GETTING STARTED WITH
COMPUTATIONAL REPRODUCIBILITY
Neha Moopen
Research Data Manager
2022-05

Bianca Kramer
Subject Specialist: (Bio)medical Sciences
Text
THESE SLIDES ARE ADAPTED FROM
Best Practices in Writing Reproducible Code
@UtrechtUniversity
@NEONScience
&
WHAT IS REPRODUCIBILITY?
Computational reproducibility is when detailed information is provided about code, software, hardware and implementation details (Victoria Stodden, 2014).
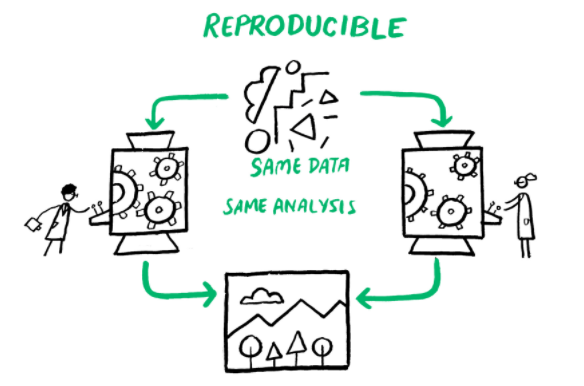
This image was created by Scriberia for The Turing Way community and is used under a CC-BY licence. The image was obtained from https://zenodo.org/record/3332808.
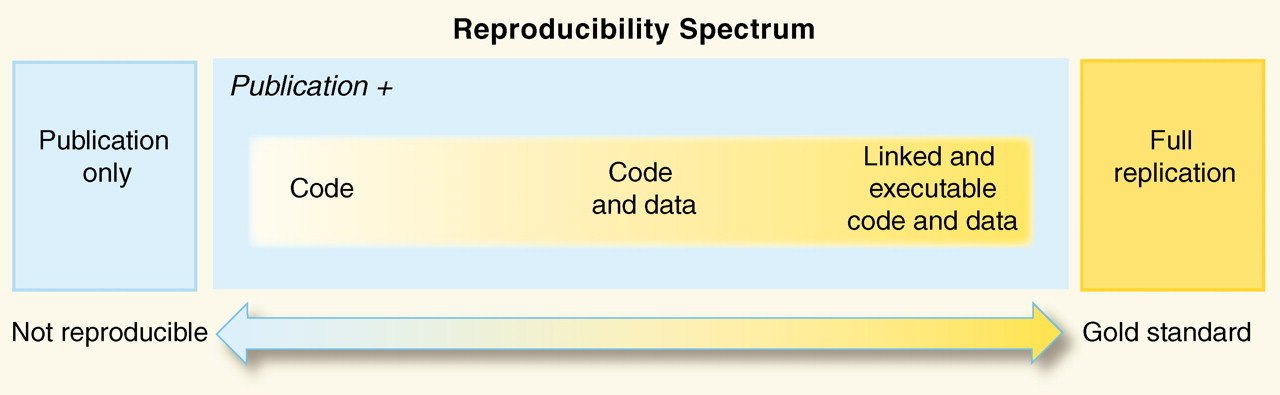
REPRODUCIBILITY SpECTRUM
A study may be more or less reproducible than another depending on what data and code are made available (Peng, 2011).
WHY IS THIS IMPORTANT?
- To show evidence of the correctness of your results.
- To enable others to make use of our methods and results.
The stakeholders involved:
- Collaborators
- Peer reviewers & journal editors
- Broad scientific community
- The public
- You!
If you need more convincing, see also: Five selfish reasons to work reproducibly (Florian Markowetz, 2015)
BEING REPRODUCIBLE

IS THIS ENOUGH?
- Access to the code
- Access to the data
-
(And let's assume we can replicate the environment)
How confident do you feel?

We need to do more: we need to inspire trust.
- The code is correct (and I have made it easy for you/someone to check);
- My workflow is robust;
- My workflow itself is accessible, and I will be guiding you through it.
THE FOUR FACETS OF REPRODUCIBILITY
ORGANIZATION
DOCUMENTATION
AUTOMATION
DISSEMINATION
THE FOUR FACETS OF REPRODUCIBILITY
ORGANIZATION
DOCUMENTATION
AUTOMATION
DISSEMINATION
ORGANIZATION
ORGANIZATION
WHY DO YOU NEED IT?
-
Your future self will be able to quickly find files.
-
Colleagues will be able to more quickly understand your workflow.
- Machine-readable names can be quickly and easily sorted and parsed.
ORGANIZATION
HOW CAN YOU DO IT?
Address folder structure:
-
Contain your project in a single recognizable folder.
-
Distinguish folder types, name them accordingly:
- Read-only: (raw) data, metadata
- Human-generated: code, paper, documentation
- Project-generated: clean data, figures, models...

Source: Wilson et al. (2017)
ORGANIZATION
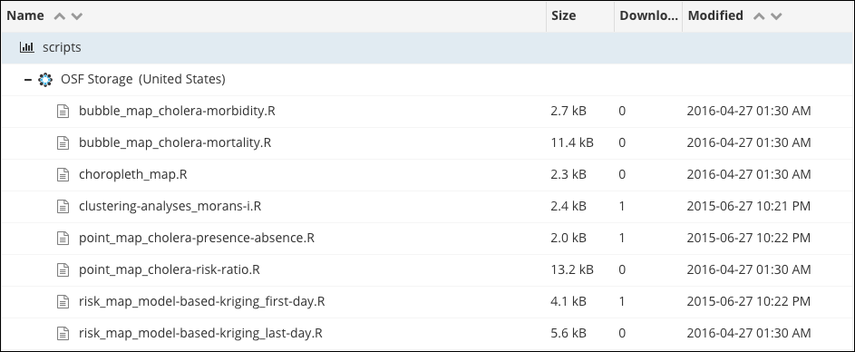
Machine-readable and Human-readable
names ->
<- Names that support sorting
source: OSF's File naming Guide
Address file & folder naming:
ORGANIZATION
File organization should:
- Reflect inputs, outputs and information flow
- Preserve raw data so it's not modified
- Carefully document & store intermediate & end outputs
- Carefully document & store data processing scripts
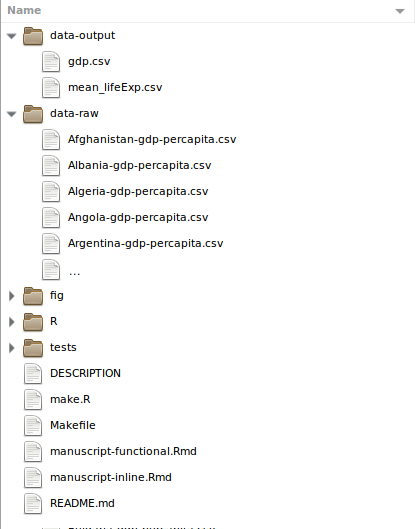
source: Intro to Reproducible Science @NEONScience
ORGANIZATION
File renaming tools:
-
Bulk Rename Utility (for Windows)
-
Renamer (for MacOS)
-
PSRenamer (for MacOS, Windows, Unix, Linux)
-
WildRename (for Windows)
ORGANIZATION
VERSION CONTROL!
FOR A Living project: git & GIThub
(or other social coding platform):
-
synergistic with version control software git
-
makes history public and accessible (eek!)
-
allows publication of different releases
-
provides a platform for interaction and collaboration
FOR ARCHIVING RELEASES: ZENODO


ORGANIZATION
WHAT IS GIT?
-
Allows you to log updates, branch your work (so you can experiment without losing the original!), keep all backups, while efficiently using your storage
-
Gives the user a lot of control on what to track, and adds a narrative to changes ('commit comments')
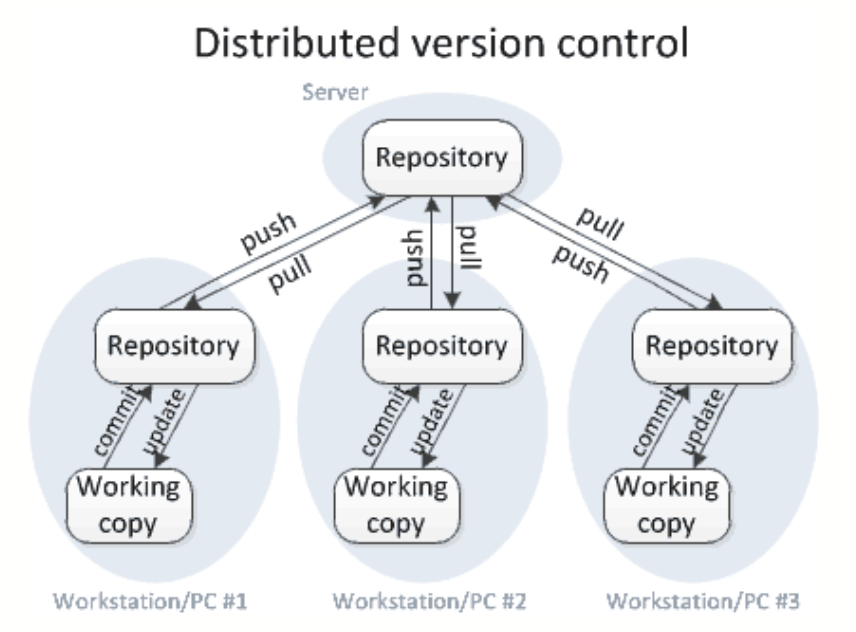
ORGANIZATION
DO: Commits should be atomic: comprehensive 'units' of changes.
DON'T: edit for a full day and put this in a single commit (or worse: forget to...)
Commits should have informative messages so you (and others) can trace your steps
Track most files; .gitignore those files you don't.
Explore new ideas with branches, keep a stable version on master
HOW TO GIT?

source: https://xkcd.com/1296/
ORGANIZATION
WHY DO YOU NEED VERSION CONTROL?

source: phdcomics.com
ORGANIZATION: DISCUSSION
Are there changes you can make in your file & folder naming/organization?
Has anyone use Git/GitHub to version control, share, find code?
THE FOUR FACETS OF REPRODUCIBILITY
ORGANIZATION
DOCUMENTATION
AUTOMATION
DISSEMINATION
DOCUMENTATION
DOCUMENTATION
WHY DO YOU NEED IT?
-
You want yourself to understand how code written some time ago works
-
You want others to understand how to (re-)use your code
- Future 're-analysis' of your data is more efficient.
DOCUMENTATION
HOW CAN YOU DO IT?
-
Explain your code with comments.
-
Explain what to install and how to get started in your README.
-
Explain in-depth use of your code in a notebook.
DOCUMENTATION
Comments are annotations you write directly in the code source.
They:
-
are written for users who deal with your source code
-
explain parts that are not intuitive from the code itself
-
do not replace readable or structured code
-
(in a specific structure) can be used to directly generate documentation for users.
Comic source: Geek & Poke
DOCUMENTATION
The README page is the first thing your user will see!
The contents typically include one or more of the following:
- Configuration instructions
- Installation instructions
- Operating instructions
- A file manifest (list of files included)
- Copyright and licensing information
- Contact information for the distributor or programmer
- Known bugs
- Troubleshooting
- Credits and acknowledgments
Reference: Wikipedia's README page
DOCUMENTATION
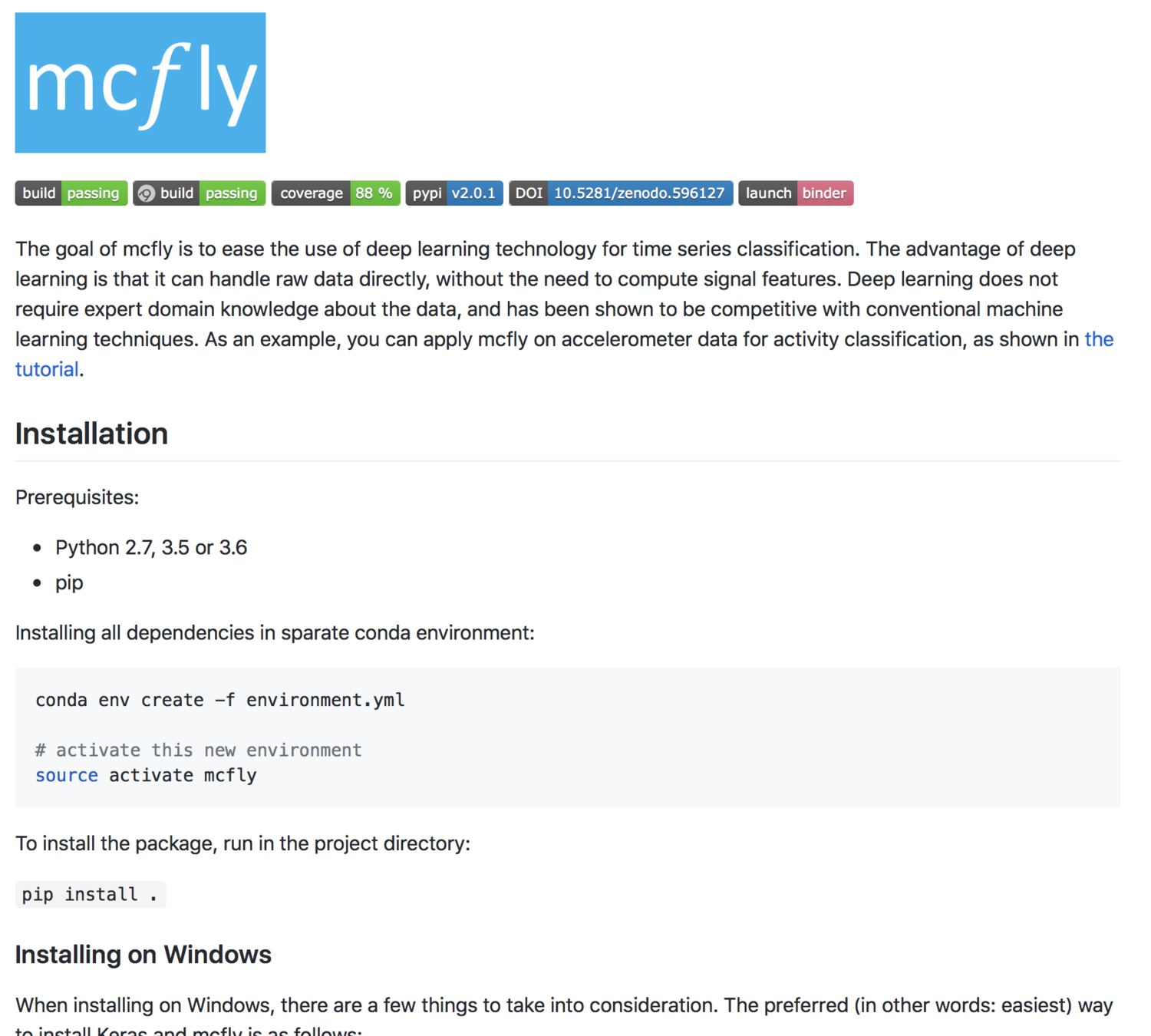
An example README:
DOCUMENTATION
Check out Make a README:
DISCUSSION: DOCUMENTATION
Is your code well-annotated? Would future you, a direct colleague, an external colleague/peer understand it?
Do you have README files already?
THE FOUR FACETS OF REPRODUCIBILITY
ORGANIZATION
DOCUMENTATION
AUTOMATION
DISSEMINATION
AUTOMATION
AUTOMATION
WHY DO YOU NEED IT?
- More efficient to modify and repeat an analysis down the road.
- Easier for reviewers and colleagues to see every aspect of your methods.
- Self-documenting methods: your future self will forget small steps.
AUTOMATION
HOW DO YOU DO IT?
-
SCRIPTING VS. POINT & CLICK
Script = more time spent up front, but will save time in the long run.
-
DRY & FUNCTIONALIZE EVERYTHING!
Don't Repeat Yourself: if your analysis is composed of scripts, with repeated code throughout, it will be more time consuming to maintain and update.
Instead use modularity: use functions to write code in reusable chunks
AUTOMATION
source: Best Practices in Writing Reproducible Code @UtrechtUniversity
AUTOMATION
Functions are smaller code units reponsible of one task.
-
Functions are meant to be reused
-
Functions accept arguments (though they may also be empty!)
-
What arguments a function accept is defined by its parameters
Functions do not necessarily make code shorter (at first)! Compare:

source: Best Practices in Writing Reproducible Code @UtrechtUniversity
AUTOMATION
It's better to think in building blocks:

source: Best Practices in Writing Reproducible Code @UtrechtUniversity
DISCUSSION: AUTOMATION
Do you have a tendency to rewrite or repeat code?
Do you see opportunities to write more functions?
THE FOUR FACETS OF REPRODUCIBILITY
ORGANIZATION
DOCUMENTATION
AUTOMATION
DISSEMINATION
DISSEMINATION
DISSEMINATION
WHY DO YOU NEED IT?
- Funding agency / journal requirement
- Community expects it
- Increased visibility / citation
- More efficient, less redundant science
DISSEMINATION
HOW DO YOU DO IT?
-
Document workflow: R Markdown / Jupyter Notebook
-
Collaborate with Colleagues / Version Control : GitHub
-
Publish Data Snapshot: FigShare, Dryad, Zenodo, etc
-
Share workflow: Notebook Viewer, Binder
DISSEMINATION
But first, some notes on software licenses.
-
Copyright is implicit; others cannot use your code without your permission.
-
Licensing gives that permission, and its boundaries and conditions.
-
Choosing a license early on means being aware of your license as the project proceeds (and not creating conflicts).
DISSEMINATION
But first, some notes on software licenses.
-
There are over 80 OSI-approved licenses (and many, many others) to choose from. This is one that's often used:

DISSEMINATION
But first, some notes on software licenses.
DISSEMINATION
Archive your project on Zenodo, and get a DOI!
GitHub & Zenodo have a great integration that makes it easy to archive a whole repository.

DISSEMINATION
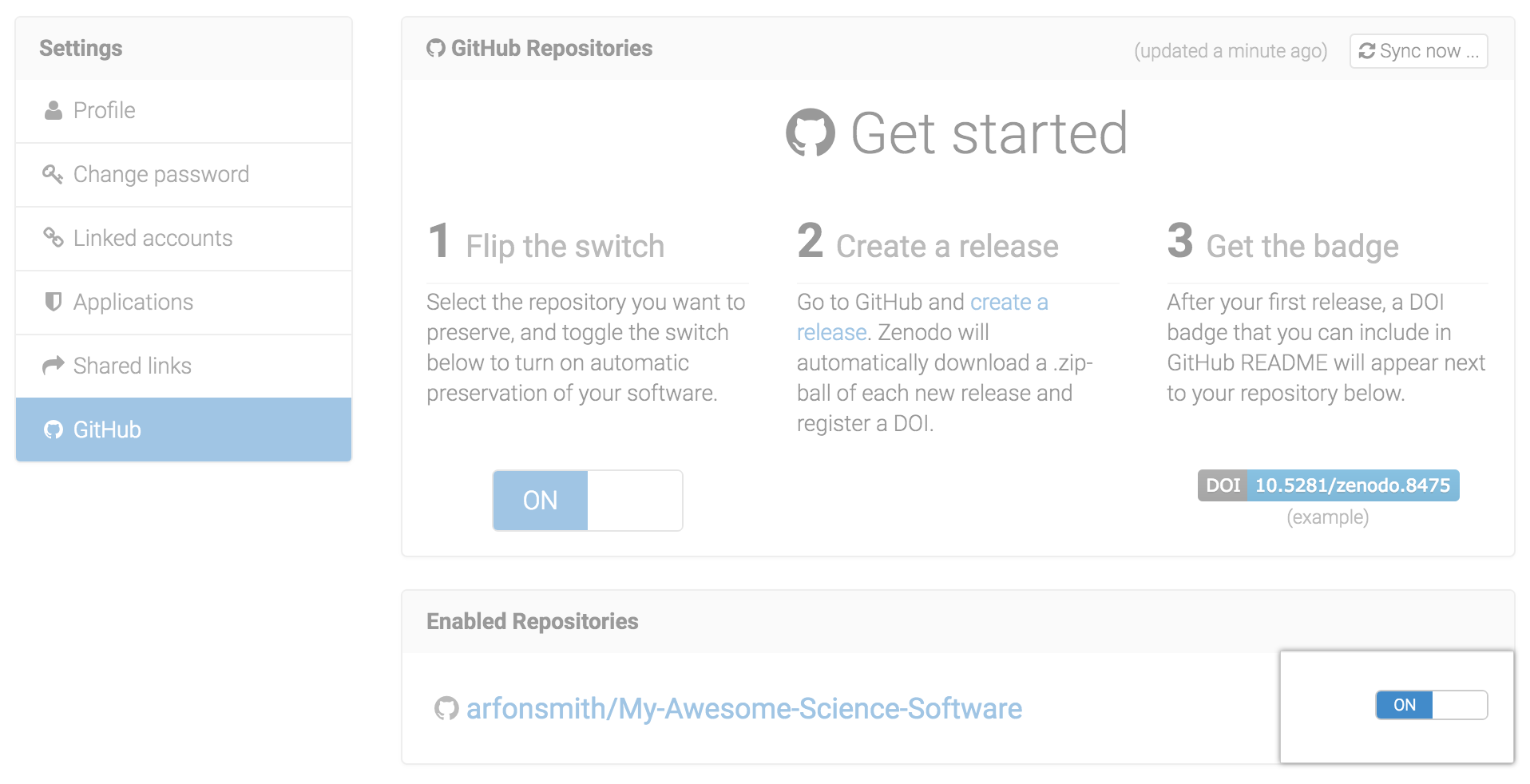
First, select your repository:
source: Best Practices in Writing Reproducible Code @UtrechtUniversity
DISSEMINATION
Second, release your project and follow the workflow:
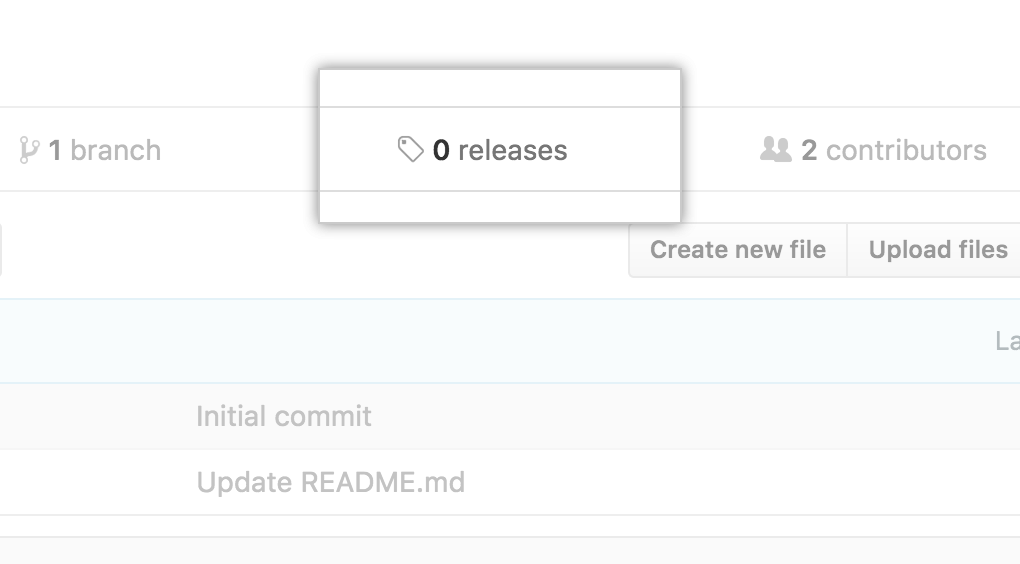
source: Best Practices in Writing Reproducible Code @UtrechtUniversity
DISSEMINATION
Last,
- Add some final descriptions
- Click 'publish'
- Voilá!
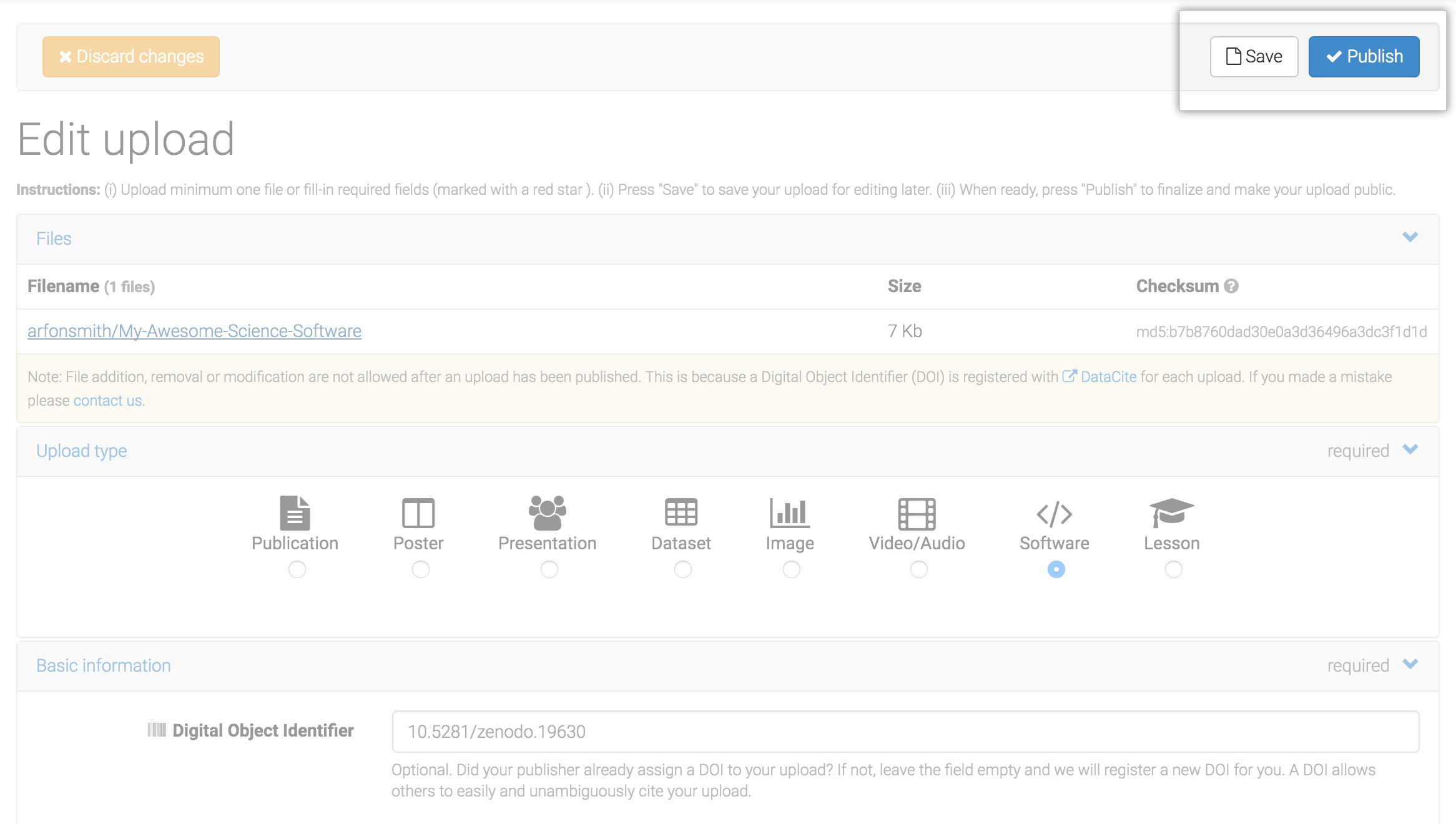
source: Best Practices in Writing Reproducible Code @UtrechtUniversity
DISSEMINATION
As a final touch, take your DOI and place it as a badge in your GitHub README!

source: Best Practices in Writing Reproducible Code @UtrechtUniversity
DISSEMINATION
Use Binder to make your code immediately reproducible by anyone, anywhere!
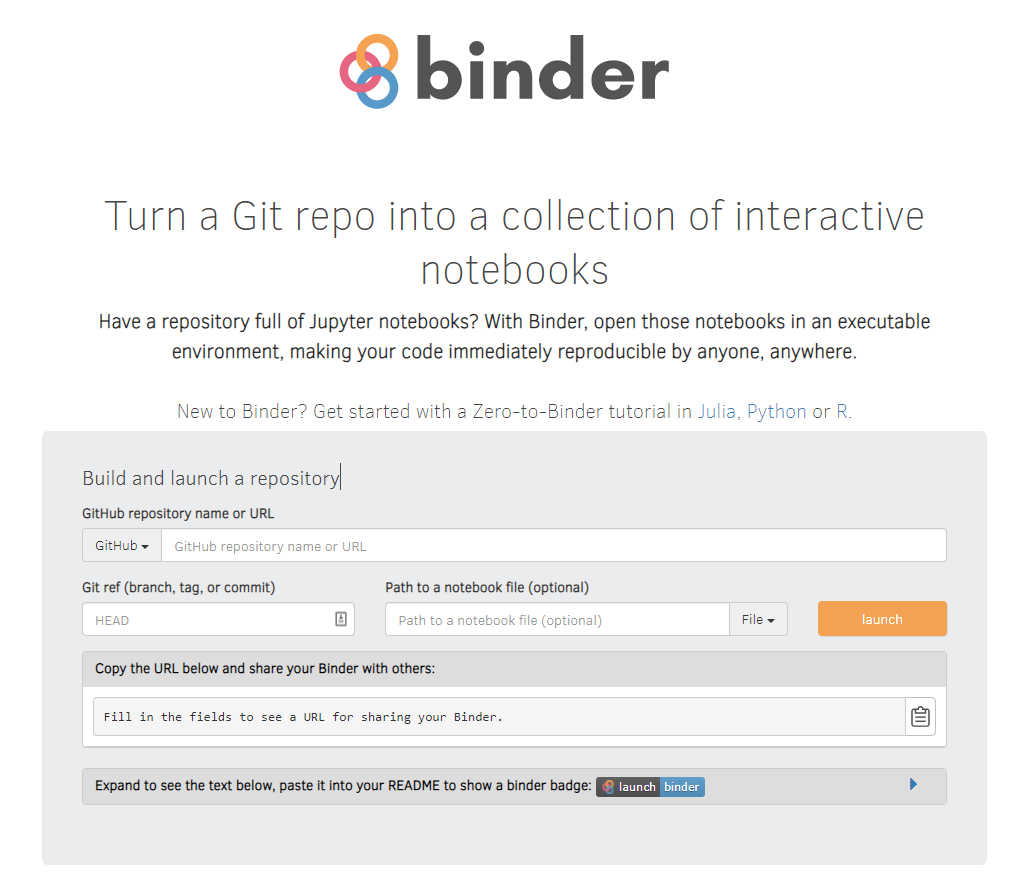
DISSEMINATION
How does it work? Binder is a virtual, executable environment that runs the code in your GitHub repository.

DISSEMINATION
How does it work? Binder is a virtual, executable environment that runs the code in your GitHub repository.

DISCUSSION: DISSEMINATION
When do you think is the best time to publish your code / place it online? Why?
Have you already published your code? Where?
What software license did/would you choose? Why?
THE FOUR FACETS OF REPRODUCIBILITY
ORGANIZATION
DOCUMENTATION
AUTOMATION
DISSEMINATION
TIME TO WRAP UP!
TAKE-HOME MESSAGE!
You get more efficient, less redundant science: others can build upon our work!

ANOTHER WAY TO LOOK AT IT...

...YOUR REPRODUCIBILITY JOURNEY!
This image was created by Scriberia for The Turing Way community and is used under a CC-BY licence. The image was obtained from https://zenodo.org/record/3332808.
LEARN MORE!
LEARN MORE!

BONUS:
RECOMMENDATIONS FOR FAIR SOFTWARE
Getting Started With Computational Reproducibility
By bmkramer_uu
Getting Started With Computational Reproducibility
Presentation for the Open Science Module at the Graduate School of Life Sciences, Utrecht University (22-02-2021).
- 66