A community approach to taking out the brain from a vat and putting it back in the body

| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |
Hanover, 07.11.2025

Faculty of Psychology and Neuroscience, Maastricht University, Maastricht, The Netherlands; Open Science Special Interest Group (OHBM); physiopy (https://github.com/physiopy)




I have no financial interests or relationship to disclose with regard to the subject matter of this presentation.
Disclaimers
1. I am an "open" scientist. I have a bias toward the
core tenets of Open Science as better practices.
2. I am a co-founder and current member of the
physiopy community.
This is a new chapter
This is a new chapter
Take home #0
This is a take home message
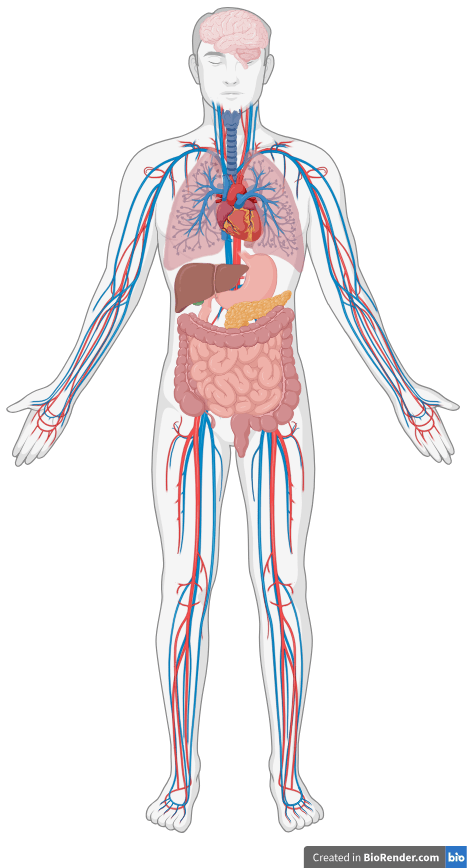
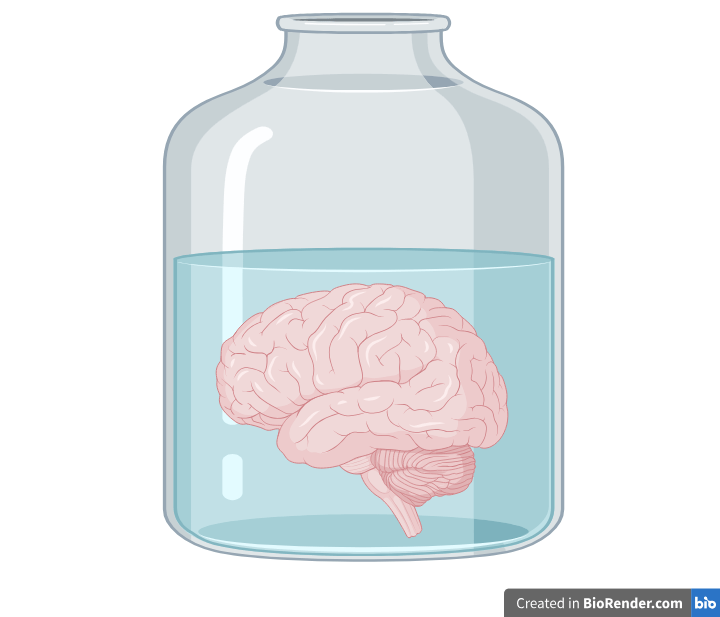
Brains in vats?
No, brains in bodies

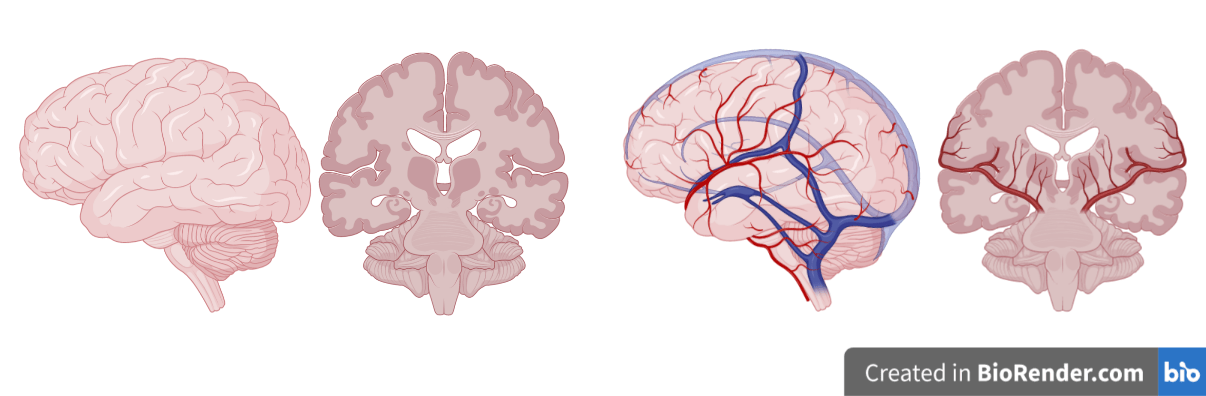
Neurovascular Coupling
Image courtesy of Martin Havlicheck and Dimo Ivanov






Changes in
Haemodynamics
Neurovascular
Coupling
Changes in Oxygen Metabolism
Changes in
BOLD signal
(Venous) Vasculature
Arrows indicate causal influence
Impact of physiology on data variance
1. Bianciardi et al., 2009 (Magn. Reson. Imaging.); 2. Triantafyllou et al., 2005 (NeuroImage);
3. jorge et al., 2013 (Magn. Reson. Imaging.), Reynaud et al., 2017 (Magn. Reson. Med.)
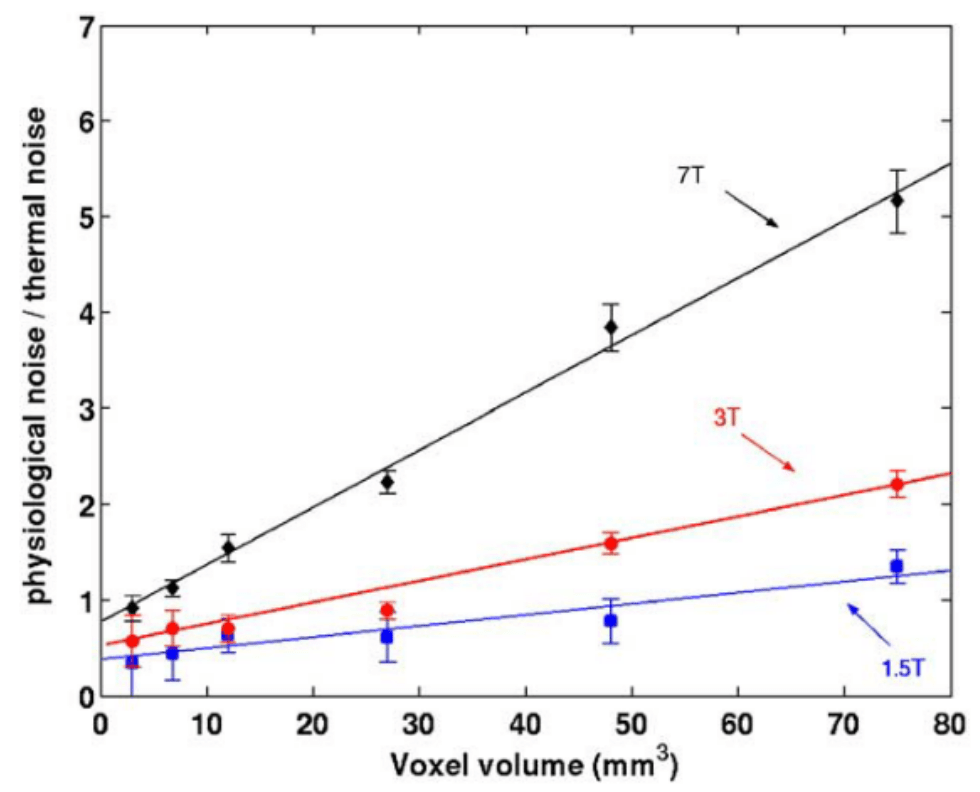
Physiology-related variance varies:
- By voxel size¹ ² and position¹
- By field strength²
- By sequence type and TR³
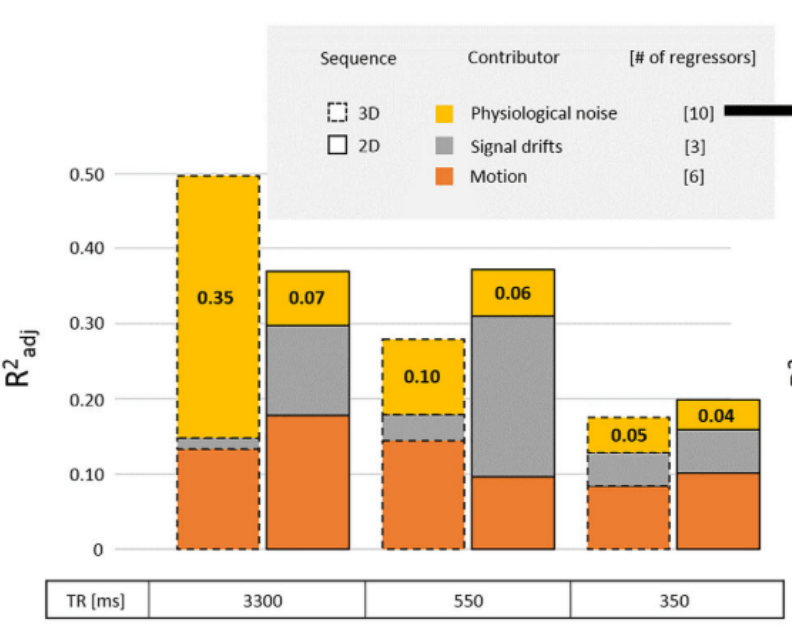
Impact of physiology on data variance
1. Krentz et al., 2023 (bioRxiv), Carlton et al., 2024 (bioRxiv), Moia et al., 2024 (bioRxiv);
2. Birn et al., 2009 (NeuroImage), image courtesy of Jingyuan Chen; 3. Lee et al., 2023 (HBM)
Physiology-related variance varies:
- By individual & session¹
- By task (task-locked)²
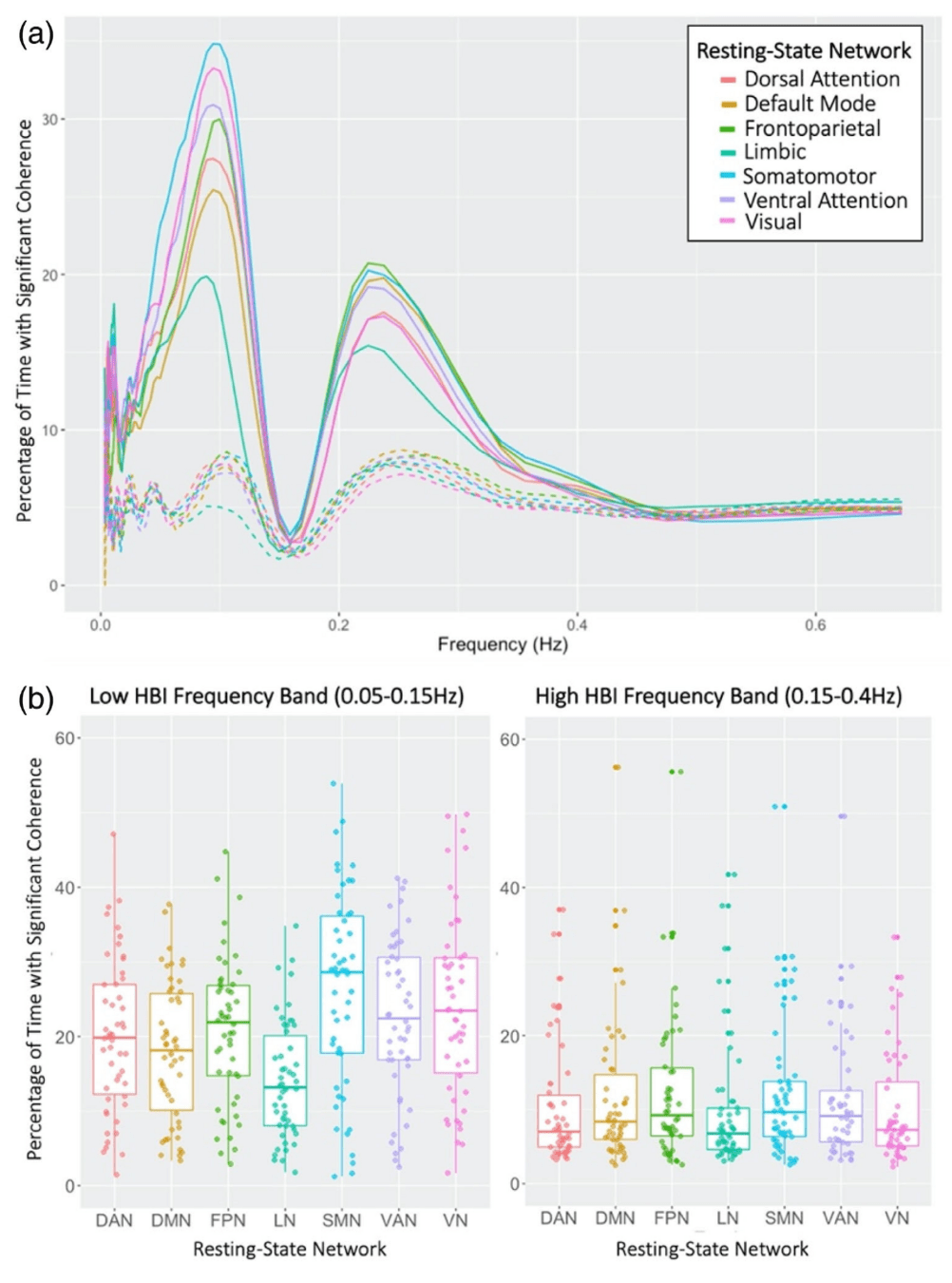
- By Resting State Network³
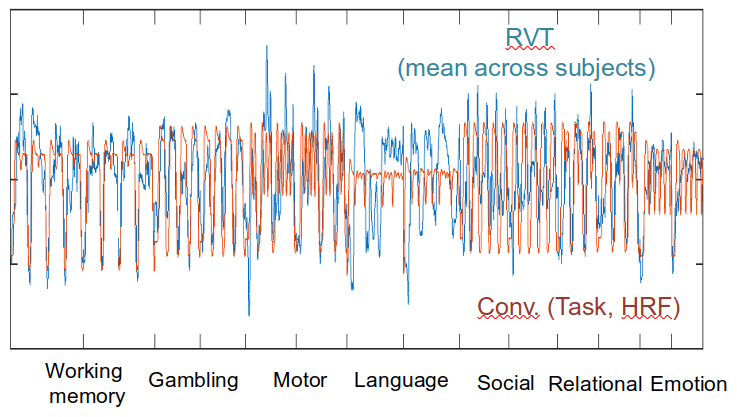
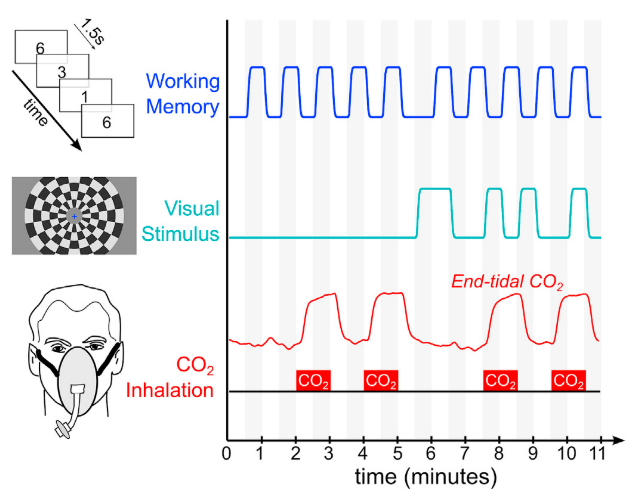
Ventilation
Task (convolved)
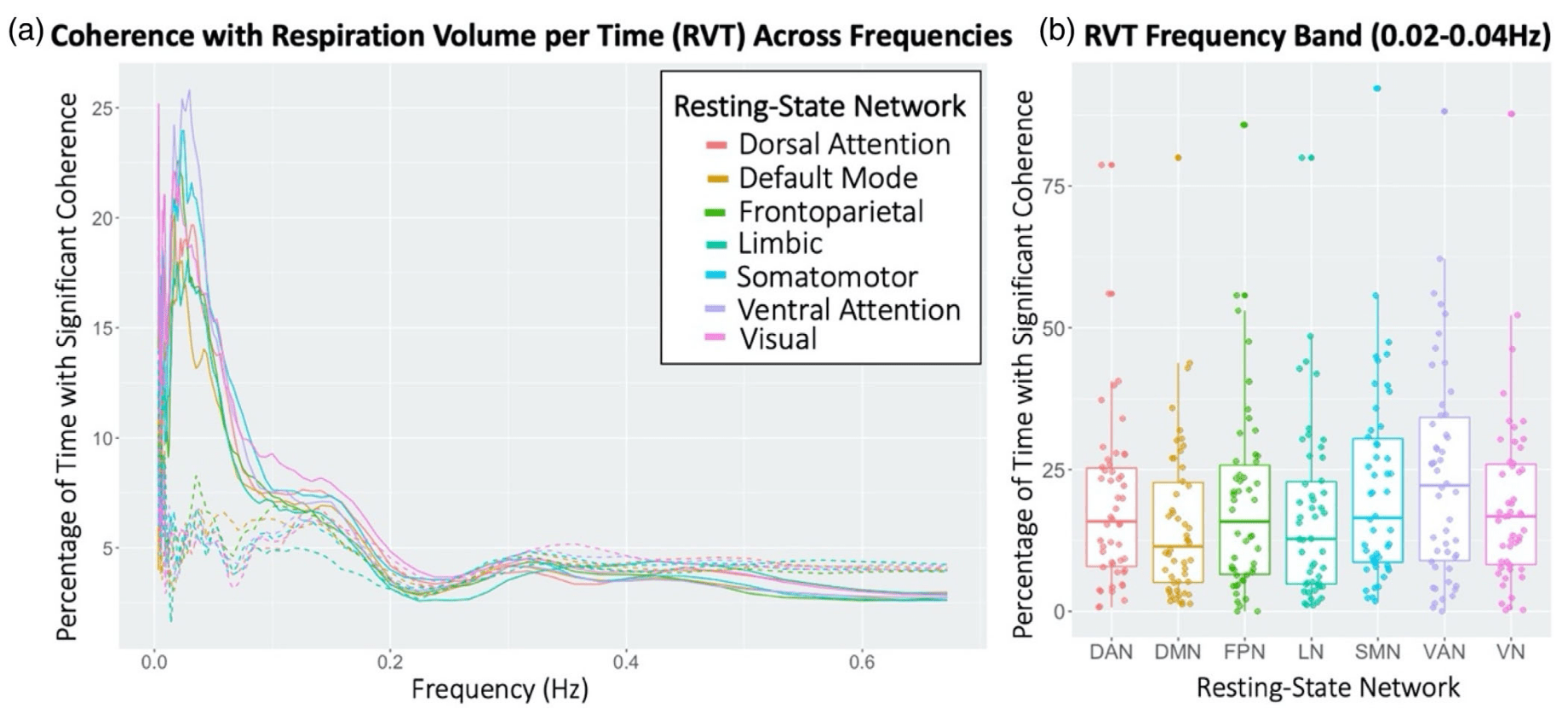

RETROICOR variability
Denoising is strongly linked to interpretation
Are you interested in individual or group level effects?
Are physiological responses part of your interpretation?
(e.g. affective neuroscience)
Should your interpretation include autonomic nervous system changes?
[...]
Are your comparisons robust to physiological responses?
Physiological data processing
Raw data
BIDSification
phys. data preprocessing
(peak detect.)
phys. denoising
phys. imaging
Data acquisition
QA/QC
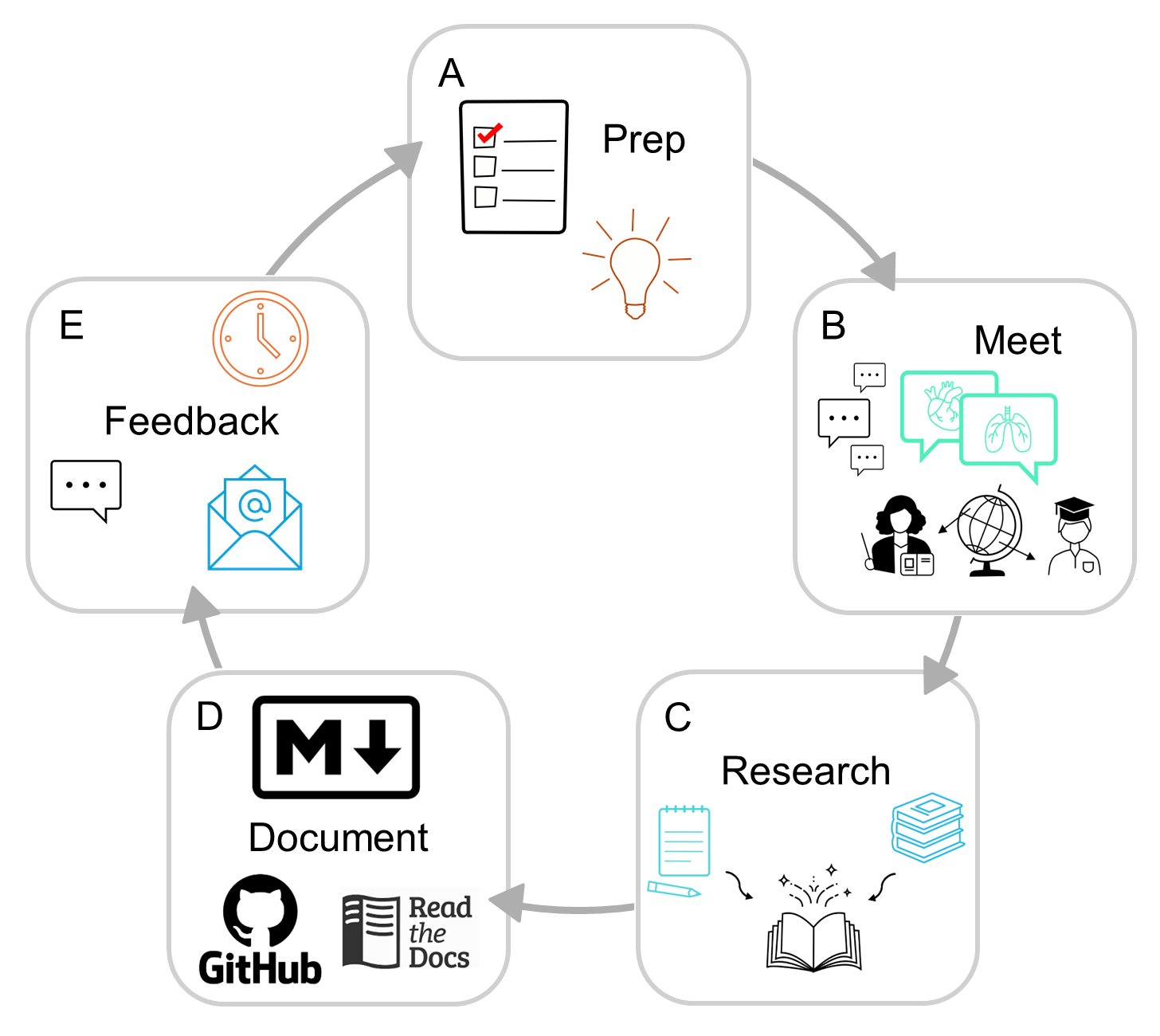
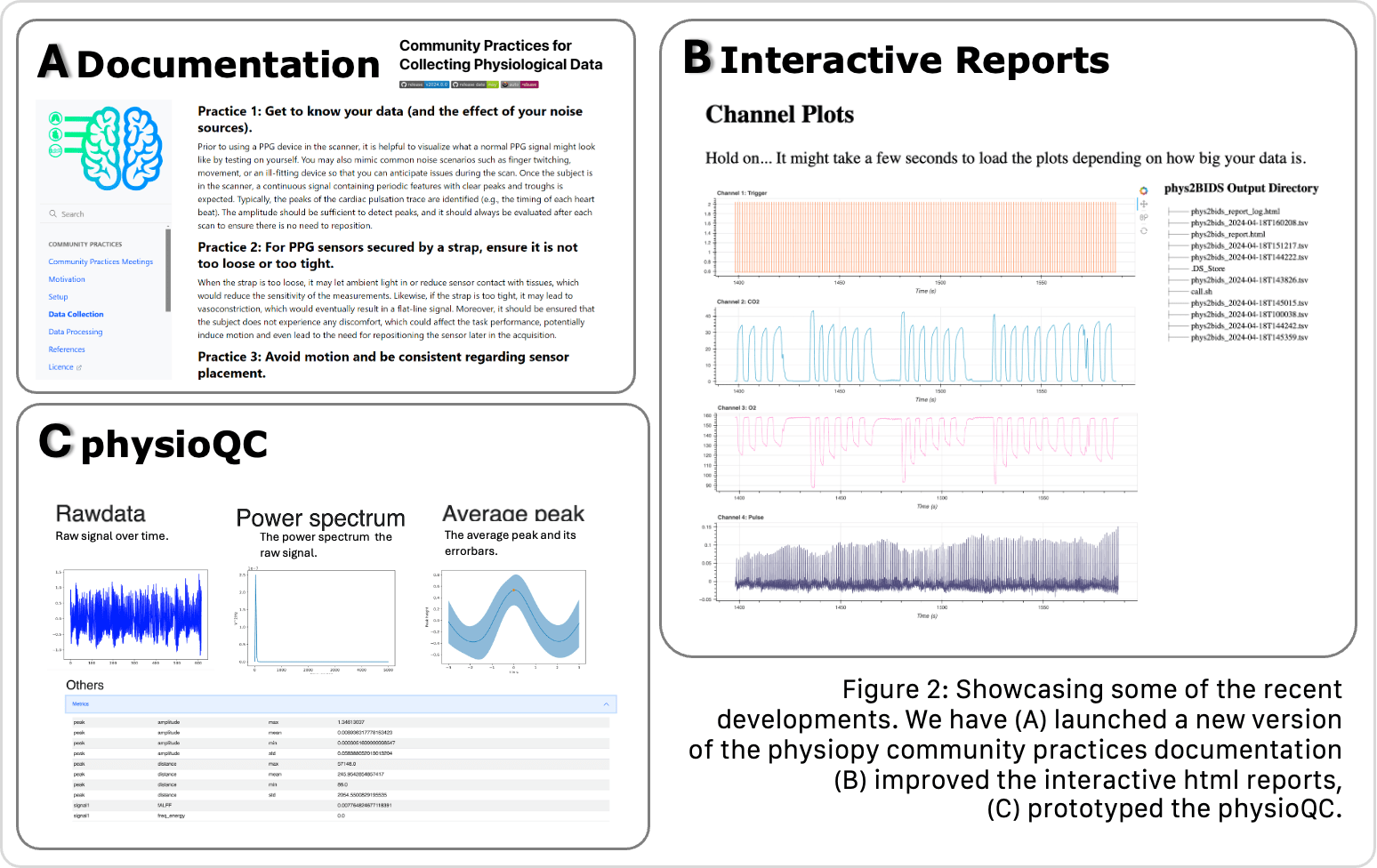
Why physiopy
In neuroimaging, integration of physiological measures to data collection and analyses are still a niche topic. By raising awareness, we can inspire researchers and clinicians become interested in the topic.
*Open Source Software Development is the idea of developing a software publicly, sharing it from the beginning of the development, fostering a democratic community of contributors in support of the project, using version control and software testing.
physiopy adopts a Community driven, BIDS-based, Open Development* approach, aimed at bringing governance back to the users.
Sharing physiological data, toolboxes, and documentation following the concepts of Open Science could improve the exposition to this topic.
Community practices meetings, community consensus, and community guidelines.
Spark interest!
The more we share, the better it becomes
This is (not)
the way!
Of the People, by the People, for the People
Core components
A set of easily adoptable toolboxes
Community of users, developers, and researchers interested in physiology
Clear and approachable documentation
Community practices based on consensus
Core components
physiopy's status
- Highly inspired by the tedana community and The Turing Way
- Founded in 2019 by 9 people in 3 labs (during the DC Code Convergence 2019!)
- It now counts over 35 contributors from over 25 institutions (not all currently active)
- All contributors are recognised via all-contributors schema and authorship, alphabetically
- All volunteers are... volunteering (so far) - and development spikes during Brainhacks
- We got a new governance based on the Minimal Viable Governance model
- We are also restructuring contributors' guidelines, to make them more friendly!









Raw data
BIDSification
phys. data preprocessing
(peak detect.)
phys. denoising
phys. imaging
Data acquisition
QA/QC
physiopy's projects
physiopy's projects
Raw data
BIDSification
phys. data preprocessing
(peak detect.)
phys. denoising
phys. imaging
Data acquisition
Process description
QA/QC
physiopy's projects
Raw data
phys2bids
peakdet
phys2denoise
phys. imaging
Data acquisition
physiopy's documentation
&
Coordinated testing suite
BIDS Extension Proposal
physioQC
Physiopy's Community Practices
physiopy's projects
Raw data
phys2bids
peakdet
phys2denoise
phys. imaging
Data acquisition
physiopy's documentation
&
Coordinated testing suite
BIDS Extension Proposal
physioQC
Physiopy's Community Practices
prep4phys
How to record physiological data
Ventilation
Respiration (O2/CO2)
Pulse
Electrodermal Activity
Community practices
physiopy-community-guidelines.readthedocs.io

Community practices
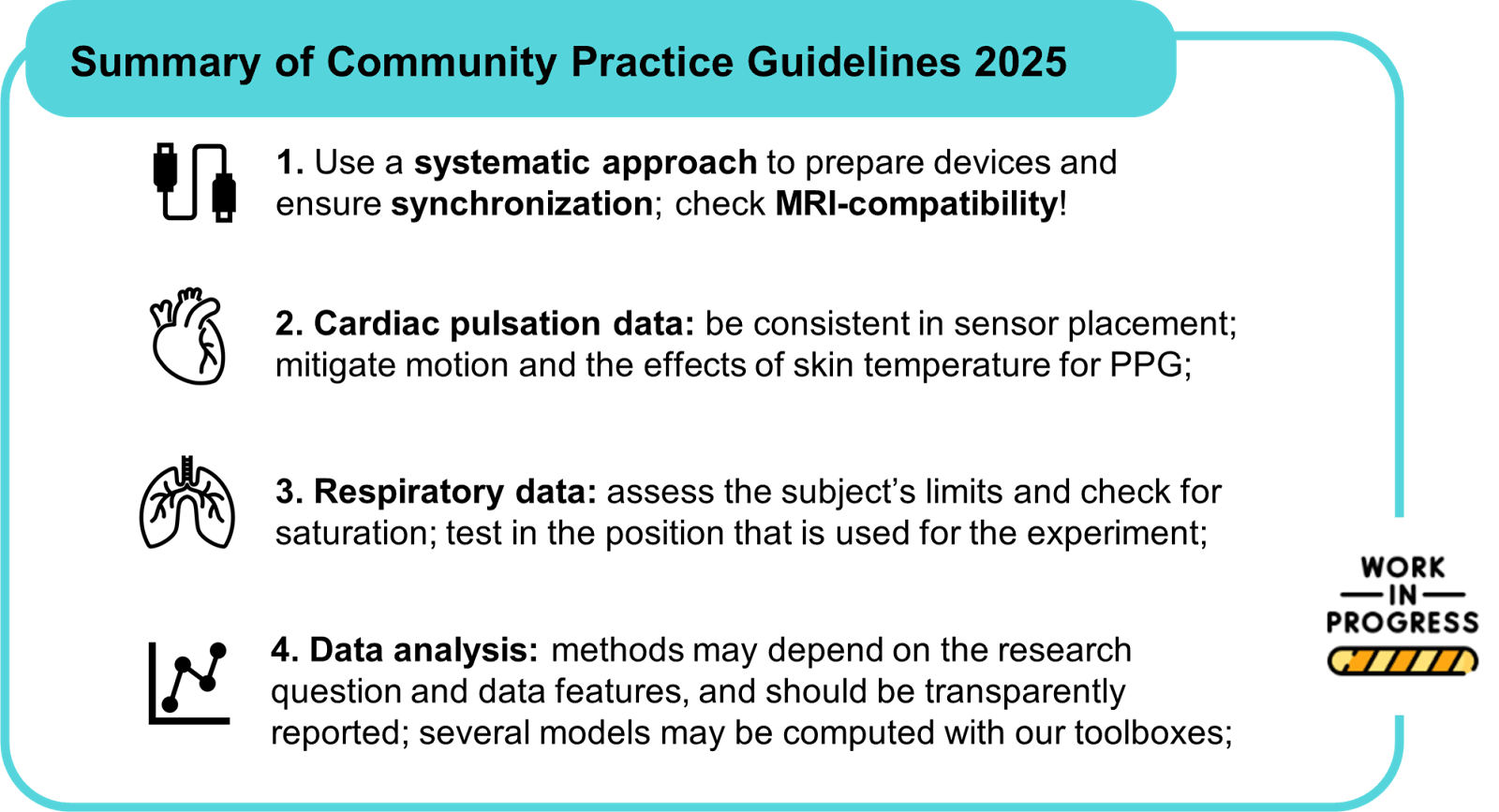
Community practices
More Community Practices!
Response functions and convolutions
Physiology and vigilance
Physiological confounds in resting state
ECG, PPG, and Heart Rate Variability
Blood Pressure and Cerebral Autoregulation
Brain-Body Interaction
Physiological workflows

Open meetings
every 3rd Thursday of the month
at 17h00 UTC
New cycle starting soon!
Data standardisation (BIDSification)

Alcalà et al., 2023 (Zenodo)
BIDS Extension Proposal(s)

- Physio as independent modality & multimodal concurrency
- Identification of jey metadata
- Provenance of derivatives
- Guidance for formatting output files
- Flexibility and specificity for different processing stages
https://github.com/physiopy/bids-specification-physio/discussions/1

Open meetings
every 2 weeks
at 14h00 UTC
Quality Control
images courtesy of Kristina Zvolanek, Elenor Morgenroth, and César Caballero-Gaudes
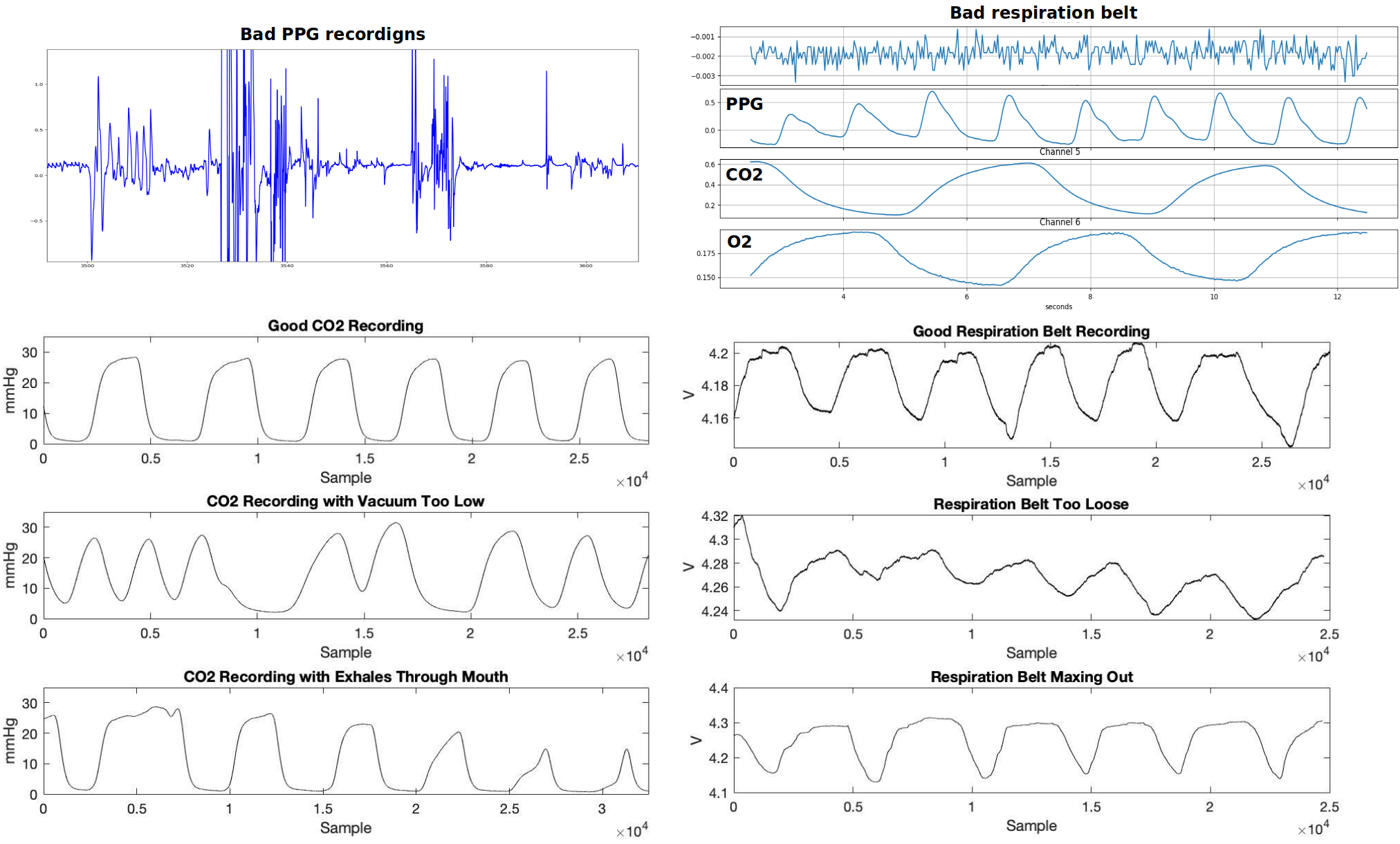
Quality Assessment/Control
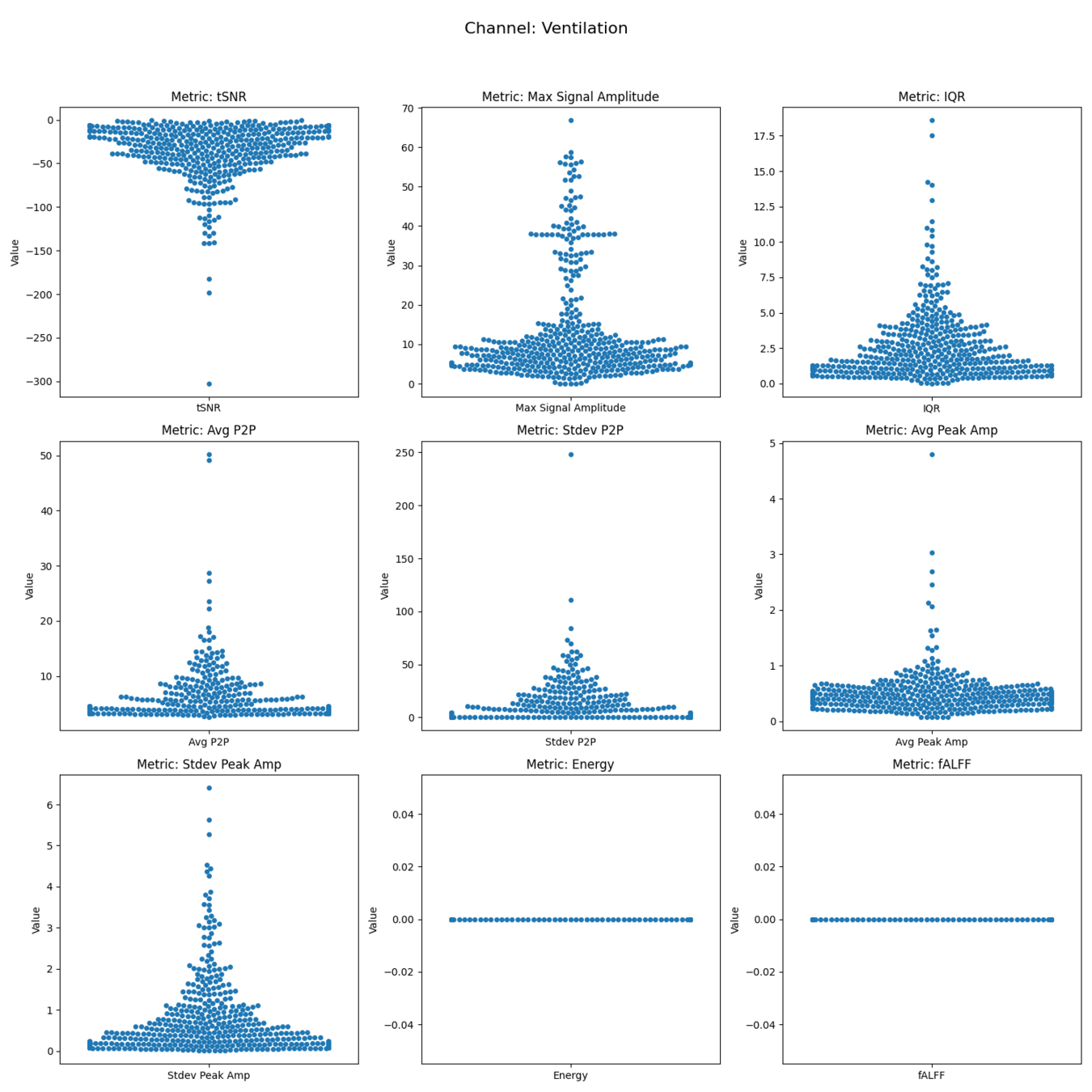
Goodale et al., 2024 (Zenodo)
"Denoise" the "noise"
Bottenhorn et al., 2023 (Aperture Neuro)
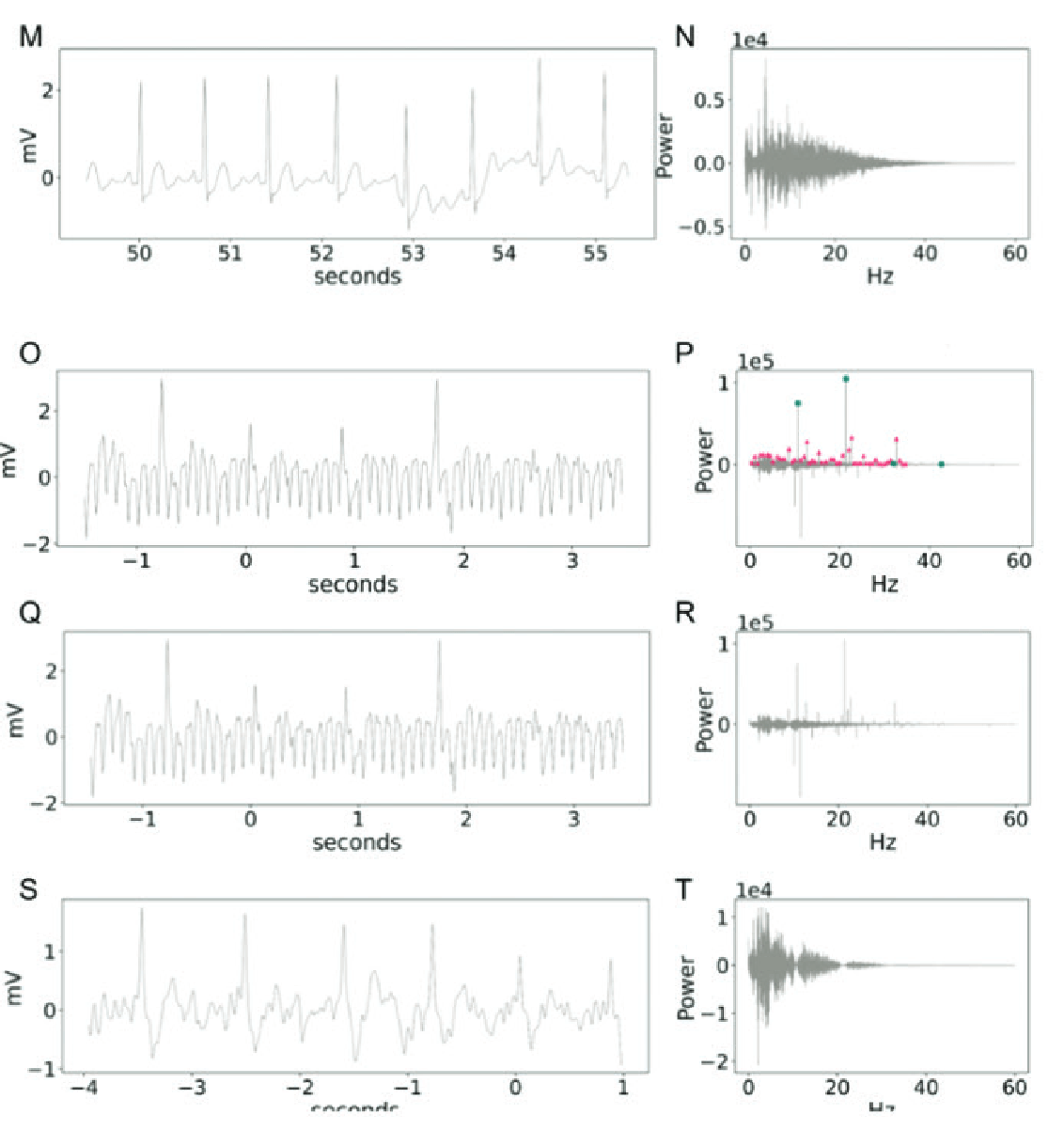

ECG before scanner
ECG during fMRI
ECG after Bottenhorn filter
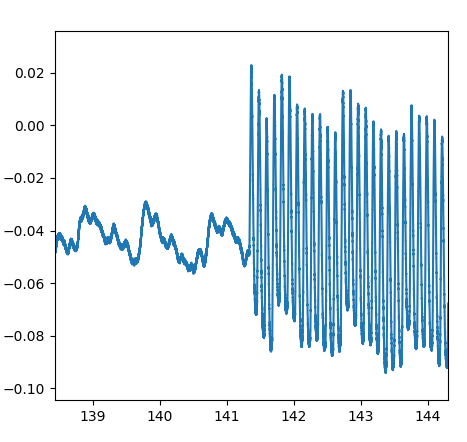
Scanner off
Scanner on
PPG
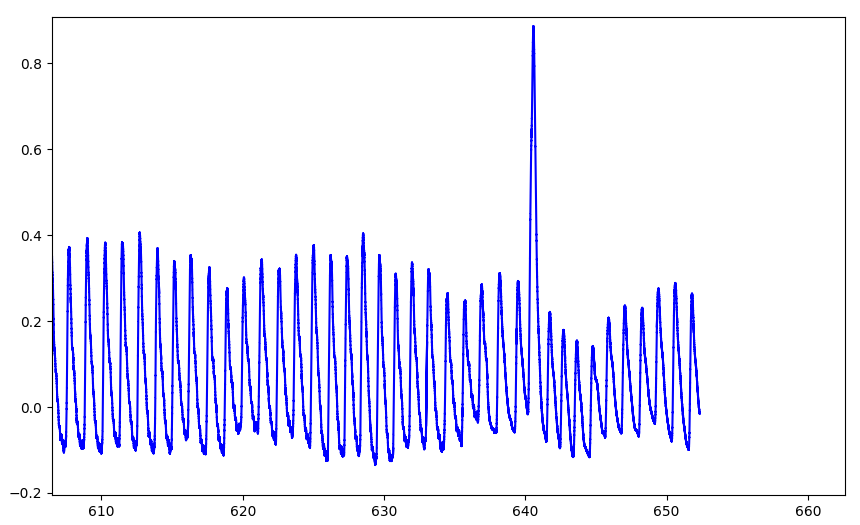
Make sure you're removing what you should
data = np.genfromtxt('sub-007_ses-05_task-rest_run-01_physio.tsv.gz', usecols=[0, 1, 3])
ph = peakdet.Physio(data[:, 1], fs=10000, suppdata=data[:, 2])
ph = peakdet.operations.peakfind_physio(ph, thresh=thr, dist=dist)
ph = peakdet.operations.edit_physio(ph)
DuPre et al., 2024 (Zenodo)
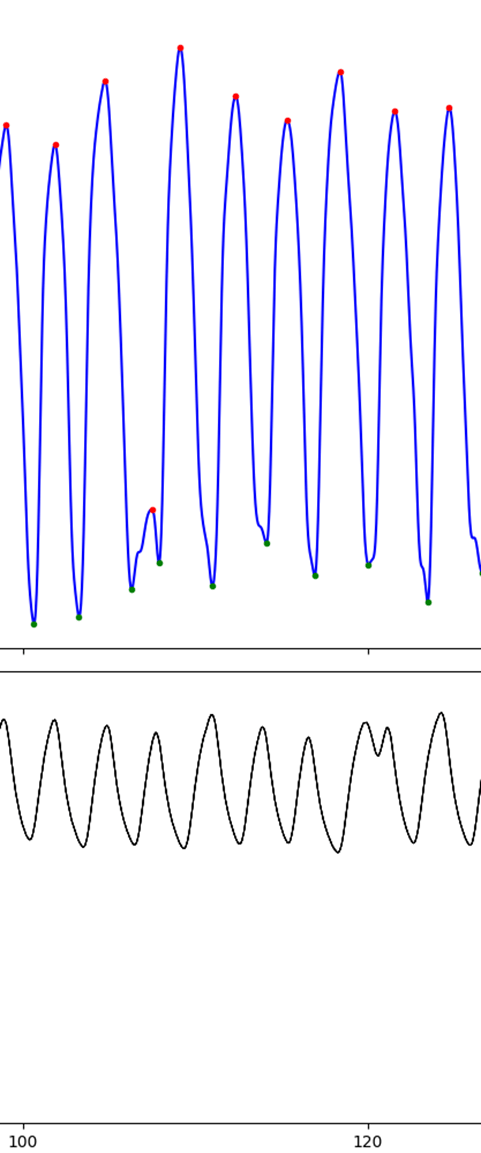
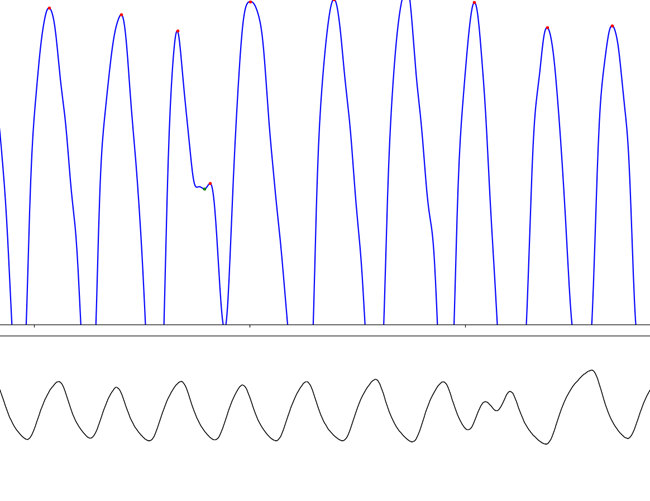
PPG
Ventilation
CO2
Phase changes over physiological cycles
image courtesy of Marta Bianciardi
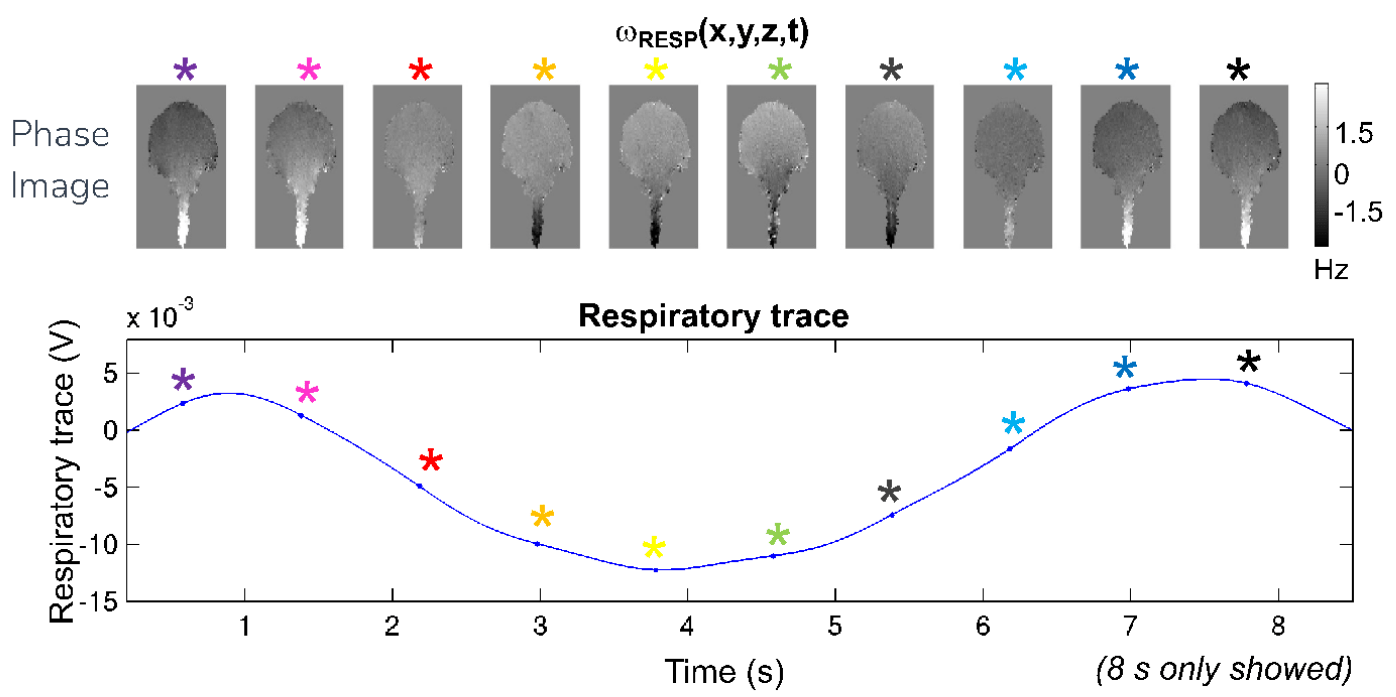
Denoising with physiological data: RETROICOR¹
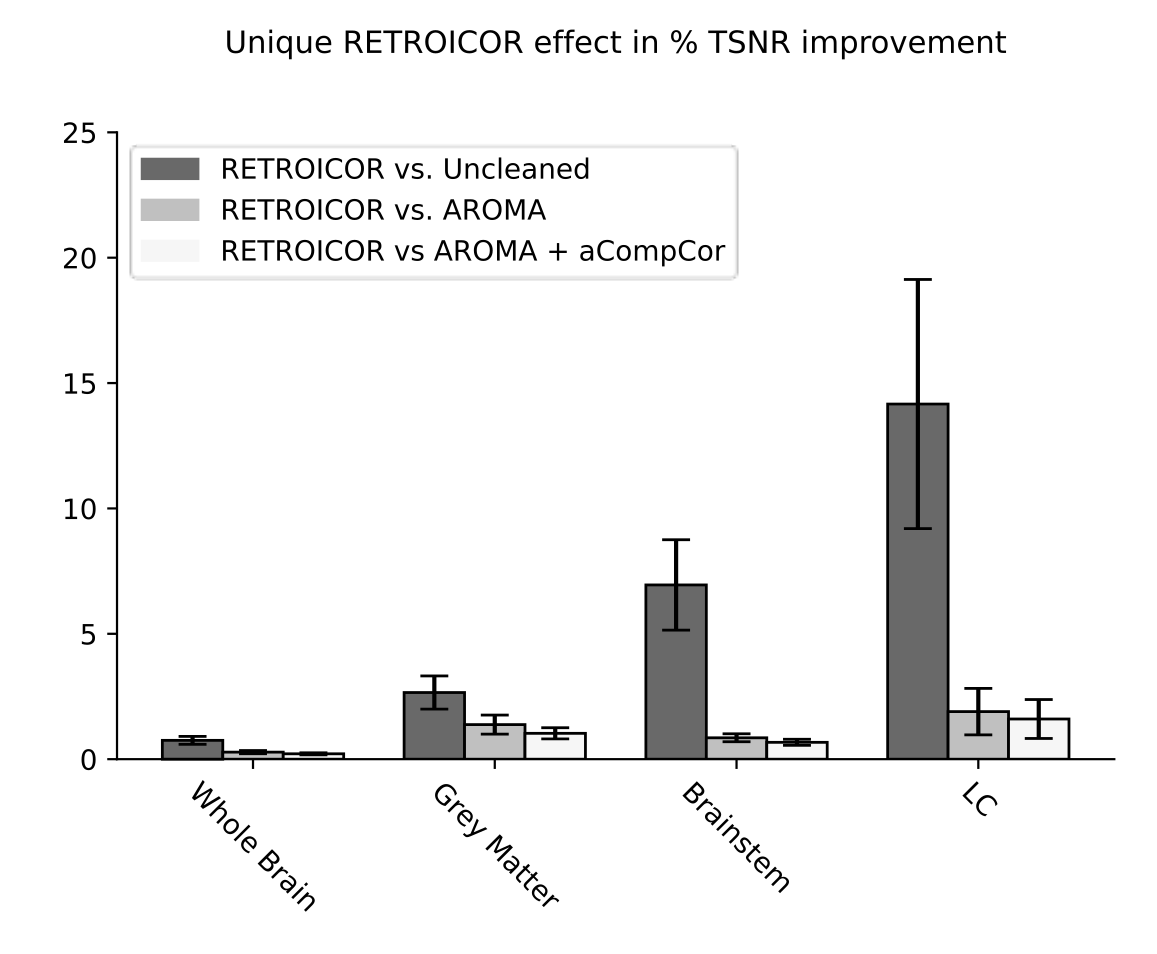
1. Glover et al., 2000 (Magn. Reson. Med.); 2. Kasper et al, 2017 (J. Neurosci. Methods); 3. Krentz et al., 2023 (bioRxiv)
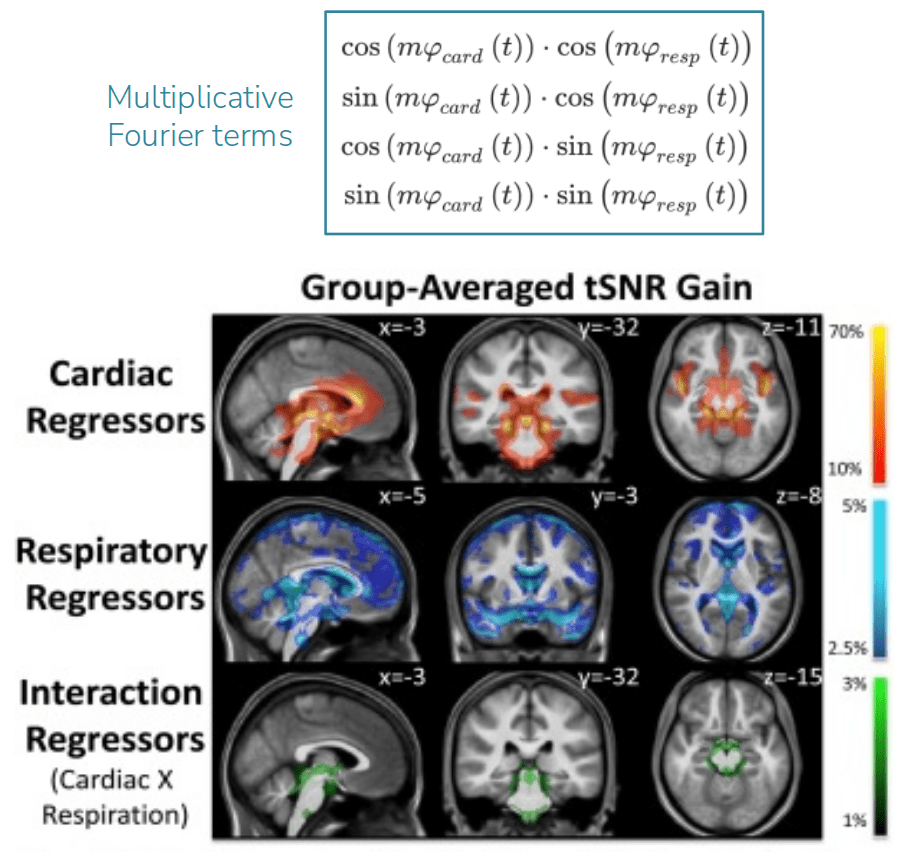




Whole
Brain
GM
Brainstem
LC
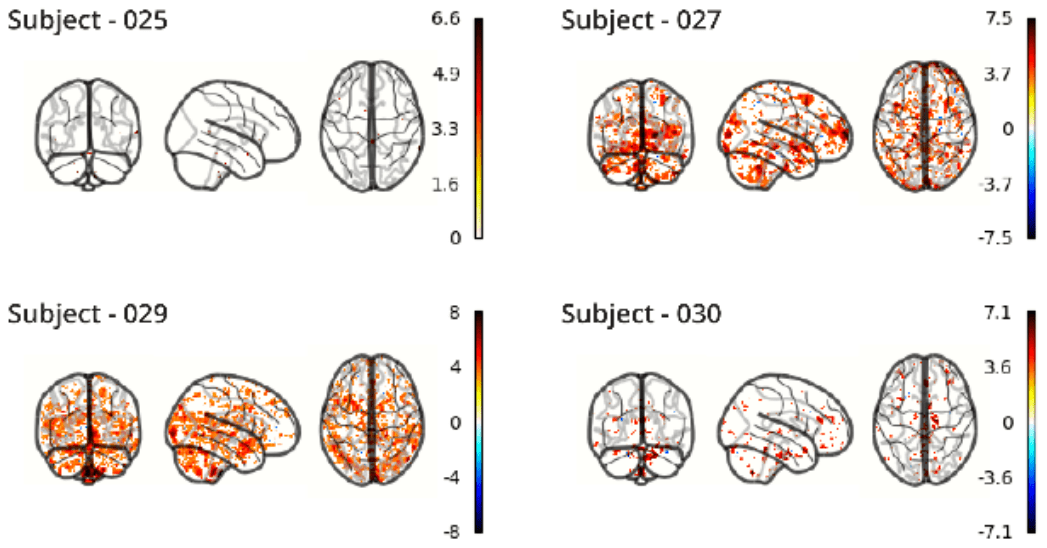
RETROICOR variability across subjects³
²
Physiological harmonics
Image courtesy of Blaise Frederick
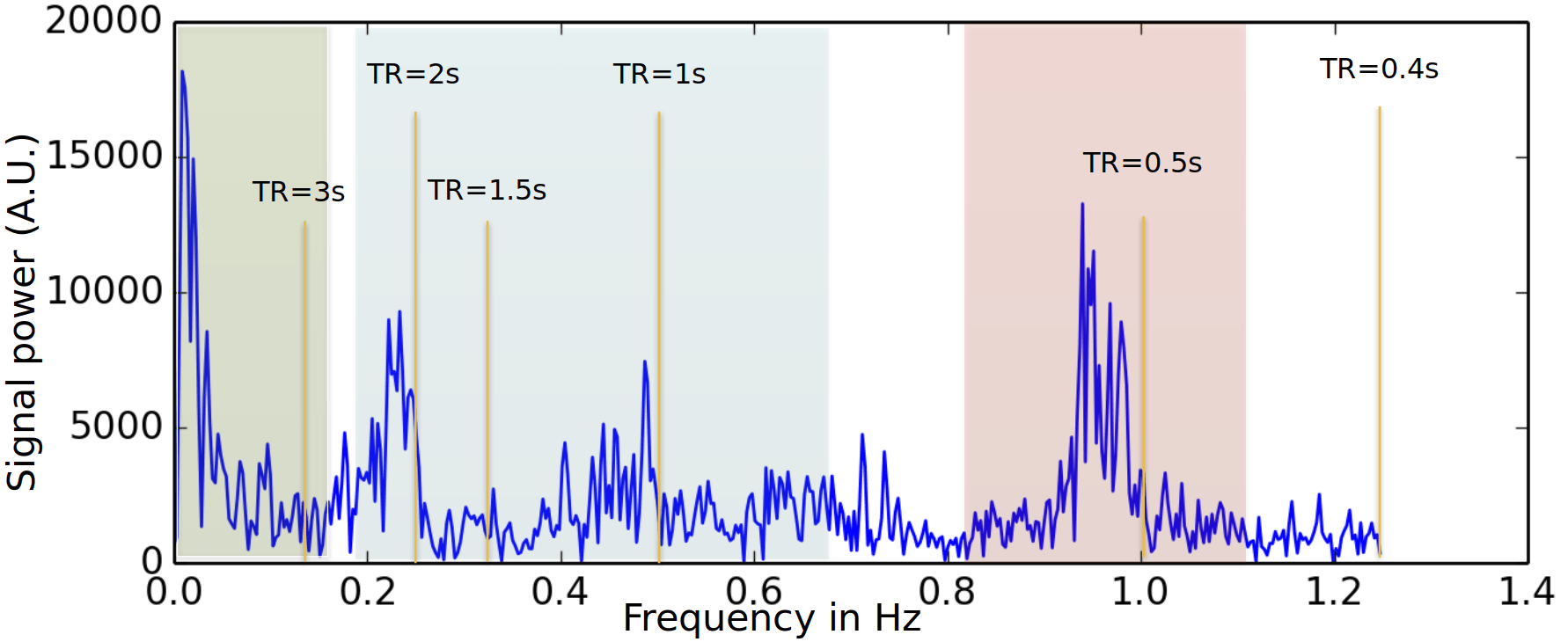
Cardiac
Respiratory
Denoising with physiological data: slow fluctuations
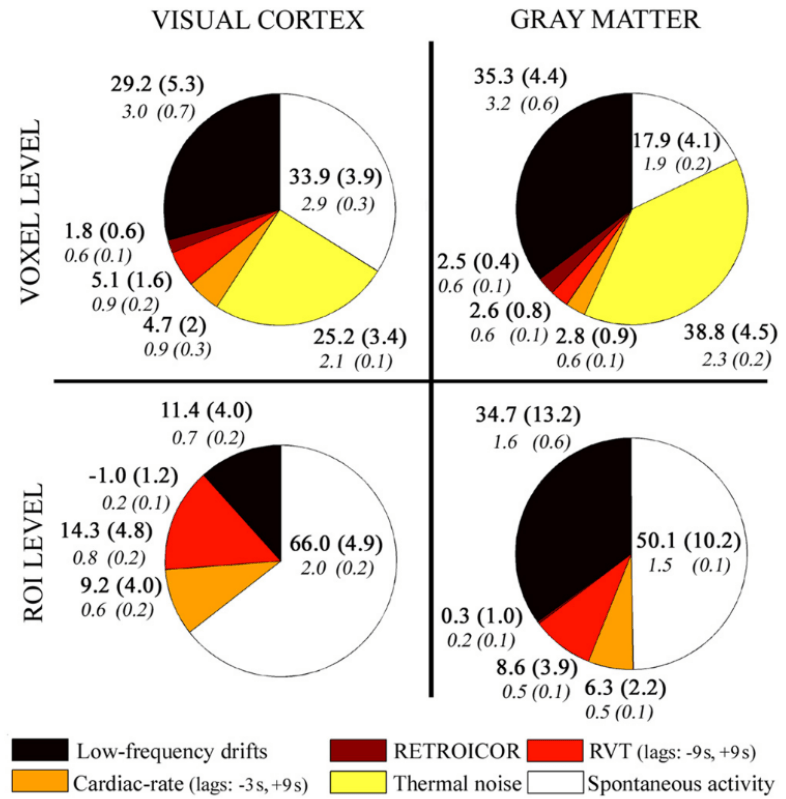
HRV: Shmueli et al., 2007 (NeuroImage); HBI: Chen et al., 2020 (NeuroImage), img from Lee et al., 2023 (HBM)
phys2denoise: Bottenhorn et al., 2024 (Zenodo); phys2cvr: Moia et al., 2024 (Zenodo); PhysIO Toolbox: Kasper et al., 2017 (J. Neurosci. Methods)
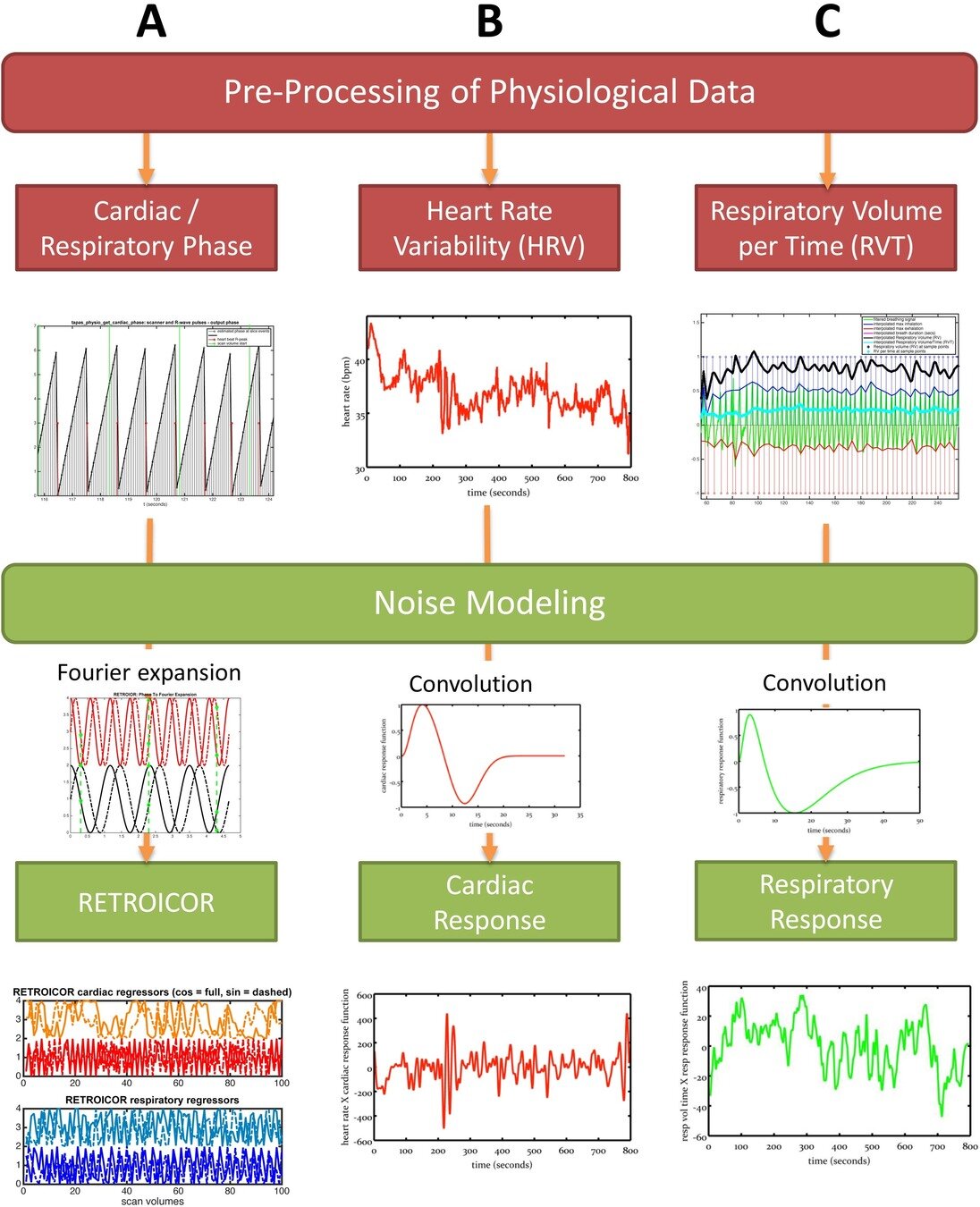

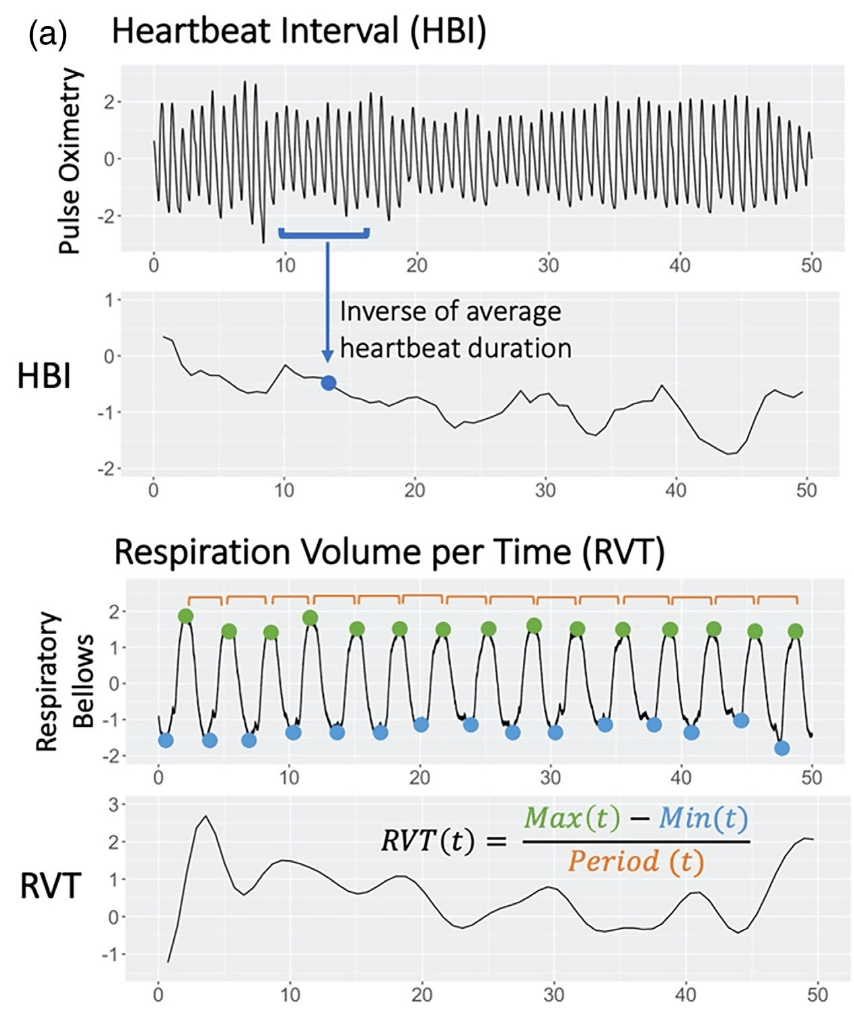

RVT: Birn et al., 2006 (NeuroImage), img from Lee et al., 2023 (HBM); RV: Chang & Glover, 2009 (NeuroImage), img from Chen et al., 2010 (NeuroImage)
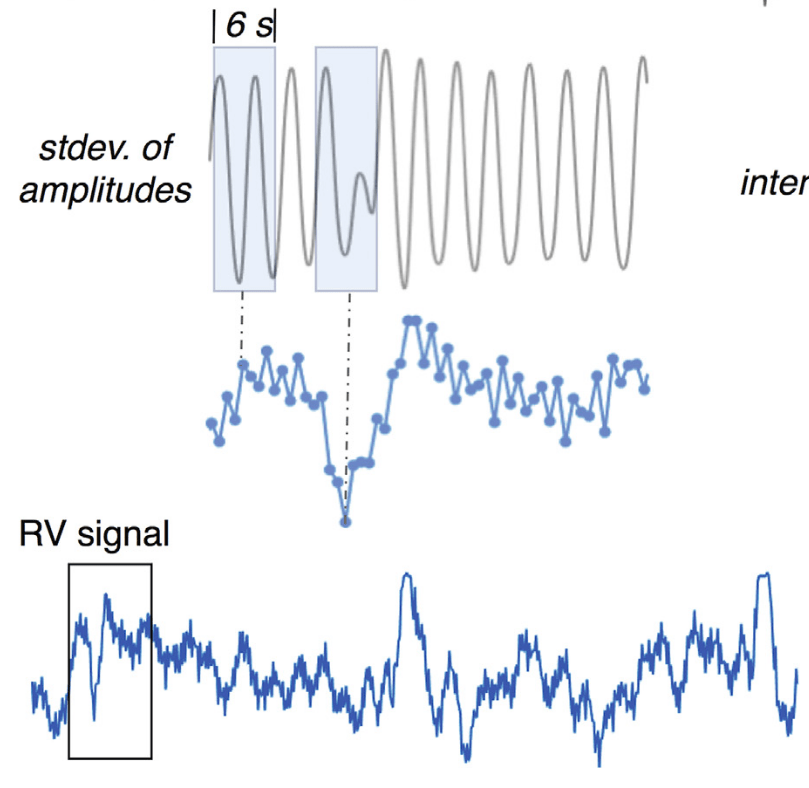
Respiratory Variance (RV)
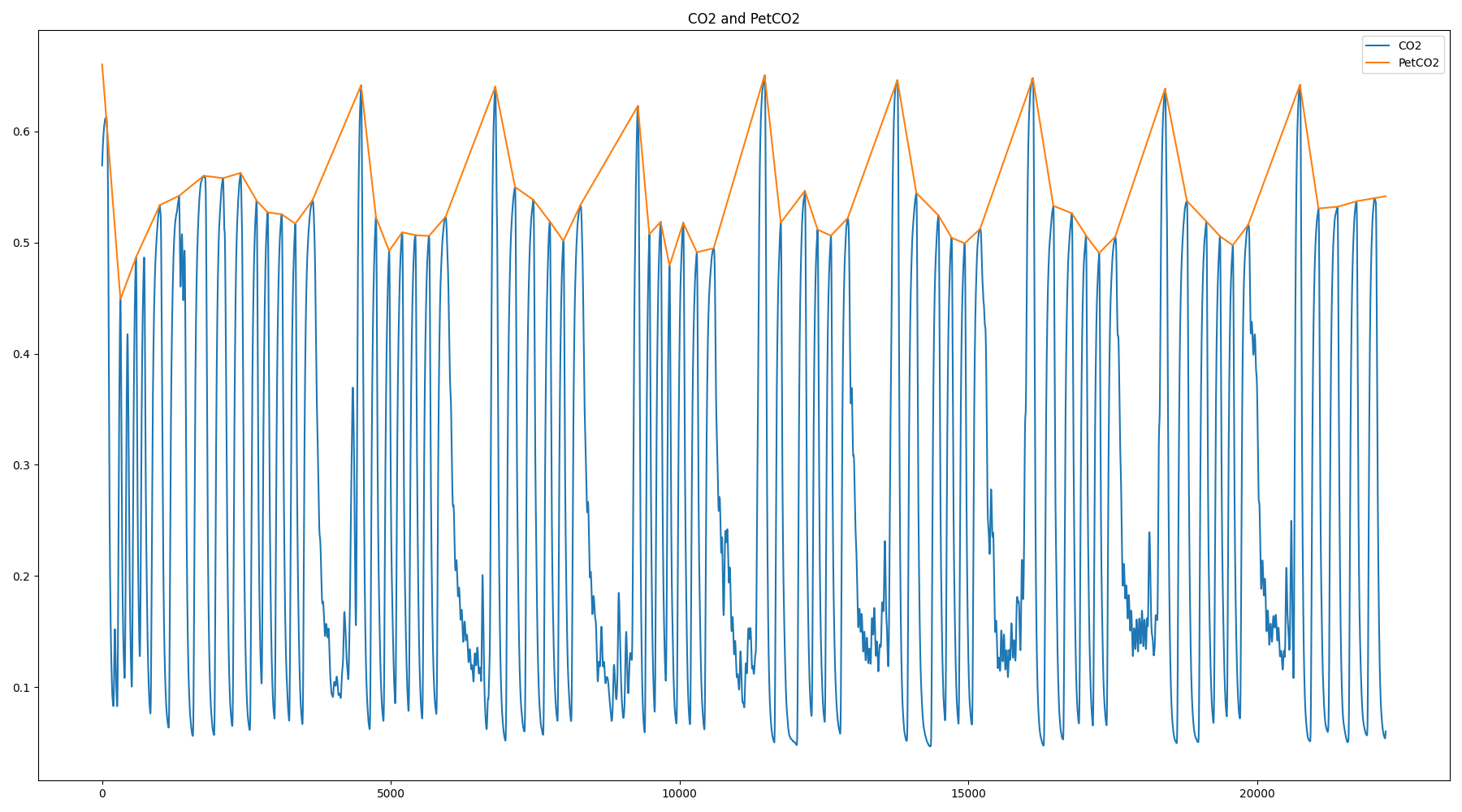

Pressure of End Tidal CO2 (PetCO2)
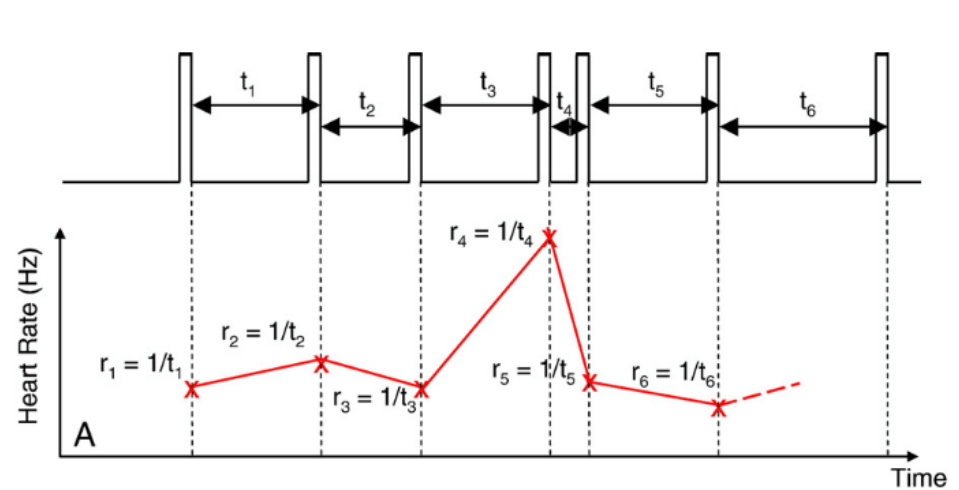
Heart Rate Variability (HRV)
PhysIO toolbox
Physiological denoising in ICC
HRV: Shmueli et al., 2007 (NeuroImage); HBI: Chen et al., 2020 (NeuroImage), img from Lee et al., 2023 (HBM)
phys2denoise: Bottenhorn et al., 2024 (Zenodo); phys2cvr: Moia et al., 2024 (Zenodo); PhysIO Toolbox: Kasper et al., 2017 (J. Neurosci. Methods)
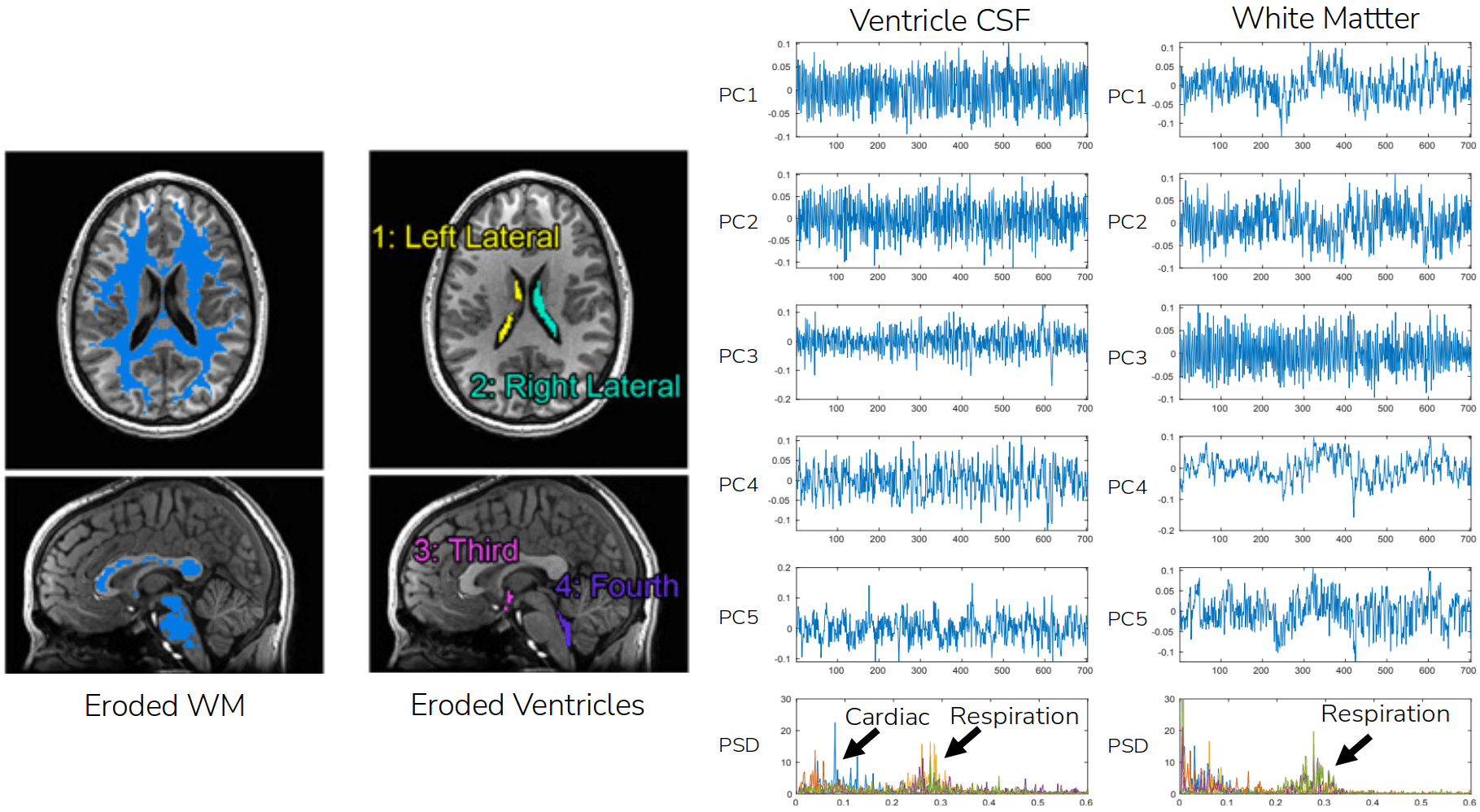
Denoising through data decomposition: CompCorr¹
1. Behzadi et al., 2007 (NeuroImage); image courtesy of César Caballero-Gaudes
Impact of physiological denoising on data reliability
Belli, 2025 (UM)

Motion regressors
Motion + Physio regressors
Motion + tissue-based regressors
& BandPass
Motion regressors & BandPass
Motion + Physio regressors
& BandPass

Reconstructing physiological data from fMRI
1. Aslan et al., 2019 (Neuroimage); 2. Wang, Xu, et al., 2024 (arXiv)
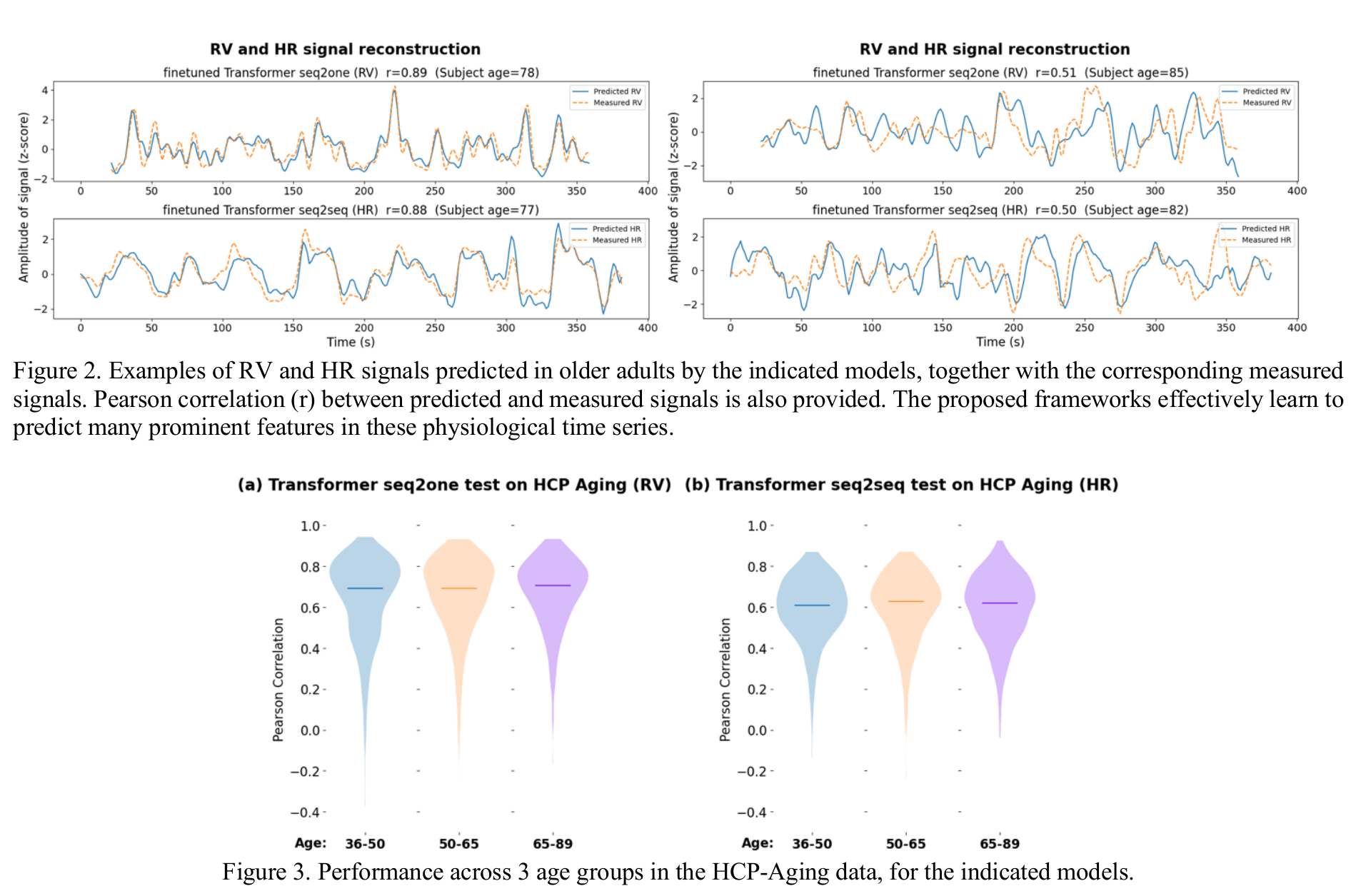


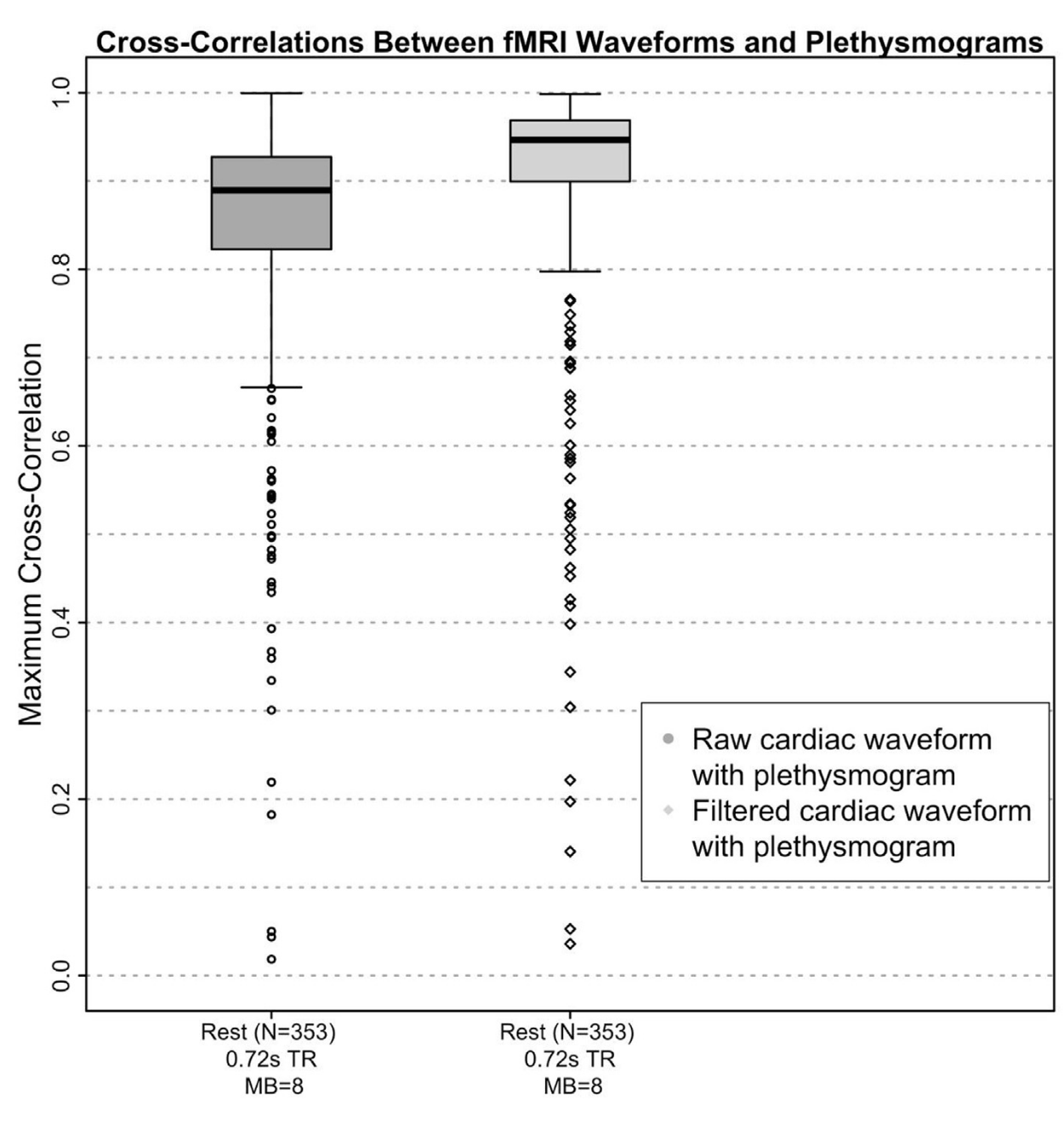
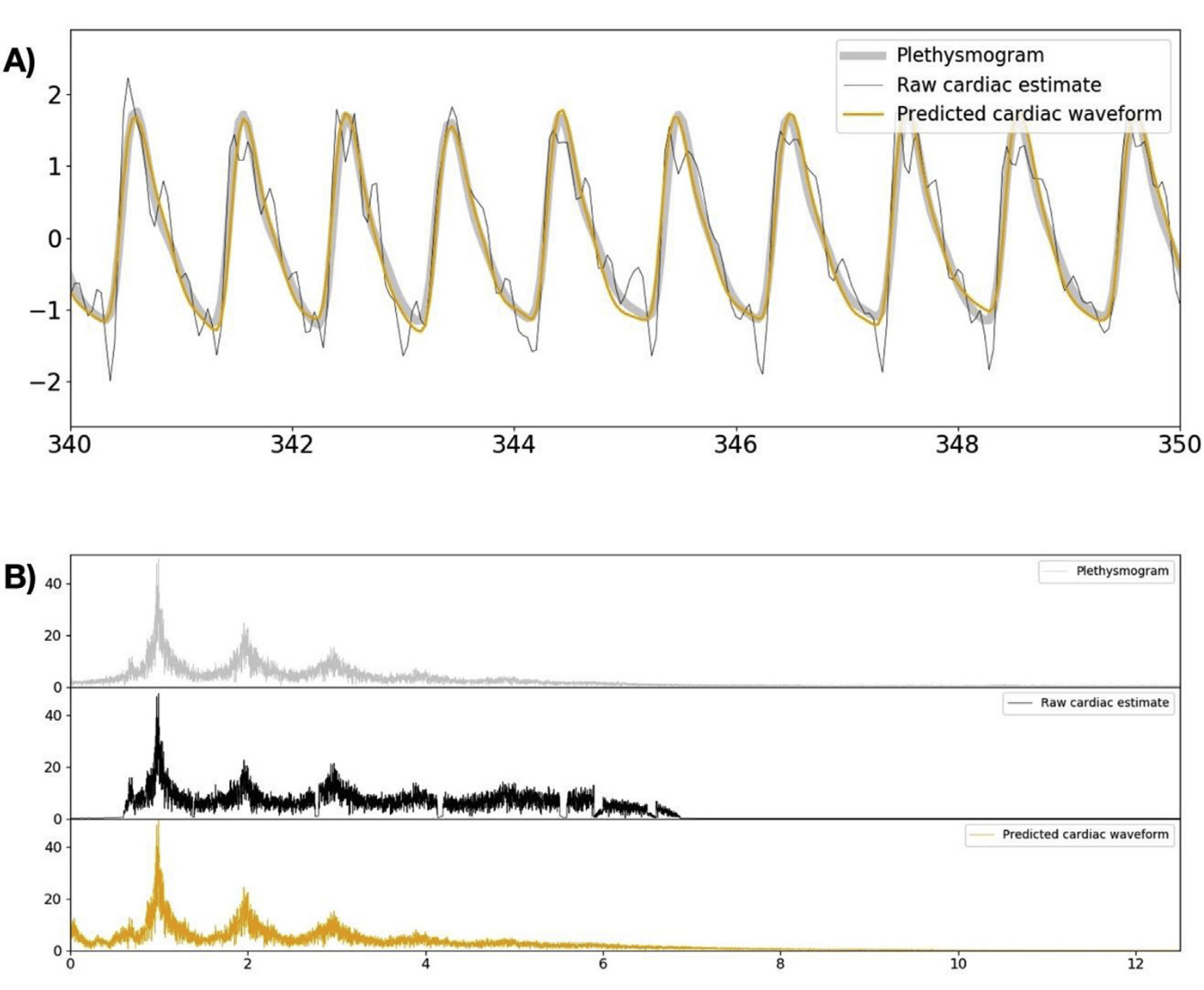
Cardiac waveform from multislice data¹
RV and HRV from parcellated BOLD fMRI²
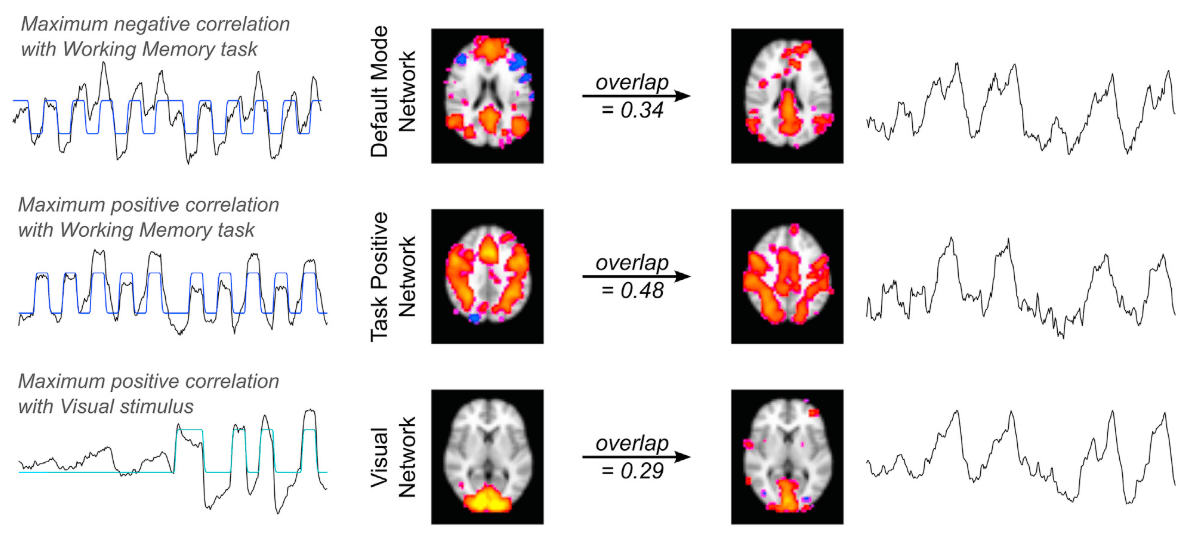

1. Bright et al. 2020 (NeuroImage), Chen et al., 2020 (NeuroImage)
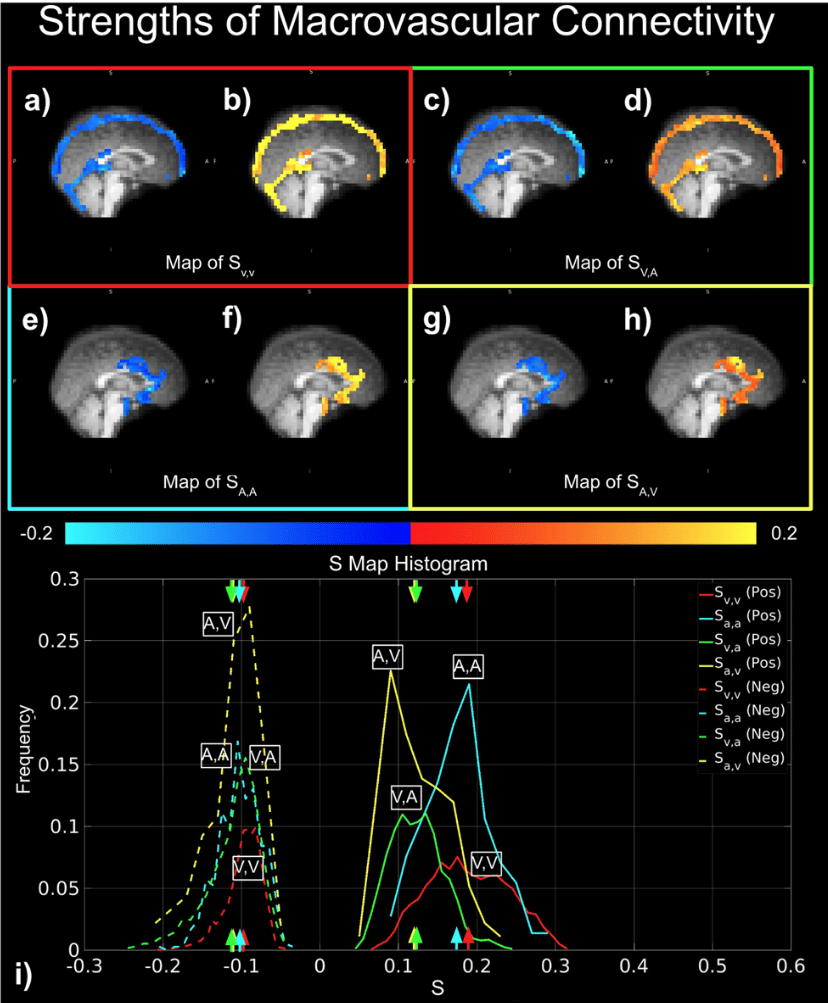
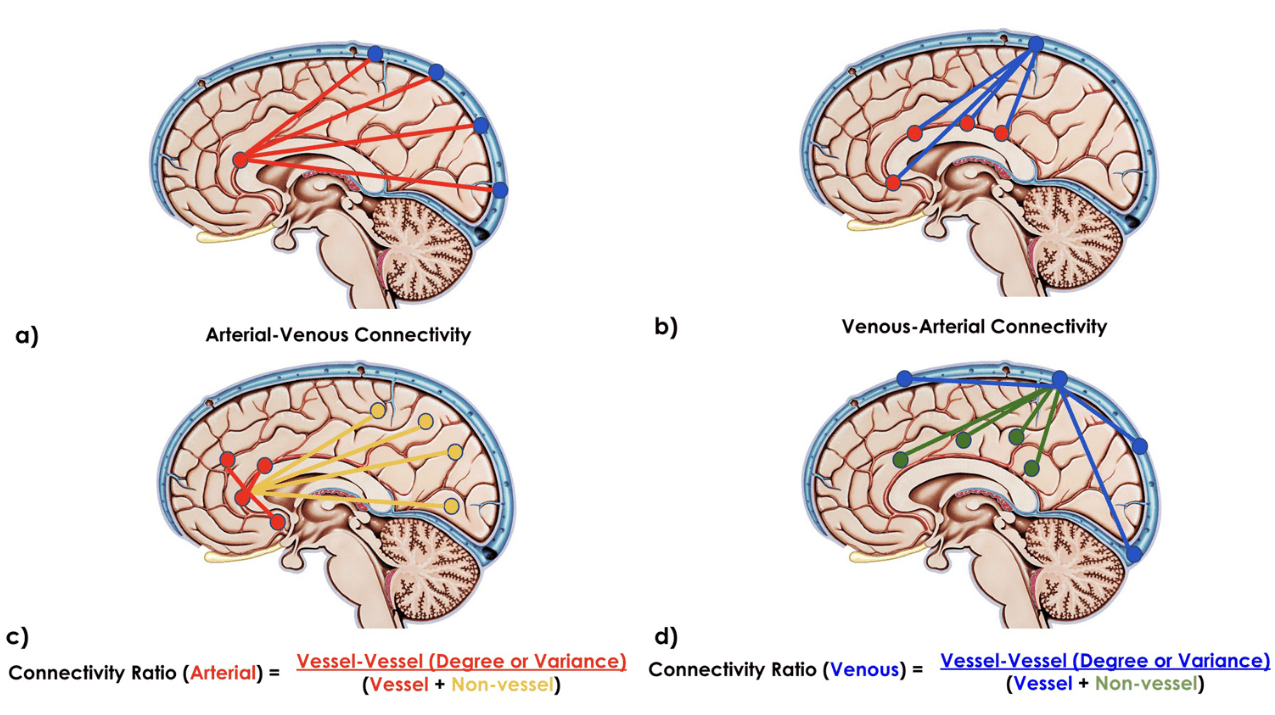
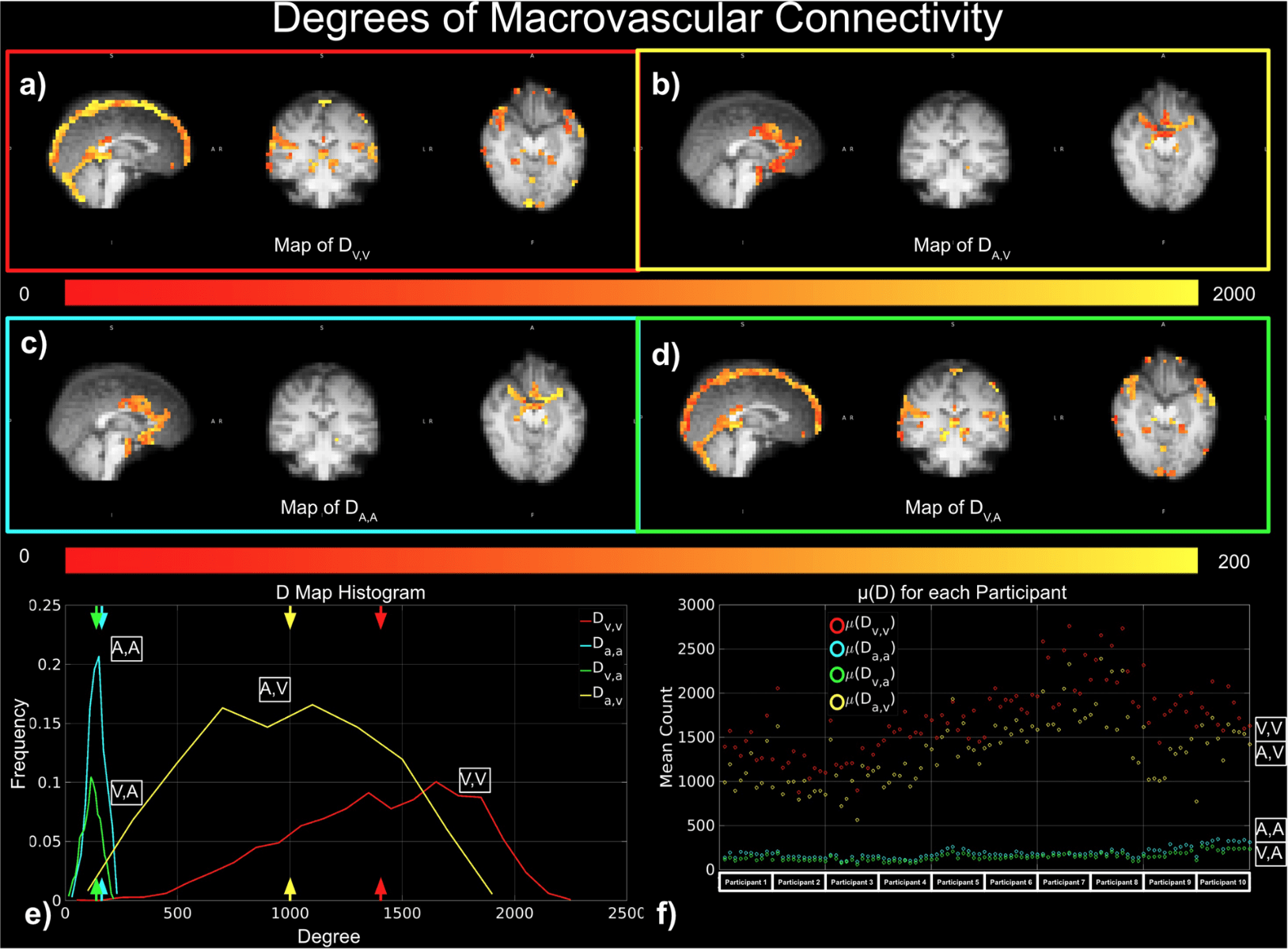
BOLD-based fMRI, in conjunction with physiological signals, reveals the existence of physiological and vascular brain networks¹.
"Static" physiology: vessels
1. Gulban et al., in prep., Gulban et al., 2025 (bioRxiv), image courtesy of Faruk Gulban; 2. Zhong et al., 2024 (Imaging Neurosci.)

- Meso-scale vessels introduce partial volumes effects
in the grey matter¹ - Macro-scale vessels affect connectivity
in the grey matter²
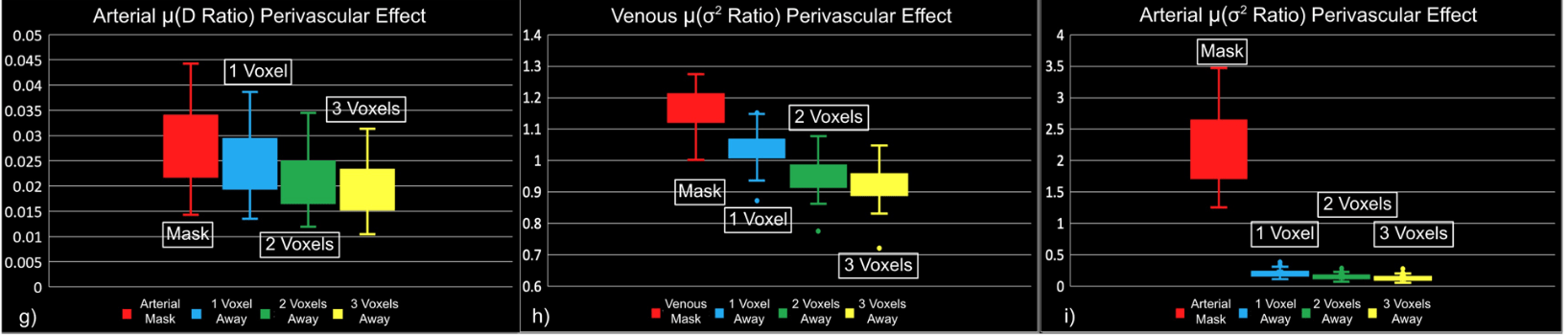
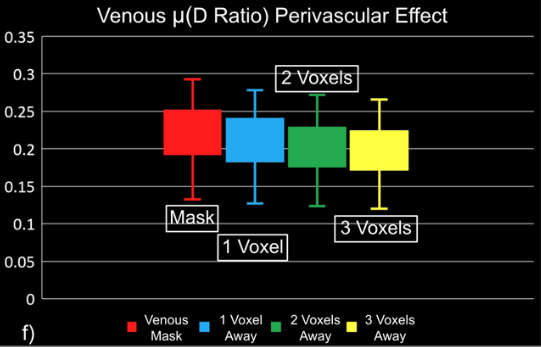
"Static" physiology: vessels
Zhong et al., 2024 (Imaging Neurosci.)













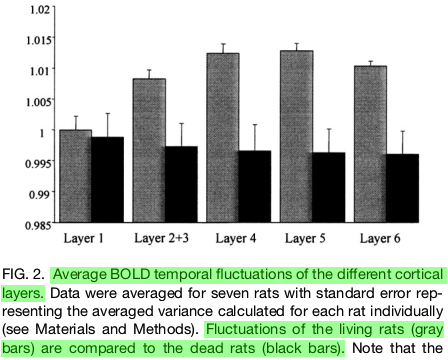
Denoising is strongly linked to interpretation

More voices & more diverse expertise improve our community

(and we're not here to tell you what to do - we want to figure it out with you)
Join us!
Send an email to:
physiopy.community@gmail.com
(Even if you are not a physiology/python guru!)
That's all folks!











Find the presentation at:
slides.com/smoia/hanover2025/scroll





| smoia | |
| @SteMoia | |
| s.moia.research@gmail.com |
Supporters: Eunice Kennedy Shriver NICHD / NIH (K12HD073945), EUs Horizon 2020 R&I program (Marie Skłodowska-Curie 713673), La Caixa Foundation (ID 100010434, fellowship LCF/BQ/IN17/11620063), Spanish E&C Ministry (Ramon y Cajal Fellowship, RYC-2017- 21845), Spanish State Research Agency (BCBL “Severo Ochoa” excellence accreditation, SEV- 2015-490), Basque Government (BERC 2018-2021 and PIBA_2019_104), Spanish SI&U Ministry (MICINN; FJCI-2017-31814)





Any question [/opinions/objections/...]?
Take home messages
- Denoising is linked to interpretation
- Physiological signals should be considered when comparing sequences, subjects, tasks, networks
- Physiological must be quality controlled to avoid bad denoising
- RETROICOR explains part of the data variance that other models do not explain
- When lacking (good) physiological data, data-driven approaches can be used
- Physiological data can be partially retrieved from BOLD MRI
- Brain vessels may bias your functional data

Physiological BIDS
extension proposal(s)

Physiopy Community Guidelines

Find the presentation at:
slides.com/smoia/hanover2025/scroll

Enhancements!
- ECG vs PPG
- EDA
- Eye Tracking

Add guidelines on
- Siemens
- Bitalino
- non-MRI modalities
Extend support to
- NeuroKit
- NiPreps
- AFNI & FSL
Integrations
- (Semi-)Automation
- BIDS Apps
- Integrated workflows
More functionalities
Contributions and communities
- Development is a straight(er) line
- Time effective (on the short run)
- Low(er) engagement
- Less mentors
- Small(er) user base
- Less tests
- Consensus not guaranteed
- Clear attribution
- Development takes its own path
- (More) Time consuming
- High(er) engagement
- More mentors
- Big(ger) user base
- More tests
- Consensus based
- Fuzzier attribution
A community approach to taking out the brain from a vat and putting it back in the body
By Stefano Moia
A community approach to taking out the brain from a vat and putting it back in the body
CC-BY 4.0 Stefano Moia, 2025. Images are property of the original authors and should be shared following their respective licences. This presentation is otherwise licensed under CC BY 4.0. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/
- 76



