Introduction to Antigenic Evolution
Sidney M. Bell, PhD (@sidneymbell)
Chan Zuckerberg Initiative
Mini workshop: California Departments of Public Health
June 10, 2021
Outline
- How do viruses evolve to escape the immune system?
- What does this mean for vaccine escape?
- How does genomic surveillance help with vaccine updates?
- Discussion and questions

Grubaugh, Nature Micro, 2019
Viruses acquire mutations as they replicate and transmit from person to person.
SARS-CoV-2
(~7 day serial interval, ~25 subs/year)
ACTG
ACTT
AGTT



Antigenic variation
Genetic
variation

A very small minority of mutations may change the way the virus "looks" to the immune system
Antigenic variation is usually caused by a mutation in a "surface protein"
For some viruses, antigenic variation can help them escape from pre-existing immunity





Measles
Antigenically uniform
Flu
Antigenically
diverse


Lifelong protection
Temporary cross-protection



What does this mean for vaccine updates?
Influenza & SARS-CoV-2

Influenza evolves very rapidly
Clades don't stick around very long because they're constantly being outcompeted by new clades

Influenza evolves very rapidly

Flu

Dengue

Population susceptibility:


Previously circulating:





Population
immunity:













Clade growth:


Population immunity can influence viral evolution
**In flu, this is driven by natural infection, not vaccination
Wren, Bell et al., 2018
Population immunity drives flu evolution, necessitating a vaccine update every ~2 years

Influenza & SARS-CoV-2 have a similar number of substitutions per year in surface proteins
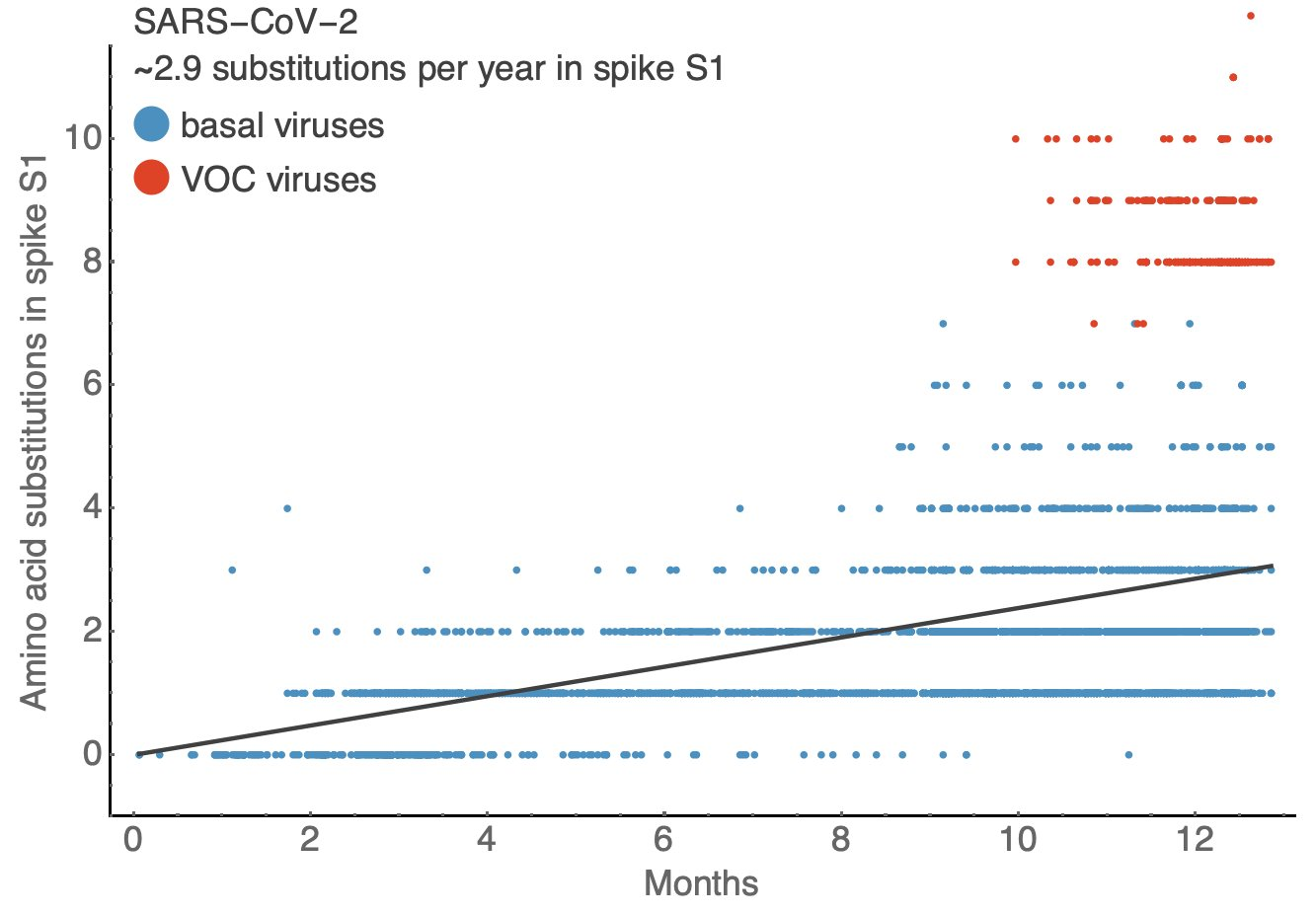

BUT! Need to consider...
-
Rate differences
SARS-CoV-2 spike S1 gene is ~2x as long as Flu's HA1 gene
-
Is the last year typical for SARS-CoV-2?
Not enough data to define "normal" for SARS-CoV-2
-
"Scariants are innocent until proven guilty"
How much does each of these mutations actually
matter for immune escape
Measuring antigenic change


Neutralization?

+






+

Replication
How much serum (neutralizing antibodies) is necessary to block replication?
Neutralizing flu requires about 2x as much serum after ~1 year's worth of antigenic change



+

Replication
Virus A
Antibodies from patient recovered from virus A


+
Replication
Virus B, which showed up ~1 year later
Antibodies from patient recovered from virus A



Neutralizing B.1.351 requires about 8x as much serum compared to the O.G.



+

Replication
O.G. SARS-CoV-2
Antibodies from patient recovered from O.G.
SARS-CoV-2


+
Replication
B.1.351
Antibodies from patient recovered from O.G. SARS-CoV-2









Some VoCs can escape antibodies from convalescent patients

Antigenic similarity = how much sera is required to neutralize viral replication?
Serum



Test viruses






1x
2x
4x
(relative to autologous)
X
X
X
X
X
X



--
Log2
0
1
2
Titer
Flu, per year
3
O.G. vs B1.351 SARS-CoV-2
Four reasons why we're not panicking about vaccine escape yet
- Immunity is broader after vaccination than after natural infection, especially for mRNA vaccines
- Antibodies aren't the whole story / immunity is a multi-layer defense system
- Usually requires a cluster of mutations to enable immune escape
- This has been a weird year for everyone -- host and virus alike
- Adapting to a new host species
(selective pressure for replication and transmission) - Totally naive population
(selective pressure for immune escape is a new thing)
- Adapting to a new host species
But, we will need to update the vaccines at some point
- Likely a similar or slightly lower frequency than flu vaccine updates
- Likely a simpler and faster process to update
SARS-CoV-2 mRNA vaccines than to update the egg-grown flu vaccine

How genomic surveillance can inform eventual vaccine updates
So, which mutations matter?
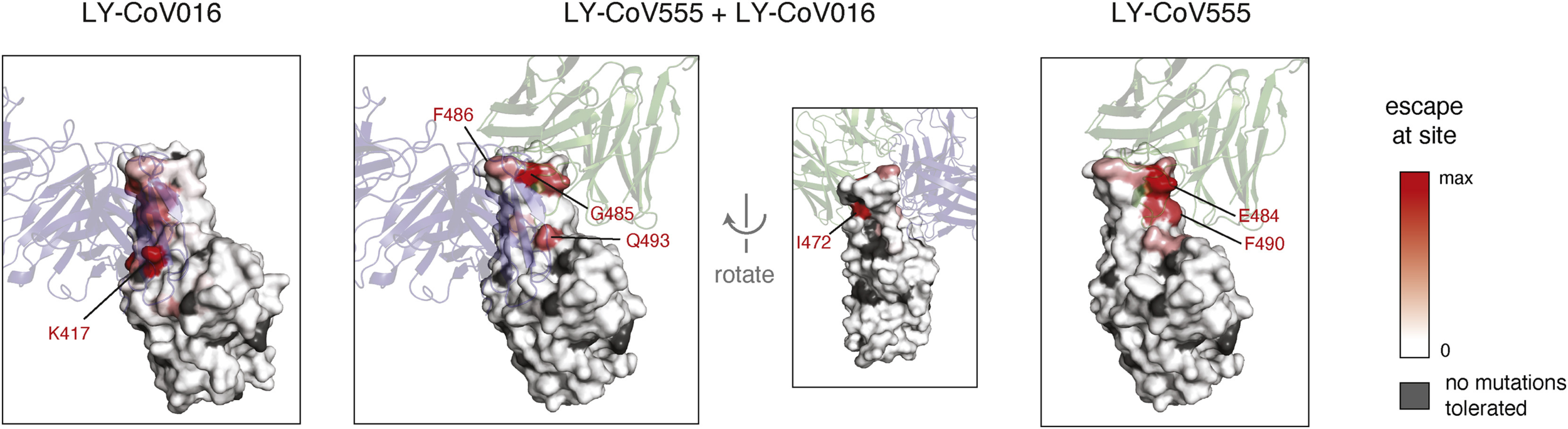
Starr et al, Cell Reports, 2021
Sequencing vaccine breakthrough cases helps us identify which [combinations of] mutations are important for vaccine escape, in the real world

Values from Juraska et al, PNAS 2018
Which clades / "variants" should we
vaccinate against?

John Huddleston
Antigenic change
Surface protein mutations
Recent clade expansion
Impact on protein function
+
+
+
Representative sampling helps us model and predict which clades ("variants") will become more common
Growth rate @
next year

Titers
Sequence
Trees
Sequence + modeling
Clade growth rates are well-predicted
John Huddleston

Clade growth vs decline is also well-predicted
John Huddleston

Representative sampling helps us model and predict which clades ("variants") will become more common
John Huddleston

Takehomes
- SARS-CoV-2 will change antigenically, and we'll need to update our vaccines at some point, but we're not panicking
- Individual mutations don't usually cause antigenic change on their own
- Genomic surveillance is one of the best ways to ensure we're ready to update the vaccines when the time comes
- SARS-CoV-2 is here to stay, but it won't ever sweep through a totally naive global population like this again. We're still learning what antigenic evolution "normally" looks like in this virus.
Questions?
@sidneymbell



Many thanks to the Bedford lab
& the Nextstrain team
Advanced methods reading
Antigenic cartography
Visualize antigenic space via a low-dimensional embedding
Evolution-based models
Assign antigenic change to branches or substitions, interpolate across dataset
Population dynamics
Antigenic fitness & clade turnover
cdph-webinar-antigenic-evolution-intro-2021
By Sidney Bell
cdph-webinar-antigenic-evolution-intro-2021
Introduction to how viruses evolve to escape the immune system and what this means for public health.
- 1,205



