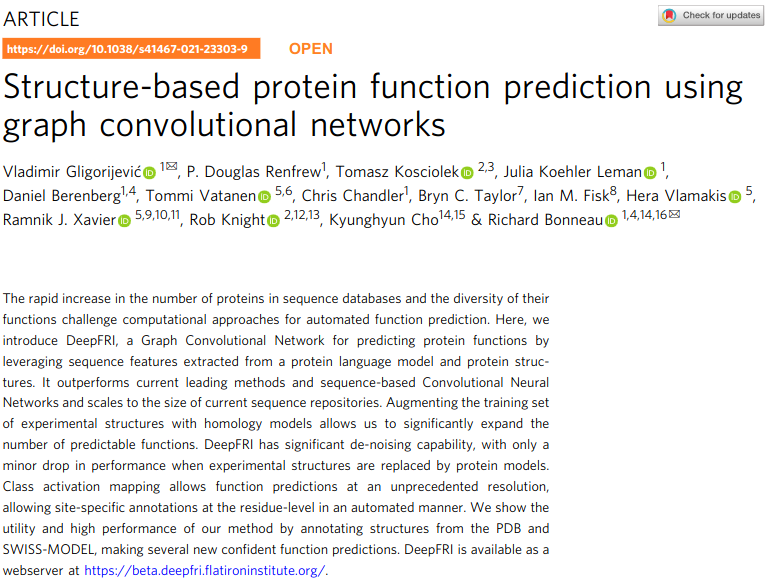

Bileschi, 2022,
DOI: 10.1038/s41587-021-01179-w
Gligorijević, 2021,
DOI: 10.1038/s41467-021-23303-9
Protein function annotation is a standardized label for what a protein does

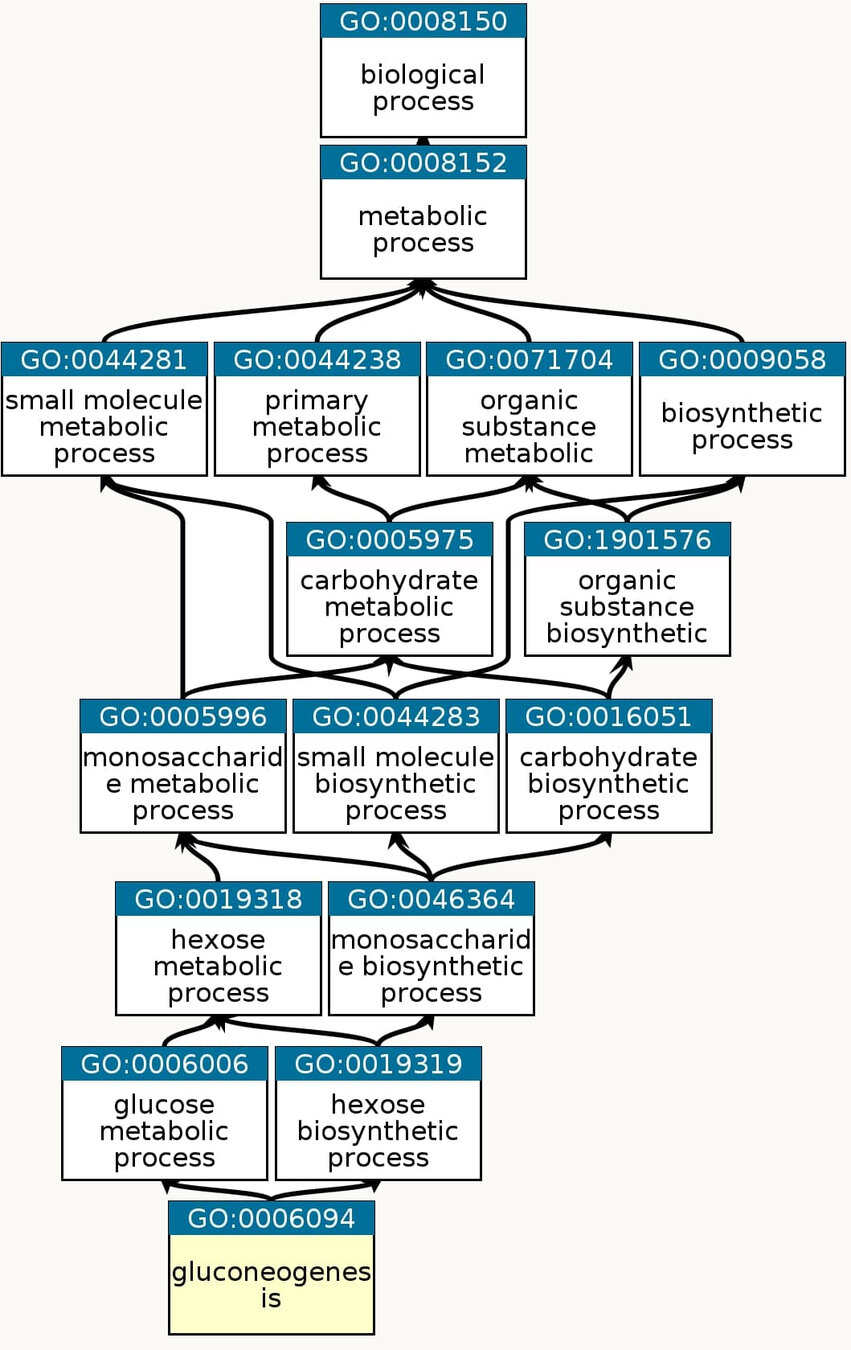
Gluconeogenesis: The formation of glucose from noncarbohydrate precursors, such as pyruvate, amino acids and glycerol
More information is here
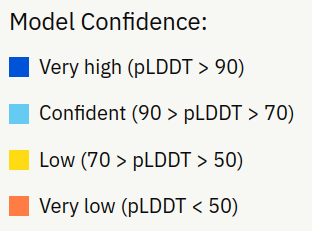
Glucose-6-phosphate isomerase from Acinetobacter baumannii has unknown structure

AlphaFold structure prediction
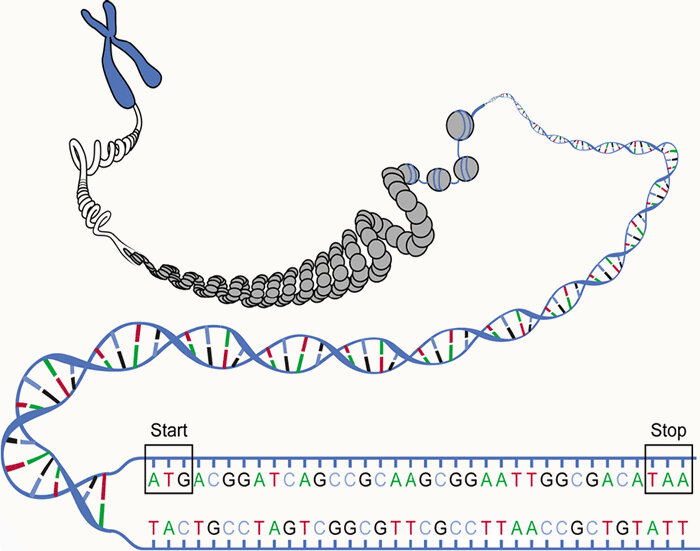
How do we figure out what a new protein-coding gene does?

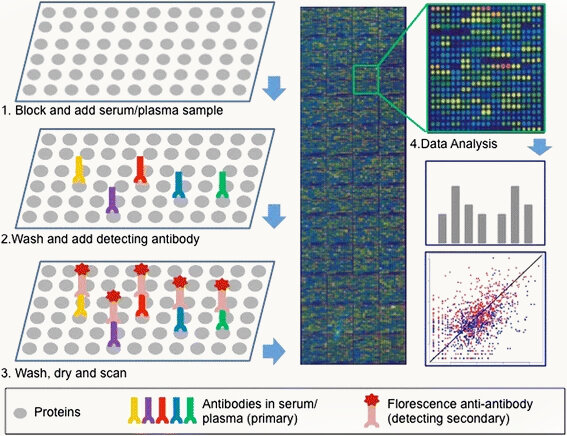
Protein microarrays can (expensively) indicate function through protein-protein interactions

Yuan, 2016, DOI: 10.1186/s40425-016-0106-4
Computational prediction has a shorter time to answer but still eventually needs experimental validation

Convolutional neural networks use parameterized filters
Graph neural networks
03-Methodology
By aalexmmaldonado
03-Methodology
- 289



