Towards Foundation Models for Science
Siavash Golkar
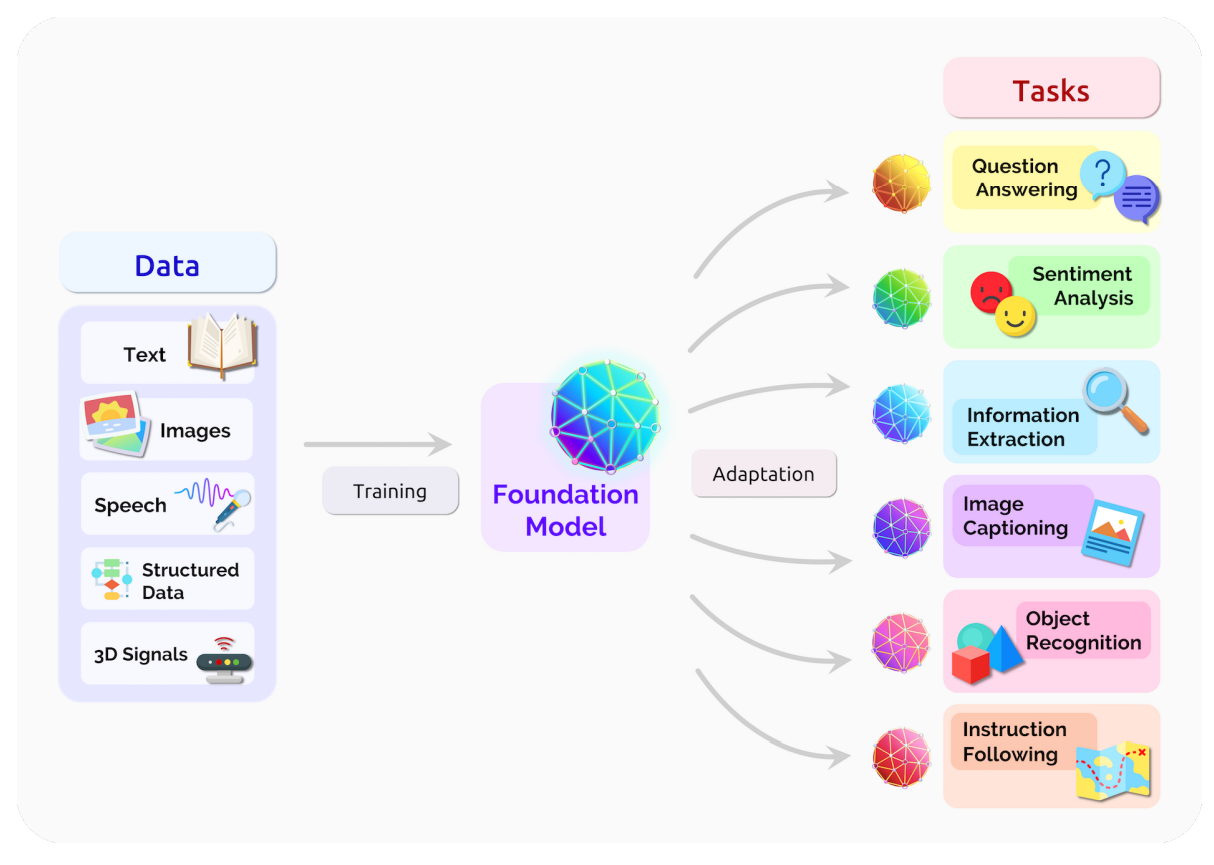
What are foundation models?
Large models pre-trained
on massive datasets
They can perform a variety of downstream tasks (zero-shot generalization)
They are good starting points for fine-tuning on data poor domains (carry useful inductive bias)
The Key Ideas
- Large models pre-trained on task-agnostic objectives on massive & diverse datasets
- They can perform a variety of downstream tasks (zero-shot generalization)
- They are good starting points for fine-tuning on data poor domains (carry useful inductive bias)

Can we translate these innovations into a paradigm shift in machine learning for scientific applications?
Polymathic
Advancing Science through Multi‑Disciplinary AI
Our mission: to usher in a new class of machine learning for scientific data, building models that can leverage shared concepts across disciplines."
Meet the Polymathic AI Team
|
Colm-Cille
Caulfield University of Cambridge
|
Leslie Greengard Flatiron Institute New York University |
David Ha Sakana AI |
Yann LeCun Meta AI New York University |
|---|---|---|---|
|
Stephane Mallat École Normale Supérieure Collège de France Flatiron Institute |
David Spergel Simons Foundation |
Olga Troyanskaya Flatiron Institute Princeton University |
Laure
Zanna New York University
|
Our Resources
SCIENTIFIC ADVISORY GROUP
COMPUTING RESOURCES
- Internal H100 GPUs resources at the Flatiron Institute
- External 500k GPU hours (V100 and A100)
- In the process of securing additional O(10^2) dedicated H100 GPUs

Thanks Ian!
The Foundation Model Spectrum
Scientific Reasoning
Multi-Modality
Generalization to Data-Limited Domains
How can we build foundation models that can have these properties?
- How many assumptions should we make about the structure of our inputs?
-
What kind of embedding schemes should we use?
Different choices lead to foundation models with different strengths and weaknesses.
more efficient, less general
more general, less efficient
Language-like/less structured
Structured-data
The Foundation Model Spectrum
Language-like/less structured
Structured-data

xVal
A Continuous Number Encoding for LLMs

AstroCLIP
Cross-Modal Pretraining for Astronomical data


MPP
Multiple Physics Pretraining for Physical Surrogate Models

Scientific Reasoning
Multi-Modality
Generalization to Data-Limited Domains
more efficient, less general
more general, less efficient
MPP
Multiple Physics Pretraining for
Physical Surrogate Models


Project led by Michael McCabe, Bruno Régaldo, Liam Parker, Ruben Ohana, Miles Cranmer
Oral presentation at the NeurIPS 2023 AI4Science Workshop






Context
- Previous works on large domain-specific pretrained models: chemistry, medicine, astrophysics, climate, ...
→ extension on surrogate modeling of spatiotemporal physical systems - Spatiotemporal predictions motivated by faster surrogates for PDE solvers, systems that are hard to simulate with current models/hardware
- Situations where data is expensive…




Time
Ex: N-body simulation
Springel et al. 2005
The Foundation Model Spectrum
Language-like/less structured
Structured-data

xVal
A Continuous Number Encoding for LLMs


AstroCLIP
Cross-Modal Pretraining for Astronomical data


MPP
Multiple Physics Pretraining for Physical Surrogate Models

Scientific Reasoning
Multi-Modality
Generalization to Data-Limited Domains
Can we build a domain specific foundation model that would be finetunable on a few training examples?
Background
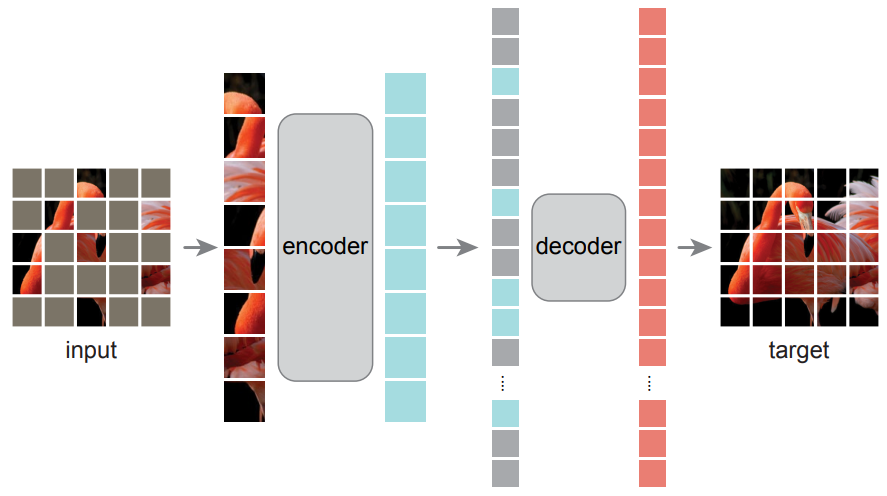
- Main pretraining strategies
- autoregressive prediction
- masked reconstruction
- contrastive learning - No conditioning on physical parameters
- Spatiotemporal physics: PDEs from a physical system with typically conservations and symmetries…
→ Suggests that there are some learnable shared features
Natural choice for physical surrogate modeling
Physical Systems from PDEBench
Navier-Stokes
Incompressible
Compressible
Shallow Water
Diffusion-Reaction
Takamoto et al. 2022
Compositionality and Pretraining


Balancing objectives during training


Normalized MSE:
Architecture for MPP
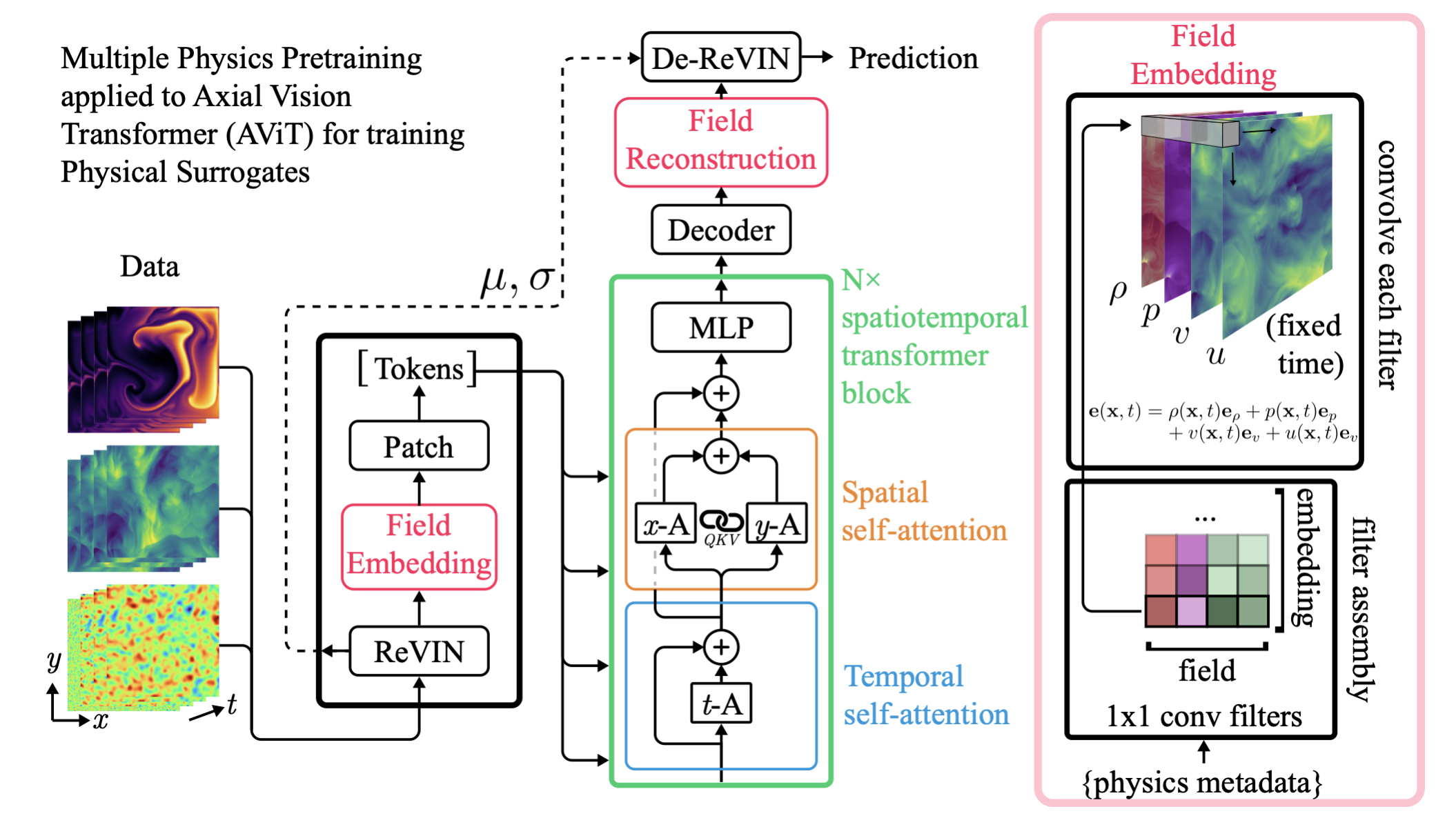
Experiment 1: Performance on Pretraining Tasks

Context size: 16 frames

Experiment 2: Transfer


Compressible Navier-Stokes
M = 0.1
M = 1.0
Fun Fact: Finetuning with VideoMAE works quite well…

Tube masking
ViT
ViT
ViT

Trained on reconstructing masked pixels on natural videos (SSV2 and K400)
Tong et al. 2022
Experiment 3: Broader Downstream Tasks
Regression Problems on Incompressible Navier-Stokes

Mixed results
Long time predictions on Navier-Stokes?
Conclusion
- A single pre-trained transformer model matches/outperforms specialized baselines for each in-distribution task.
- Demonstrated transfer capabilities were (both near and far).
- Early days. We are in the process of scaling up to more diverse data.
- Open code and pretrained models: https://github.com/PolymathicAI/multiple_physics_pretraining

xVal
A Continuous Number Encoding for LLMs


Project led by Siavash Golkar, Mariel Pettee, Michael Eickenberg, Alberto Bietti
Accepted contribution at the NeurIPS 2023 AI4Science Workshop





The Foundation Model Spectrum
Language-like/less structured
Structured-data

xVal
A Continuous Number Encoding for LLMs


AstroCLIP
Cross-Modal Pretraining for Astronomical data


MPP
Multiple Physics Pretraining for Physical Surrogate Models

Scientific Reasoning
Multi-Modality
Generalization to Data-Limited Domains
The problem: existing LLMs are not suitable for reliable zero-shot numerical operations.


arXiv:2305.18654 [cs.CL]
arXiv:2109.03137 [cs.CL]
They make erratic, discontinuous predictions.
Even fine-tuning exhaustively does not grant out-of-distribution generalization abilities.
Can we build an LLM better suited for numerical data analysis?
xVal in a Nutshell: a continuous numerical encoding for language models

xVal in a Nutshell: a continuous numerical encoding for language models
This encoding strategy has 3 main benefits:
-
Continuity
-
The model is now end-to-end continuous by construction.
(Standard LLMs are discontinuous both at the input and output stage.)
-
-
Interpolation
-
It makes better out-of-distribution predictions than other numerical encodings.
-
-
Efficiency
-
By using just a single token to represent any number, it requires less memory, compute resources, and training time to achieve strong results.
-
xVal in a Nutshell: a continuous numerical encoding for language models

xVal shows improved predictions for out-of-distribution values.
xVal in a Nutshell: a continuous numerical encoding for language models
When evaluated on multi-digit multiplication tasks, xVal performs comparably well, and is less prone to large outliers:


And when evaluated on compound operations of basic arithmetic, xVal shows the strongest performance:
xVal in a Nutshell: a continuous numerical encoding for language models
Future directions: improving the dynamic range of the embedded values.

Conclusion
- xVal improves numerical analysis performance of transformer models.
- Good fit for numerical-data-rich texts.
- Not great for general purpose LLMs
(limited dynamic range, can't easily handle 13, 666, 1337, ...). - Not suitable for heavy numerical computation.
- Code available: https://github.com/PolymathicAI/xVal

AstroCLIP
Cross-Modal Pre-Training for Astronomical Foundation Models


Project led by Francois Lanusse, Liam Parker, Siavash Golkar, Miles Cranmer
Accepted contribution at the NeurIPS 2023 AI4Science Workshop





The Data Diversity Challenge
- Scientific data is extremely multimodal.
- Observations from different instruments are not interchangeable.
- Metadata matters

Credit: Melchior et al. 2021


Credit:DESI collaboration/DESI Legacy Imaging Surveys/LBNL/DOE & KPNO/CTIO/NOIRLab/NSF/AURA/unWISE
The Foundation Model Spectrum
Language-like/less structured
Structured-data

xVal
A Continuous Number Encoding for LLMs


AstroCLIP
Cross-Modal Pretraining for Astronomical data


MPP
Multiple Physics Pretraining for Physical Surrogate Models

Scientific Reasoning
Multi-Modality
Generalization to Data-Limited Domains
Towards Large Multi-Modal Observational Models
Most General
Most Specific
Independent models for every type of observation
Single model capable of processing all types of observations
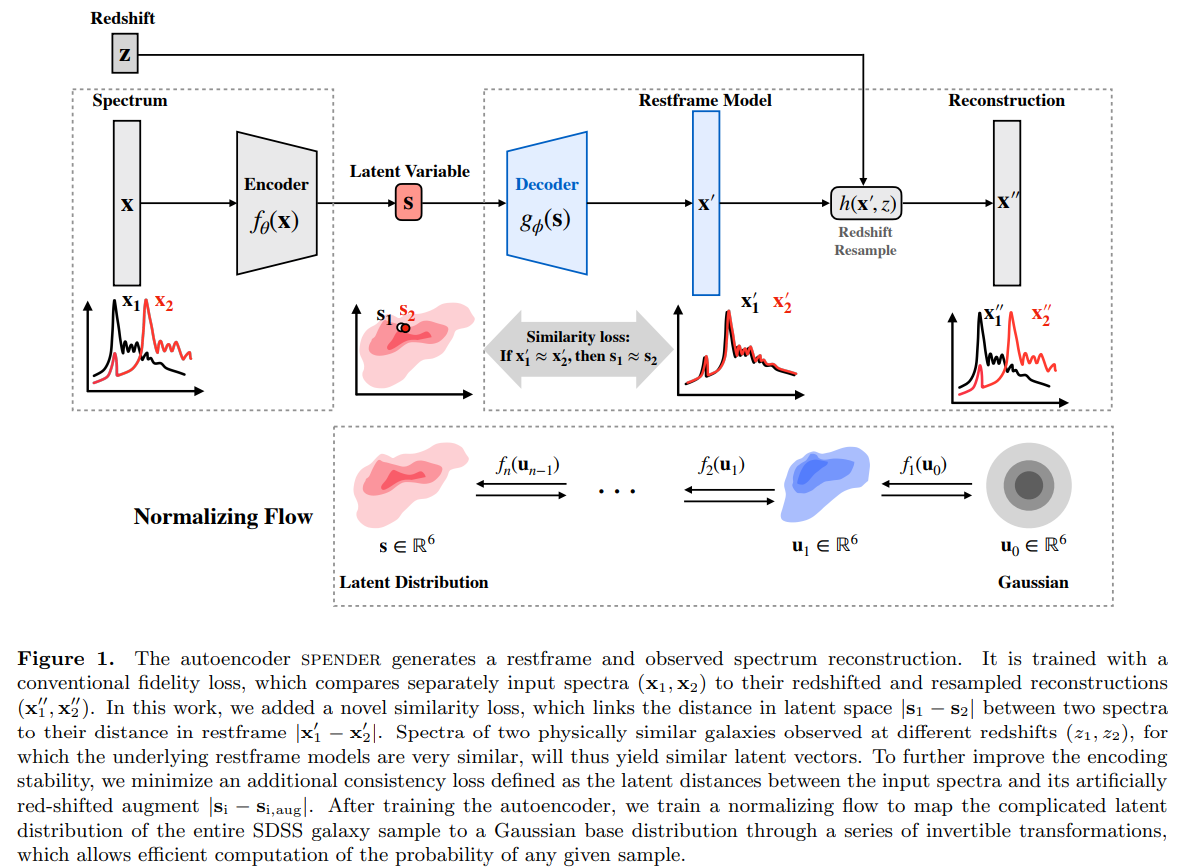
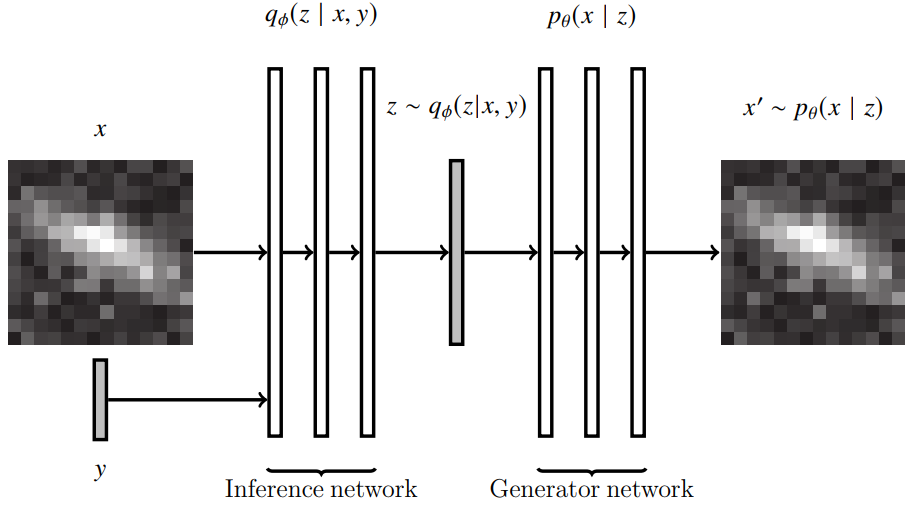
Towards Large Multi-Modal Observational Models
Most General
Most Specific
Independent models for every type of observation
Single model capable of processing all types of observations


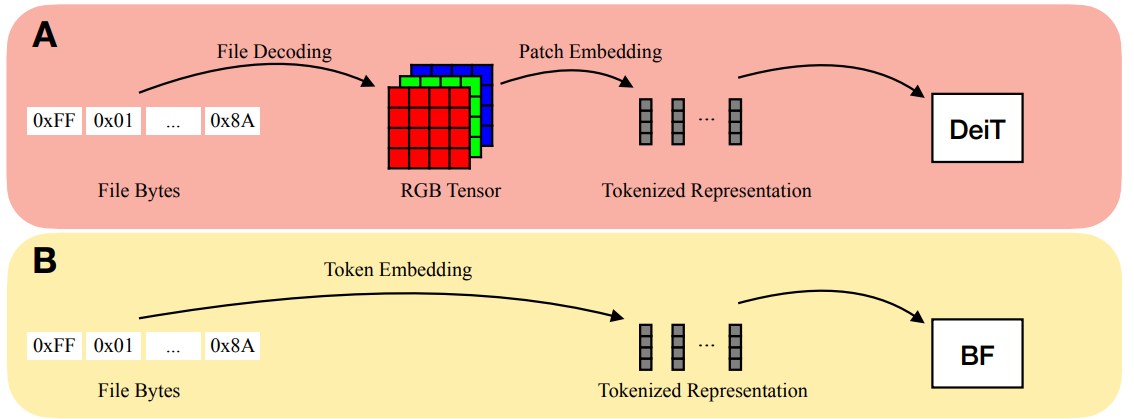
Bytes Are All You Need (Horton et al. 2023)
Towards Large Multi-Modal Observational Models
Most General
Most Specific
Independent models for every type of observation
Single model capable of processing all types of observations



Bytes Are All You Need (Horton et al. 2023)
AstroCLIP
Can we build a baseline multimodal by bringing together specialized models trained on each modality?
Contrastive Learning in Astrophysics

Self-Supervised similarity search for large scientific datasets (Stein et al. 2021)
How do we add in other modalities?
(e.g. spectral information?)
Example of Science Application: Identifying Galaxy Tidal Features

What is CLIP?
Contrastive Language Image Pretraining (CLIP)
(Radford et al. 2021)
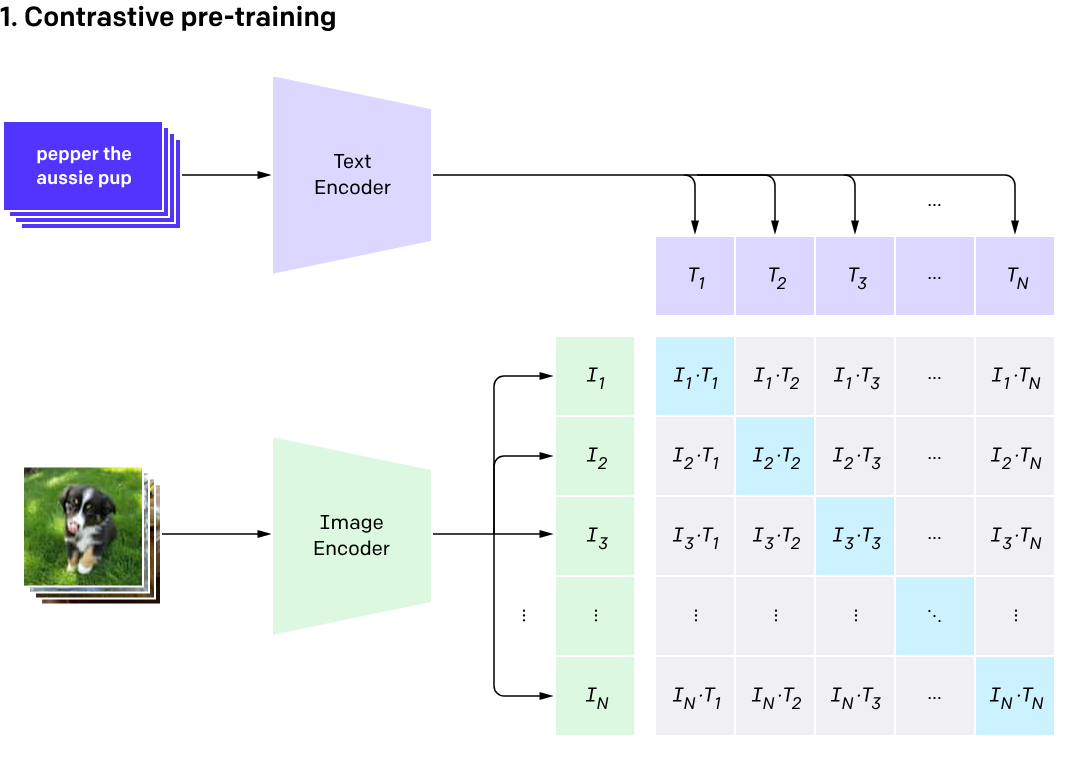
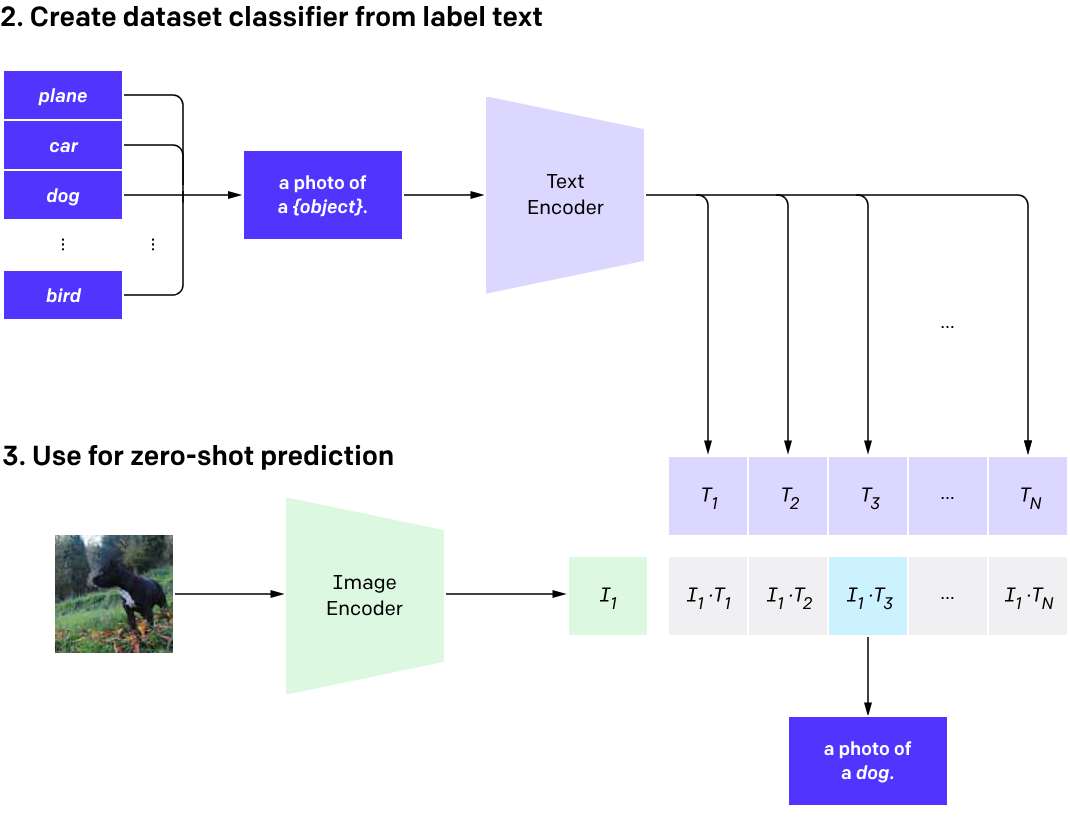
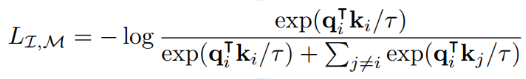
One model, many downstream applications!
Flamingo: a Visual Language Model for Few-Shot Learning (Alayrac et al. 2022)


Hierarchical Text-Conditional Image Generation with CLIP Latents (Ramesh et al. 2022)
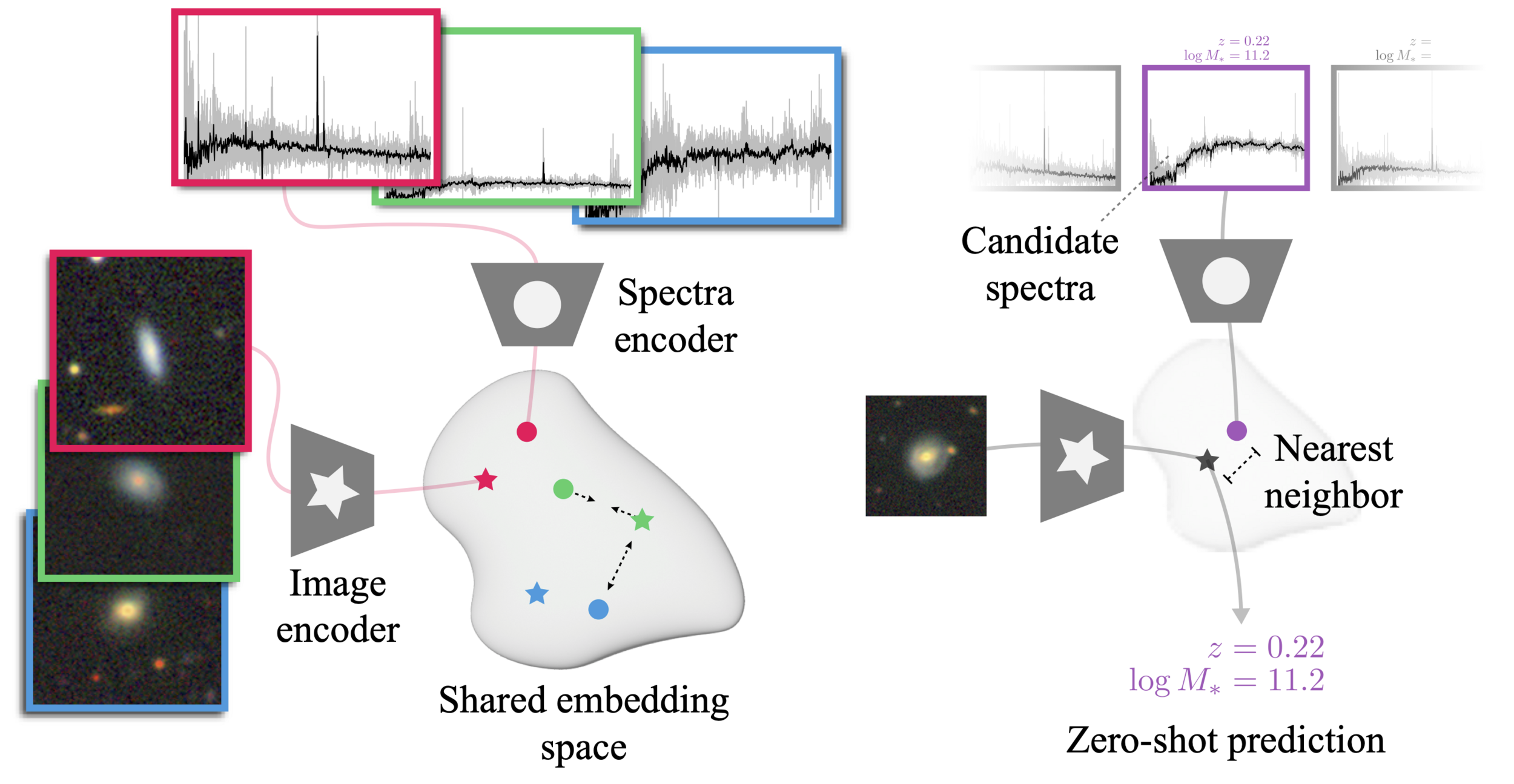
The AstroCLIP approach
- We use spectra and multi-band images as our two different views for the same underlying object.
- DESI Legacy Surveys (g,r,z) images, and DESI EDR galaxy spectra.
- Once trained, we can do example retrieval by nearest neighbor search.



How we do it
We take a two steps approach:
- Build self-supervised model separately for images and spectra
- Images: Start from pre-trained ResNet 50 from Stein et al. (2021)
- Spectra: Pretrain by mask modeling a GPT-2 like transformer on spectra
- Train an embedding module on top of each backbone under InfoNCE loss
- Images: Simple MLP
- Spectra: Cross-attention module

More examples of retrieval


Image Similarity
Spectral Similarity

Image-Spectral Similarity
Visualizing properties of the embedding space


UMAP representation of spectra embeddings
Testing Structure of Embedding Space by k-NN Regression







Comparison to image-only SSL (from Stein et al. 2021)




The Information Point of View
- The InfoNCE loss is a lower bound on the Mutual Information between modalities
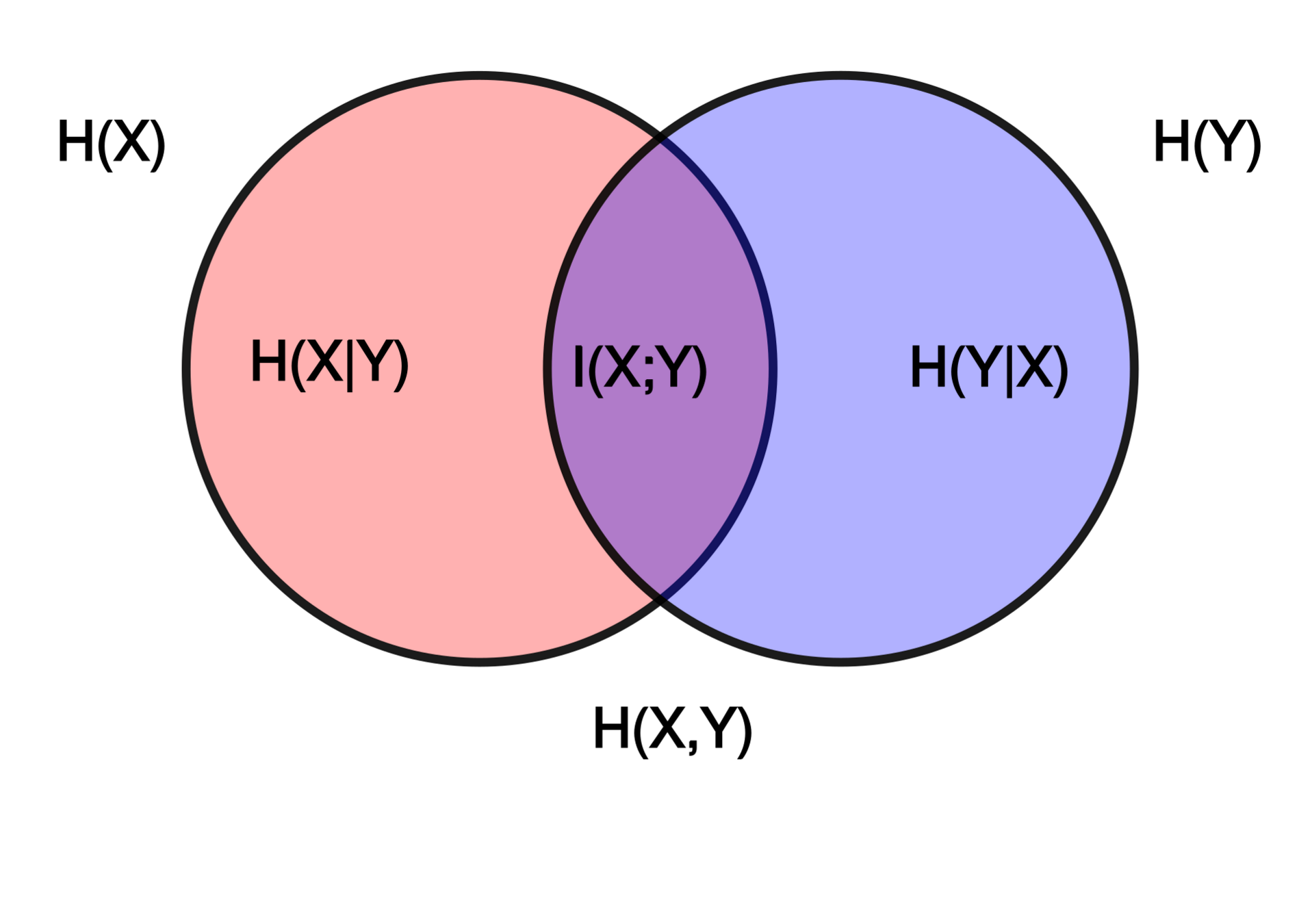
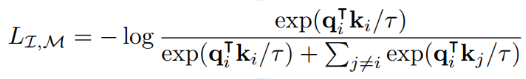
Shared physical information about galaxies between images and spectra
Thinking about data from a hierarchical Bayesian model point of view

=> We are building summary statistics for the physical parameters describing an object in a completely data driven way
What comes next!
- We want to build a shared embedding space for all astrophysics observation modalities.
- Next steps would be, embedded data from different instruments, different filters, etc... to build a universal embedding for types of galaxy observations
- Deeper interaction between the modalities beyond alignment.
- Next steps would be, embedded data from different instruments, different filters, etc... to build a universal embedding for types of galaxy observations
Teaser for what comes next


We are just getting started!
Thank you for listening!
Grand Challenges for Foundation Models in Science
- Scientific data is massively multi-modal (not finite). Training a specialist model for each modality is not feasible as we grow the scope of our models.
- Transformers have good inductive bias for many tasks (vision, language, etc.)
But they are not necessarily the best models for scientific foundation models.
(Provably inefficient in specific cases.)
- Many fundamental challenges remain (especially in physics).
(Uncertainty estimates, interpretation, concept discovery, etc)
"large data + large transformer" does not solve everything.
Towards Foundation Models for Science
By Siavash Golkar
Towards Foundation Models for Science
Overview talk of the Polymathic AI Initiative
- 811
