Generic solutions for specific problems: Snakemake, Datavzrd, and Varlociraptor in 2025
Johannes Köster




Snakemake
Snakemake
>1 million downloads since 2015
>3000 citations
>14 citations per week in 2023
affiliated


Mölder, F., Jablonski, K.P., Letcher, B., Hall, M.B., Tomkins-Tinch, C.H., Sochat, V., Forster, J., Lee, S., Twardziok, S.O., Kanitz, A., Wilm, A., Holtgrewe, M., Rahmann, S., Nahnsen, S., Köster, J. Sustainable data analysis with Snakemake. F1000Res 10, 33 (2021).
https://snakemake.github.io
dataset
results
dataset
dataset
dataset
dataset
dataset
https://snakemake.github.io
Define workflows
in terms of rules
https://snakemake.github.io
Define workflows
in terms of rules
rule mytask:
input:
"path/to/{dataset}.txt"
output:
"result/{dataset}.txt"
script:
"scripts/myscript.R"
rule myfiltration:
input:
"result/{dataset}.txt"
output:
"result/{dataset}.filtered.txt"
shell:
"mycommand {input} > {output}"
rule aggregate:
input:
"results/dataset1.filtered.txt",
"results/dataset2.filtered.txt"
output:
"plots/myplot.pdf"
script:
"scripts/myplot.R"https://snakemake.github.io
Define workflows
in terms of rules
Define workflows
in terms of rules
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
params:
someparam=0.1
conda:
"envs/sometool.yaml"
shell:
"some-tool {params.someparam} {input} > {output}"- simple paths
- helpers for branching
and aggregation - Python logic
- shell commands
- ad-hoc scripts and notebooks (Python, R, Julia, Rust, Bash)
- wrappers
https://snakemake.github.io
deployment via
conda, containers
rule mytask:
input:
"path/to/{dataset}.txt"
output:
"result/{dataset}.txt"
script:
"scripts/myscript.R"
rule myfiltration:
input:
"result/{dataset}.txt"
output:
"result/{dataset}.filtered.txt"
shell:
"mycommand {input} > {output}"
rule aggregate:
input:
"results/dataset1.filtered.txt",
"results/dataset2.filtered.txt"
output:
"plots/myplot.pdf"
script:
"scripts/myplot.R"Automatic inference of DAG of jobs

https://snakemake.github.io
Scalable to any platform



workstation
compute server
cluster






grid computing
cloud computing



https://snakemake.github.io
Self-contained HTML reports
https://snakemake.github.io
Snakemake 8: plugin architecture
https://snakemake.github.io

Simplicity and automation
Automatic documentation
https://snakemake.github.io
Datavzrd
Tables are the central entity
in data analysis


https://datavzrd.github.io
Not always a single table
oncoprint + individual variant calls
differentially expressed genes + expression matrix
https://datavzrd.github.io
Not always just a table
oncoprint + individual variant calls +
differentially expressed genes + expression matrix +
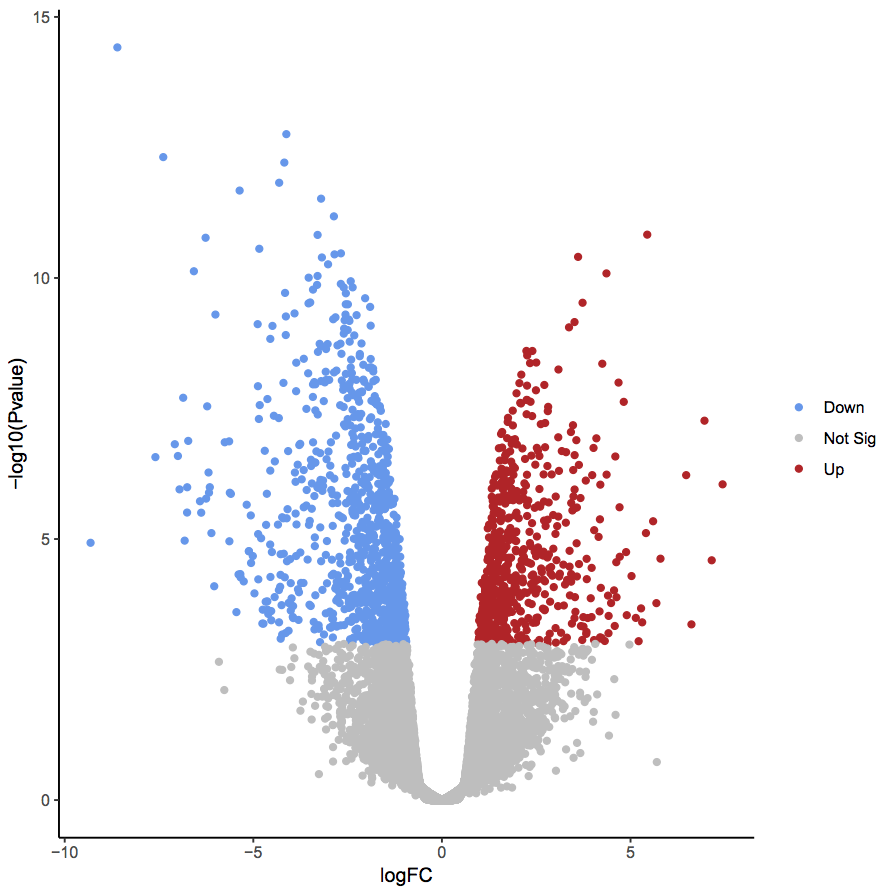

https://datavzrd.github.io
State of the art
Individual tables (tsv, excel) and plots:
- easy to publish
- limited interactivity
- no jumping between corresponding items
Web applications (custom, shiny, ...):
- running server (or local installation)
- implementation overhead
- long-term maintenance is challenging
https://datavzrd.github.io
The problem
Input:
- set of tables
- relations between tables
- set of rendering definitions
Output:
portable interactive visual presentation
https://datavzrd.github.io
datasets:
oscars:
path: "data/oscars.csv"
links:
link to oscar plot:
column: age
view: oscar-plot
link to movie:
column: movie
table-row: movies/Title
movies:
path: "data/movies.csv"
links:
link to oscar entry:
column: Title
table-row: oscars/movie
views:
oscars:
dataset: oscars
desc: |
### All winning oscars beginning in the year 1929.
This table contains *all* winning oscars for best
actress and best actor.
page-size: 25
render-table:
columns:
age:
plot:
ticks:
scale: linear
domain:
- 20
- 100
name:
link-to-url: "https://lmgtfy.app/?q=Is {name} in {movie}?"
movie:
link-to-url: "https://de.wikipedia.org/wiki/{value}"
award:
plot:
heatmap:
scale: ordinal
domain:
- Best actor
- Best actress
range:
- "#add8e6"
- "#ffb6c1"
index(0):
display-mode: hidden
regex('birth_.+'):
display-mode: detail
movies:
dataset: movies
render-table:
columns:
Genre:
ellipsis: 15
imdbID:
link-to-url: "https://www.imdb.com/title/{value}/"
Title:
link-to-url: "https://de.wikipedia.org/wiki/{value}"
imdbRating:
precision: 1
plot:
bars:
scale: linear
domain:
- 1
- 10
Rated:
plot-view-legend: true
plot:
heatmap:
scale: ordinal
color-scheme: accent
oscar-plot:
dataset: oscars
desc: |
## My beautiful oscar scatter plot
*So many great actors and actresses*
render-plot:
spec-path: ".examples/specs/oscars.vl.json"
movies-plot:
dataset: movies
desc: |
All movies with its *runtime* and *ratings* plotted over *time*.
render-plot:
spec-path: ".examples/specs/movies.vl.json"
The solution: Datavzrd
https://datavzrd.github.io
https://datavzrd.github.io
Portability
├── index.html
├── movies
│ ├── index_1.html
│ └── table.js
├── oscars
│ ├── index_1.html
│ └── table.js
├── movies-plot
│ └── index_1.html
├── oscar-plot
│ └── index_1.html
└── static
├── bootstrap.bundle.min.js
├── bootstrap.min.css
├── bootstrap-select.min.css
├── bootstrap-select.min.js
├── bootstrap-table-filter-control.min.js
├── bootstrap-table-fixed-columns.min.css
├── bootstrap-table-fixed-columns.min.js
├── bootstrap-table.min.css
├── bootstrap-table.min.js
├── datavzrd.css
├── jquery.min.js
├── jsonm.min.js
├── lz-string.min.js
├── showdown.min.js
├── vega-embed.min.js
├── vega-lite.min.js
└── vega.min.js- static HTML, no server needed
- load data via script tags
- javascript compliant compression
- \(\leq\) 30000 rows: in-memory filter/sort/paging
- \(>\) 30000 rows: paging/search projected to filesystem structure
https://datavzrd.github.io
Datavzrd + Snakemake
rule datavzrd:
input:
config="resources/{sample}.datavzrd.yaml",
table="data/{sample}.tsv",
output:
report(
directory("results/datavzrd-report/{sample}"),
htmlindex="index.html",
),
wrapper:
"v4.6.0/utils/datavzrd"https://datavzrd.github.io
Datavzrd + Snakemake
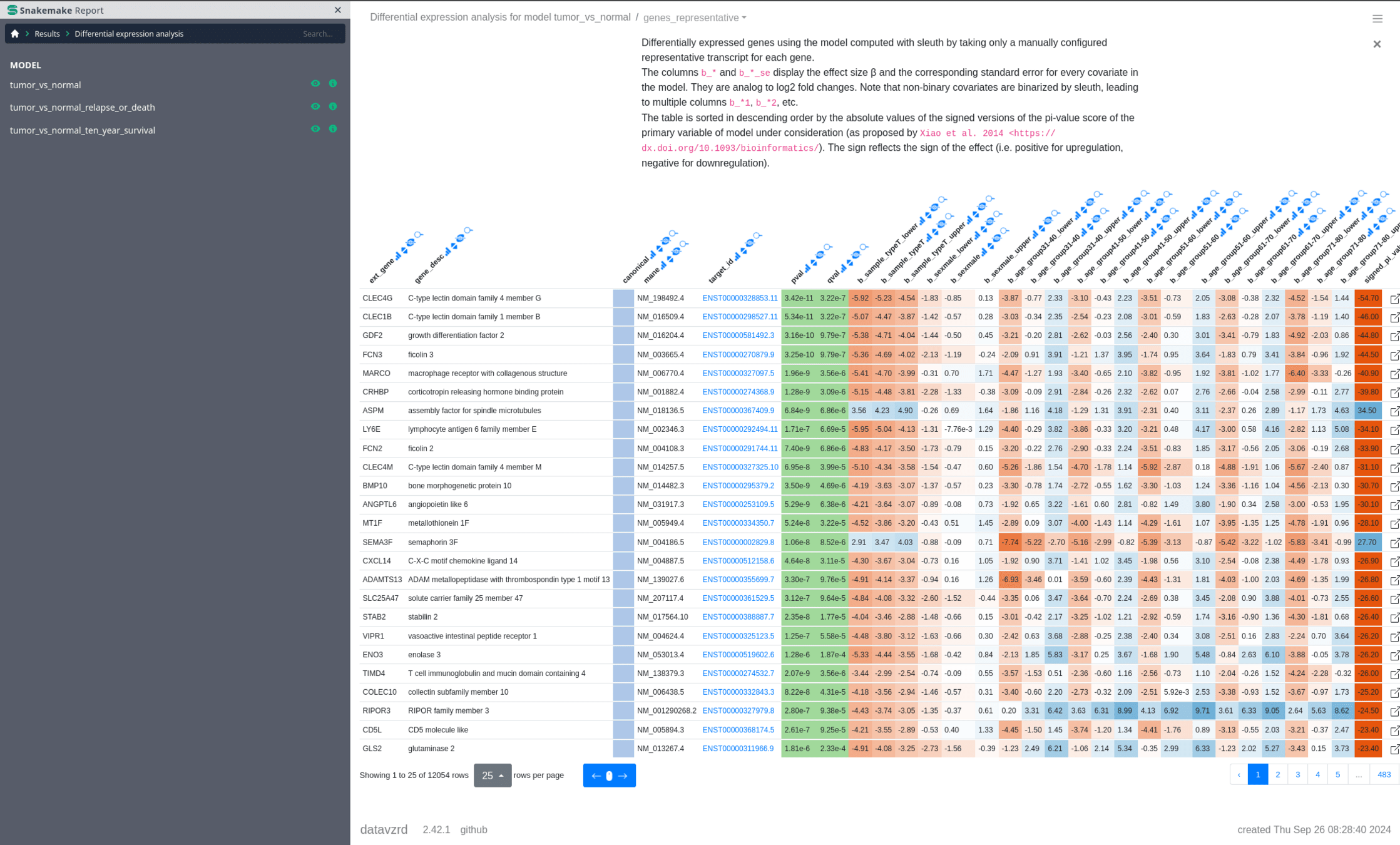
https://datavzrd.github.io
Varlociraptor
Varlociraptor
Johannes Köster, Louis Dijkstra, Tobias Marschall, Alexander Schönhuth.
Varlociraptor: enhancing sensitivity and controlling false discovery rate in somatic indel discovery. Genome Biology 21, 2020
https://varlociraptor.github.io
Traditional variant calling approach:
detection
genotyping/VAF prediction
Varlociraptor approach:
candidate variant calling
event calling
same tool
any tool
Varlociraptor

Varlociraptor
Johannes Köster, Louis Dijkstra, Tobias Marschall, Alexander Schönhuth.
Varlociraptor: enhancing sensitivity and controlling false discovery rate in somatic indel discovery. Genome Biology 21, 2020
https://varlociraptor.github.io

Varlociraptor
germline: "normal:0.5 | normal:1.0"
somatic_normal: "normal:]0.0,0.5["
somatic_tumor: "normal:0.0 & tumor:]0.0,1.0]"
absent: "normal:0.0 & tumor:0.0https://varlociraptor.github.io
Varlociraptor model building block
\(\xi_i \sim \text{Bernoulli}(\theta \tau)\)
\(\omega_i \sim Bernoulli(\pi_i)\)
\(Z_i \mid \xi_i, \omega_i=1,\beta,\delta \sim\)


\(\beta, \delta\)
- allele frequency
- sampling bias
- allele uncertainty
- biases/artifacts (strand, orientation, softclip, homopolymer, ...)
- alignment uncertainty

https://varlociraptor.github.io
Variant calling grammar
species:
heterozygosity: 0.001
ploidy:
male:
all: 2
X: 1
Y: 1
female:
all: 2
X: 2
Y: 0
samples:
jane:
sex: female
events:
present: "jane:0.5 | jane:1.0"https://varlociraptor.github.io
species:
heterozygosity: 0.001
ploidy:
male:
all: 2
X: 1
Y: 1
female:
all: 2
X: 2
Y: 0
samples:
jane:
sex: female
john:
sex: male
james:
sex: male
inheritance:
mendelian:
mother: jane
father: johnevents:
john: "john:0.5 | john:1.0"
jane: "jane:0.5 | jane:1.0"
denovo_james: "(james:0.5 | james:1.0) & !$jane & !$john"
Variant calling grammar


https://varlociraptor.github.io
species:
heterozygosity: 0.001
ploidy:
male:
all: 2
X: 1
Y: 1
female:
all: 2
X: 2
Y: 0
samples:
jane:
sex: female
somatic-effective-mutation-rate: 1e-10
tumor:
inheritance:
clonal:
from: jane
contamination:
by: jane
fraction: 0.1
somatic-effective-mutation-rate: 1e-6events:
germline: "jane:0.5 | jane:1.0"
somatic: "jane:]0.0,0.5["
somatic_tumor_low: "jane:0.0 & tumor:]0.0,0.1["
somatic_tumor_high: "jane:0.0 & tumor:[0.1,1.0]"
Variant calling grammar
https://varlociraptor.github.io
samples:
jane:
sex: female
somatic-effective-mutation-rate: 1e-10
tumor:
inheritance:
clonal:
from: jane
contamination:
by: jane
fraction: 0.1
somatic-effective-mutation-rate: 1e-6
relapse:
inheritance:
clonal:
from: jane
contamination:
by: jane
fraction: 0.2
somatic-effective-mutation-rate: 1e-6expressions:
somatic_tumor: "jane:0.0 & tumor:]0.0,1.0]"
events:
germline: "jane:0.5 | jane:1.0"
somatic: "jane:]0.0,0.5["
somatic_tumor_no_increase: "$somatic_tumor & l2fc(relapse,tumor) < 1"
somatic_tumor_increase: "$somatic_tumor & l2fc(relapse,tumor) >= 1"
somatic_relapse: "jane:0.0 & tumor:0.0 & relapse:]0.0,1.0]"
Variant calling grammar
https://varlociraptor.github.io
species:
heterozygosity: 0.001
ploidy:
male:
all: 2
X: 1
Y: 1
female:
all: 2
X: 2
Y: 0
samples:
dna_illumina:
sex: female
dna_nanopore:
inheritance:
clonal:
from: dna_illumina
rna_illumina:
universe: [0.0,1.0]events:
het: "dna_illumina:0.5 & dna_nanopore:0.5 & rna_illumina:]0.0,1.0]"
hom: "dna_illumina:1.0 & dna_nanopore:1.0 & rna_illumina:1.0"
rna_editing: "dna_illumina:0.0 & dna_nanopore:0.0 & rna_illumina:]0.0,1.0]"
Variant calling grammar
https://varlociraptor.github.io
Long read support
https://varlociraptor.github.io


Discovering nucleotide level and structural variants in cancer genome data from second and third generation sequencing technologies. Till Hartmann, PhD thesis 2024
Continuous testing
varlociraptor call variants --testcase-prefix testcase --testcase-locus CHROM:POS generic \
--scenario scenario.yaml --obs tumor=tumor.bcf normal=normal.bcfAutomatic test case generation:
145
public testcases (simulated + real benchmarks)
66
private testcases
(clinical)
https://varlociraptor.github.io

Reporting with Datavzrd
https://varlociraptor.github.io
Beyond just variant calling
Haplotype quantification:
utilizing Varlociraptor's posterior allele frequency distributions for virus variant quantification and HLA typing
Methylation calling:
joint consideration of methylation and variation, any sequencing technology
CNV calling:
joint consideration of depth and alignment evidence for arbitray scenarios
Optical mapping evidence:
extension of statistical model to consider optical mapping label positions to determine SV allele likelihoods
Phased variant impact prediction:
leveraging matching between Varlociraptor observations and variants
Fusion calling:
thorough statistical approach for fusion calling on DNA, RNA, short and long reads
Acknowledgements
Koesterlab
Laura Kühle
David Lähnemann
Felix Mölder
Adrian Prinz
Hamdiye Uzuner
Felix Wiegand
Can Özkan
Andrea Tonk
Till Hartmann (alumn.)
Snakemake Community
Christian Meesters
Michael B. Hall
Filipe G. Vieira
Morten E. Lund
Vanessa Sochat
Alexaner Kanitz
Kim-Phillip Jablonski
Brice Letcher
Michael B. Hall
Chris Tomkins-Tinch
Sven O. Twardziok
Manuel Holtgrewe
+ hundreds of contributors
Others
Sven Rahmann
Alex Schönhuth
Shirley Liu
Myles Brown
Henry Long
Louis Dijkstra
Tobias Marschall
Marcel Martin
Sven Nahnsen
Alex Schramm
Ina Pretzell
Michael Wessolly
Thomas Herold
Martin Schuler
And thanks to all the users and contributors

Copy of Invited talk at Schwarz lab
By Johannes Köster
Copy of Invited talk at Schwarz lab
- 127



