


Computational Biology
(BIOSC 1540)
Jan 23, 2025
Lecture 03B
Genome assembly
Methodology
Announcements
Assignments
Quizzes
CBytes
ATP until the next reward: 1,903
Quick homework tip
When asking for five FASTQ entries, here is what it should look like
fastq_five = """
@synthetic_read_1/f
TACGGCTAGGCATCTCGAGATCTGTGACGTTTCAGATCCCCTGCTGCGTGCGTTTGATGTCCAACTGTCGTACTCACGCCGGACGGGGAGTAACTTCTTTTCGAGCCGTAGTTGCGCGTACCCCATAAATTAGCCTTGGGAGCAGACAAGGCAAAACCCCAGAAGTTGGGTGTCCAATAAGGTTCACATCTGACCGGTCGCTTAAAAGGAAGCTTCCATTATCCGCTATATAAACGCCGGTGCTACCTAAGATGCGACACCT
+
46:47287653825380557902185865586;11784536:8>:7946436;67:04>8671293:53991474581727927476120866:4;;441889567264523394268:775;459726:6:5;865421764998273667561/6:6=987731855057787594555;4:859357388590:54:67365857865:66456:2737:3:34683758339648455674;6569;82364766;67
@synthetic_read_2/f
GACGATCGTAGCTCAGTCGGACCAACGACTCGCTGCTTACTGGAAGATCCTCGTAGACGGTTTTTTTGCGAAAGTACAGGCGACCCAGTACAAATCGGGATAGTGGTCACTTACTTATTTGGGCGGCTGCGATCATGGCCGCCGGTGGTTTAGGGTGTACGAATTCATCCTAGCCGCGAATGATTACGGACATCGTGACCATTATATGATCCGTTGTAGGCAGGTGTAACCCGACACTACTACATGACGGCTCACTCCAACA
+
GGDIHIFGEHGGIGGIHGFGIIFIHFDEFEFCCFFIIHIGIEEFIEFFICDGFHICFEICGGFFEIEEFGIFGFIHIIBDGHIGHIIGGGHFGIEHIIIDIIECAIHDHCEDECFHFBIIEIFHIIECIIIGIEIGEEFIDIHIEIFIDHFDIGEHEAGEIGDICIGFHHGGEIEDDDIGIDH?EIICDEHEHFGGIFGIGAIIIIIIIFGIEEIGCHIGIDIHIIIIICID@FIIIIEIGCIIGIHGIIIHIEGFHEEAFI
@synthetic_read_3/f
CGTAGCTGACGTAGATTCGATTTAAGAAACGCAGATATGGACATTGTCGCCGTGCCTTTATATTCCACATATCGTGGTAATCATACCGGCATAGGGTCATGTCCGCAGCTGTCCAACTATCGGTTAACGTTCCCCCTACTATCTCTGCGCGAGCCTAGAGTAAATCGATGAGTCTGAAGAACGCCTCATATCTGCTGTATGCCCGCCGCGTGAACTCTCAGTATTCGCGAACACATTGGTCTTGCTATCCTCGGTAAGGAAC
+
:=9<<:7<9::=<?<<6;;=;?;<7;9=9?6:8;8A9:=>=<:A79;=>=;:==:<4::7<9?E4<9;;:97=<7@9;8?@<7999:A9:=;6:?>:@988A?97=A>=@:;98>::<A>?7:<<A?<<5=7<7<==:4;3><=?5>><999;;=<8?;=88>>8;:;;7967>;;59=:<;8:7687856;679763<9=@;87550:627535<755461A329795;87269;45074317650467412984:51937
@synthetic_read_4/f
TACGGCTAGGCACGTTTTCAGCAATCACGCGTGAGAATGCAATACAGCTGAGTATAGGTGGCCGGGCGTACGTTTCTACGTGAGCATGTTTTTTTATTACAGAGTACCGGTAGCGGGGTTAACATGGAAGGCAGATACCGCTACGTCCGCCCCGAACGAGCCGCACATACAACCCGATCGGTACCGGTTAGTGTAAGGGGCTTTGTGAGTAGGTGTGGGCCGTGTACGGGTAGTACTCGGGAATGATGCGGATAATACCATC
+
>:A@=@=<ABB><=:==?>@=><<<9=?3:>@CHD;?=7:@?6G<8<@?AEE<=?;<;C<66B3>>>>=8488<8>?@9>43>?A?A61:@8;:6@97;825=>7>8><1<853;@B=66><A;?@5475:382@<@@<7><5-945?=7?;9=90<;B6C;;=77:?7;69-/87665=<<:>92A8AB47B5;8;3<A=8<92;7.99?65>653::@97A>6B>>11BAD>C>5:7,:A>@8<=7:<99??57:=:95<
@synthetic_read_5/f
GTACGATCGTACCTGCGTACAAAACAGTTTCGGGGTCCAAACCACGCCTCAACTGTTCTCGGTTAGTACCGTAGCTACACTCGGTCTATCTGTCAGCTGCCGTTCATTCGAGCTTTCGCGTACTTAACAGTACCAGAAGGGCGGGTCAGTATCCATGGTTGGTGCAAGCCCCTGTCCCAGTCTTACCGGATAGCGCACATTACTTCTCCTGGTTCGGGCGAATCCCCTAGCAATCCCATTTCATTACGAAACCAACGCTCCA
+
78<8675<68;9;9<72;4==:689<;95=5;?76:57<16;:4@;9.=:1:;?<49;89;0<>?6327778:8:518?7=79:6:<7><A@16:65<98:6<7446<;@9=9:<@<27:9<38@98<?8;9<5:4<3547?9>5:1;A=695=6<:8/;873?66::?45;:C857>:E:9:;9;258<:<<<79:599<<>77679<<;3<9328=53>8;35?E7=;8<A99>68;A99799?=5:6:70?<8::187;
"""After today, you should have a better understanding of
Problem formulation of genome assembly
Why we need genome assembly
DNA sequence (i.e., contig)
Overlapping reads
Assembly algorithms
TACGATCGGATTACGCGTAGGCTAGCTTACGGACTCGATGTACGATCGGATTACG
Genome assembly reconstructs a long DNA sequence from short, error-prone reads, ensuring as many reads fit into the final sequence
Recap from L03A
After today, you should have a better understanding of
Problem formulation of genome assembly
Assumptions
We make simplifying assumptions to address challenges and make assembly tractable
Reads originate from a single, contiguous genome
If we had two sources of DNA
Chance of overlap is likely, and it would be challenging to differentiate the origin of each read
+
It dramatically simplifies our problem if we assume only a single source of reads
Sequencing coverage is sufficient for redundancy and error correction
TACGATCGGATTACGCGTAGGCTAGCTTACGGACTCGATGTACGATCGGATTACGCGTAGG
Real sequencing errors
can be fixed in high-coverage areas
Real SNPs
can be confidently detected when all reads have the same base
Assume that we have
high coverage
What if your sequencing data does not meet these assumptions?
This happens all the time in science!
If you more robust options are available, using those may be required
If there is no other option, use the best approach and disclose how this could impact your results and interpretation
After today, you should have a better understanding of
Problem formulation of genome assembly
String manipulation in Python
Review: DNA sequences are represented as strings in Python
read1 = "ATCG"
read2 = "TCGA"
A DNA sequence is simply a sequence of letters: A, T, C, and G. In Python, we can represent this using quotation marks ("" or '').
read_long = """
CGTAGCTGACGTAGATTCGATTTAAGAAACGCAGATATGGACATTGTCGCCGTGCCTTTATAT
TCCACATATCGTGGTAATCATACCGGCATAGGGTCATGTCCGCAGCTGTCCAACTATCGGTTA
ACGTTCCCCCTACTATCTCTGCGCGAGCCTAGAGTAAATCGATGAGTCTGAAGAACGCCTCAT
ATCTGCTGTATGCCCGCCGCGTGAACTCTCAGTATTCGCGAACACATTGGTCTTGCTATCCTC
GGTAAGGAAC
"""
Comparing strings allows us to detect similarities or differences between DNA reads
read1 = "ATCG"
read2 = "TCGA"
# Compare two strings
print(read1 == read2) # Output: False
To compare strings, we can use the equality operator ==
read1 = "ATCG"
read2 = "ATCG"
# Compare two strings
print(read1 == read2) # Output: True
We can extract parts of a string using indices in Python
0123
ATCGUse square brackets [] to get a character by its index
read = "ATCG"
print(read[0]) # Output: A
print(read[2]) # Output: C
Use slicing with start:stop to get part of a string
print(read[0:2]) # Output: AT
print(read[1:3]) # Output: TCPython does not include the stop index
Each character in a string has an index
Example: "C" has index of 2
We can use loops to check every position in a string
Use a for loop to go through each character one by one
read = "ATCG"
for char in read:
print(char)
# Output:
# A
# T
# C
# G
read = "ATCG"
for i in range(len(read)):
# Print substrings starting at index i
print(read[i:])
# Output:
# ATCG
# TCG
# CG
# G
You can also slice inside of a for loop with an index
range(len(read))generates integers from 0 until the length of the read (in this case 4)
Comparing parts of strings allows us to find overlaps between DNA reads
read1 = "ATCG"
read2 = "TCGA"
for i in range(len(read1)):
if read1[i:] == read2[:len(read1) - i]:
print(f"Overlap found: {read1[i:]}")
break
# Output: Overlap found: TCG
Let’s find where read1 overlaps with read2
When i = 0:
-
read1[0:]gives us"ATCG"(the full string) -
read2[:4]gives us"TCGA"(first 4 characters) - Comparison:
"ATCG" == "TCGA" - Result: No match
Comparing parts of strings allows us to find overlaps between DNA reads
read1 = "ATCG"
read2 = "TCGA"
for i in range(len(read1)):
if read1[i:] == read2[:len(read1) - i]:
print(f"Overlap found: {read1[i:]}")
break
# Output: Overlap found: TCG
Next is i = 1:
-
read1[1:]gives us"TCG"(excluding'A') -
read2[:3]gives us"TCG"(first 3 characters) - Comparison:
"TCG" == "TCG" - Result: Match found! 🎉
Comparing parts of strings allows us to find overlaps between DNA reads
Once we find the overlap, we can merge the reads
read1 = "ATCG"
read2 = "TCGA"
i = 1
merged = read1[:i] + read2
print(merged)
# Output: ATCGA
We can use this approach of finding overlaps and merging reads to form a contig
This idea of finding overlaps and merging motivates our first assembly approach: the greedy algorithm
After today, you should have a better understanding of
The greedy algorithm for genome assembly
Overlaps and merges
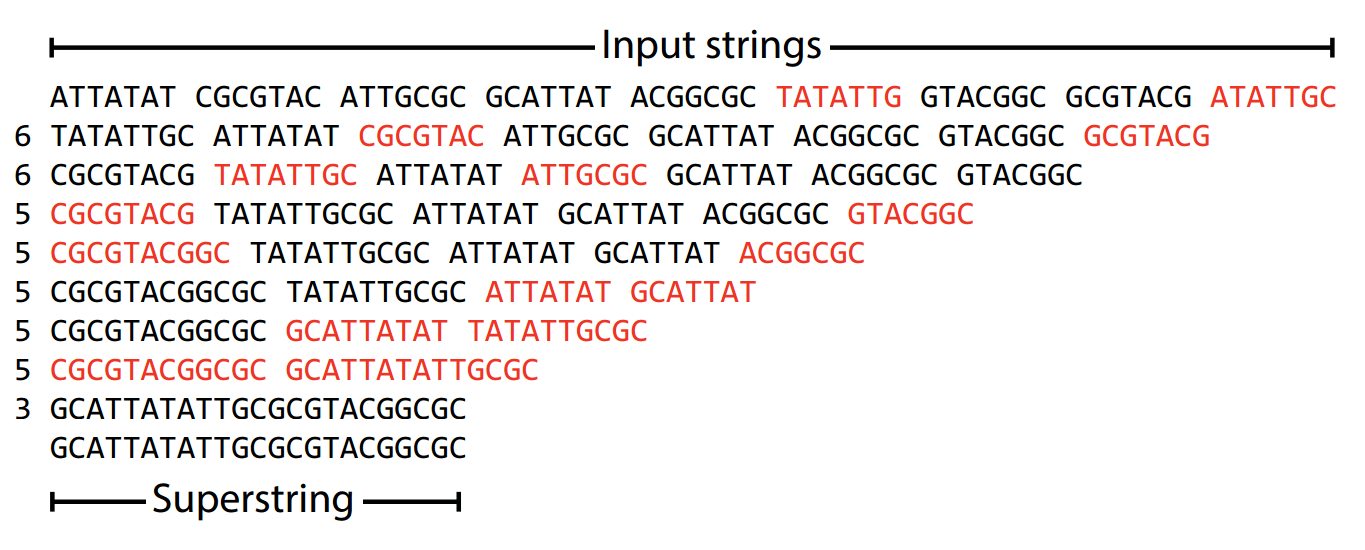
The greedy algorithm builds genome assemblies by iteratively merging the best overlaps
Algorithm
1. Check every possible read for the largest overlap.
2. Merge the two reads with largest overlap.
3. Repeat until no further merges are possible.
At the end, we have a set of contigs that represent our original DNA sequence
The greedy algorithm minimizes repeats by maximizing overlap
The greedy algorithm focuses on selecting the best immediate option (i.e., local optimal) at each step without full consideration of the overall global solution
The greedy algorithm aims to find the shortest superstring, which minimizes unnecessary duplication.
A superstring is a single string that contains all reads as substrings
Example: ACGTAC is a superstring of
ACGT, CGTA, GTAC
This means the greedy algorithm will always make the best move in the moment even if it gives the wrong final answer
Being greedy makes genome assembly tractable
After today, you should have a better understanding of
The greedy algorithm for genome assembly
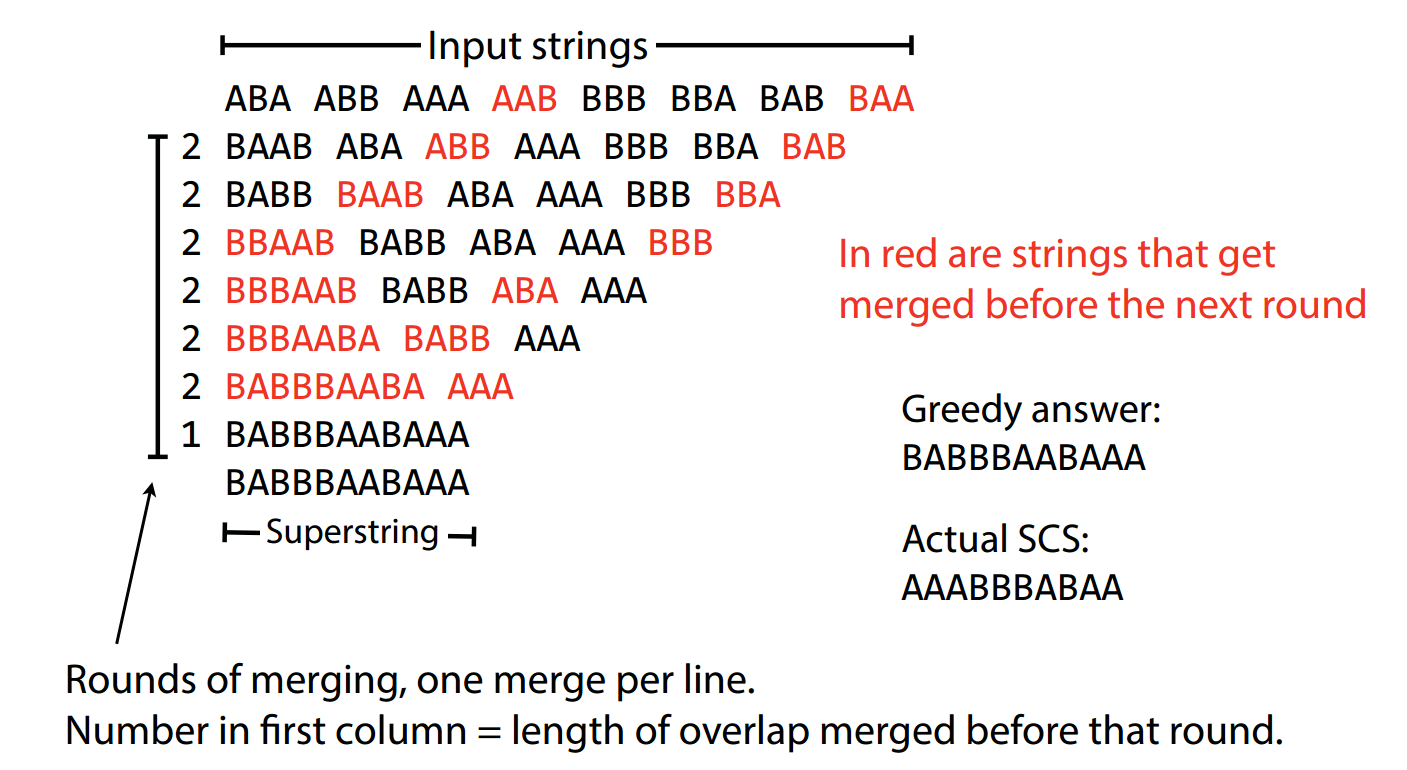
Breaking ties
Tie-breaking rules are necessary when overlaps are identical

Talk with your neighbors
Suppose we have these three reads
ACGTAA
CGTAAC
with a highest
overlap of five
We merge reads R2 and R3:
TAACGT
R1
R2
R3
TAACGT
ACGTAAC
(and keep R1)
However, now we have a problem
ACGTAACGT
TAACGTAAC
Both have a length of 9, which one is the correct move?
Overlap of 4
TAACGT
ACGTAAC
Overlap of 4
TAACGT
ACGTAAC
Tie breakers are a personal preference
First encountered, first merged
Highest quality base calls
Highest coverage
Look ahead
Exclude
The one you found first
Use sequence with highest quality
Whichever results in more coverage
Do both and evaluate consequences
Be petty and don't merge them (separate contigs)
After today, you should have a better understanding of
The greedy algorithm for genome assembly
Trouble with repeats
Greedy assembly will incorrectly collapse repeats if possible
a_long_long_long_time
We are missing a "_long". Why?
Let's take a string and cyclically permute it with k = 6


Then perform the greedy algorithm
Longer reads and genome assembly
We get the correct string back, but how did increasing our k fix this?

a_long_long_long_time
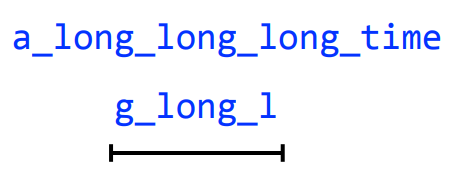
By having one read span all three "long"s, (i.e., the repeating region) we prevented a collapse
k = 8

Remember: This is why long sequencing reads are very helpful in resolving repeats!
After today, you should have a better understanding of
De Bruijn graphs and their role in assembly
K-mers
The greedy algorithm provides insights but is rarely used in modern genome assembly
The greedy approach is computationally efficient but fails for large, complex genomes.
Finding overlaps between all reads scales poorly with genome size
Full pairwise comparisons between reads require operations
where nnn is the number of reads
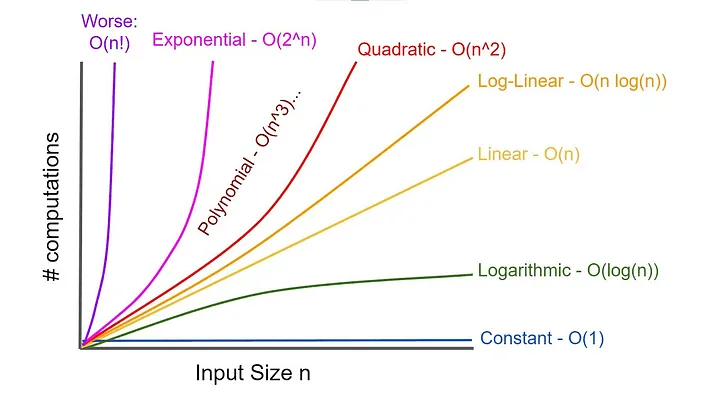
As our number of reads increases, our time to find overlaps dramatically increases
However, the number of reads also improves our assembly
k-mers break reads into manageable, fixed-length pieces
By decomposing reads into k-mers, we can:
- Represent sequences as collections of overlapping k-mers.
- Avoid comparing entire reads by focusing on k-mer matches.
- Use fixed-length k-mers to tolerate sequencing errors in overlaps.
- Number of reads does not change number of k-mers
Instead of comparing whole sequences, we can compare k-mers!
A k-mer is a substring of length kkk extracted from a sequence
Example: For the sequence ATCGT, the 3-mers are ATC, TCG, CGT.
Building k-mers from a string
GGCGATTCATCG
- Slice first k characters
- Shift right one character
- Repeat
Spectrum with k = 3
GGC
GCG
CGA
TCG
ATC
GAT
ATT
TTC
TCA
CAT
All 3-mers
k-mers are robust to sequencing errors
Sequencing errors affect only a few k-mers in a read, not the entire sequence.
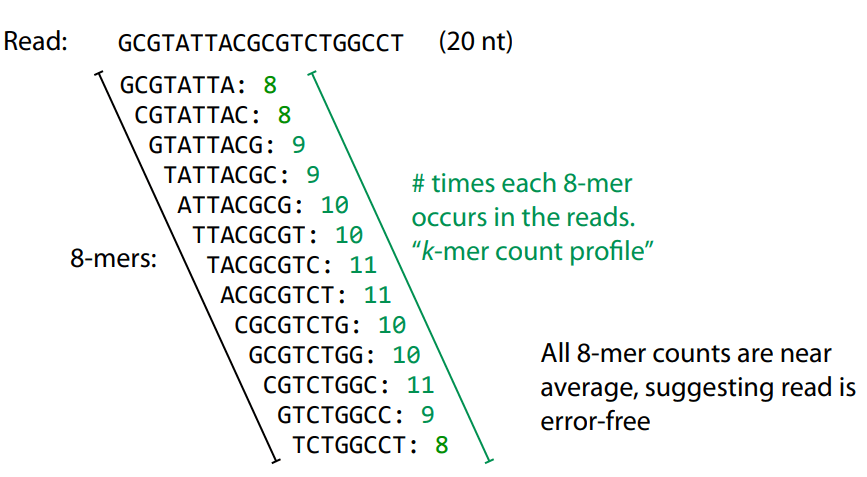
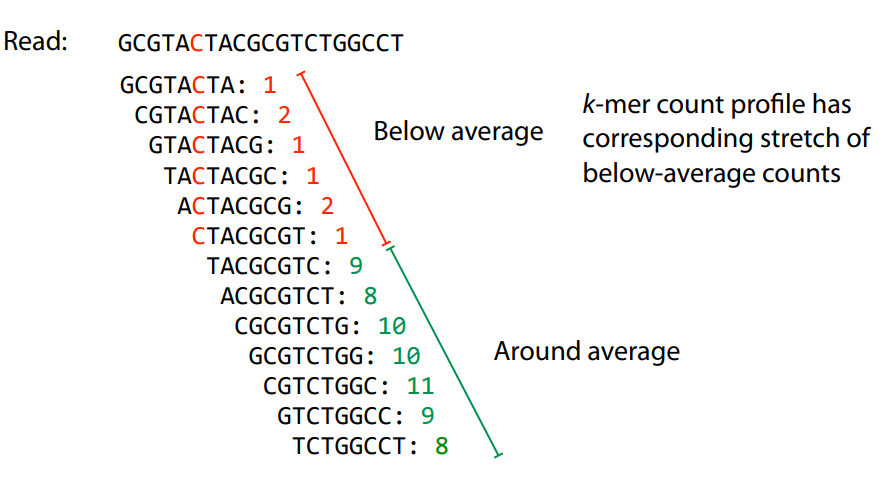
Longer k-mers provide specificity, while shorter k-mers ensure sensitivity.
Even if a single read has errors, most k-mers will match correctly to others.
After today, you should have a better understanding of
De Bruijn graphs and their role in assembly
Building graphs
Graphs is a data structure for drawing relationships between items
Node
Represents a single entity
- Person
- Location
- Protein
- Sequencing read
Edge
Represents a connection (possibly with a direction)
- Instagram follower
- Flights
- Protein-protein interaction
- Sequence overlap
Genome assembly uses direct edges to specify overlap and concatenation
"tomorrow and tomorrow and tomorrow"
Let's build a directed multigraph:
- Each unique k-mer is a node
- Add directed edges for each overlap and concatenation
K-mer is a substring of length k
(We will cheat here and write down just unique words)
tomorrow
and
Build a De Bruijn graph with k-1 nodes
AATGGCGTA
AAT
ATG
GGC
GCG
TGG
CGT
GTA
AATG
ATGG
TGGG
GGCG
GCGT
CGTA
5'
3'
Step 1:
Let's use kedge = 4
Step 2:
AATG
L
R
Step 3: Repeat
ATGG
TGGG
GGCG
GCGT
CGTA
Take left and right k-1 mer and make two connected nodes (knode = 3)
Build k-mers
Building De Bruijn graphs with a read
CGTAAAT

Build a De Bruijn graph with k = 3
CGT
GTA
TAA
AAA
AAT
CG
GT
TA
AA
AT
De Bruijn graphs with multiple reads
5' AATGGCGTA 3'
5' CGTAAAT 3'
Read 1
Read 2
Let's use nodes of length 4
AAT
ATG
GGC
GCG
TGG
CGT
GTA
TAA
AAA
Note: This is a circular genome
Frist, build the De Bruijn graph for
Read 1
Add edges and any new k-mers from
Read 2
Another example, but not circular
5' AATGGCGTA 3'
5' CGTAAAG 3'
5' TAAAGGCGAA3'
Read 1
Read 2
Read 3
AAT
ATG
GGC
GCG
TGG
CGT
GTA
TAA
AAA
AAG
CGA
GAA
AGG
We can add weights to edges instead of drawing multiple edges
5' AATGGCGTA 3'
5' CGTAAAG 3'
5' TAAAGGCGAA3'
Read 1
Read 2
Read 3
AAT
ATG
GGC
GCG
TGG
CGT
GTA
TAA
AAA
AAG
CGA
GAA
AGG
2
2
2
1
1
1
1
1
2
1
1
1
Another (another) example, but not circular
GATTAC
TACAGATT
AGATTAC
TACCGG
GGATTA
The solution is on the next slide (no peeking!)

De Bruijn graphs is one of the most missed questions on assessments, let's get some practice
Another example, but not circular
GATTAC
TACAGATT
AGATTAC
TACCGG
GGATTA
TACC
ACCG
CCGG
GATT
ATTA
TTAC
TACA
ACAG
CAGA
AGAT
GGAT
After today, you should have a better understanding of
De Bruijn graphs and their role in assembly
Characteristics
Tips are good starting points of contigs if they have high coverage
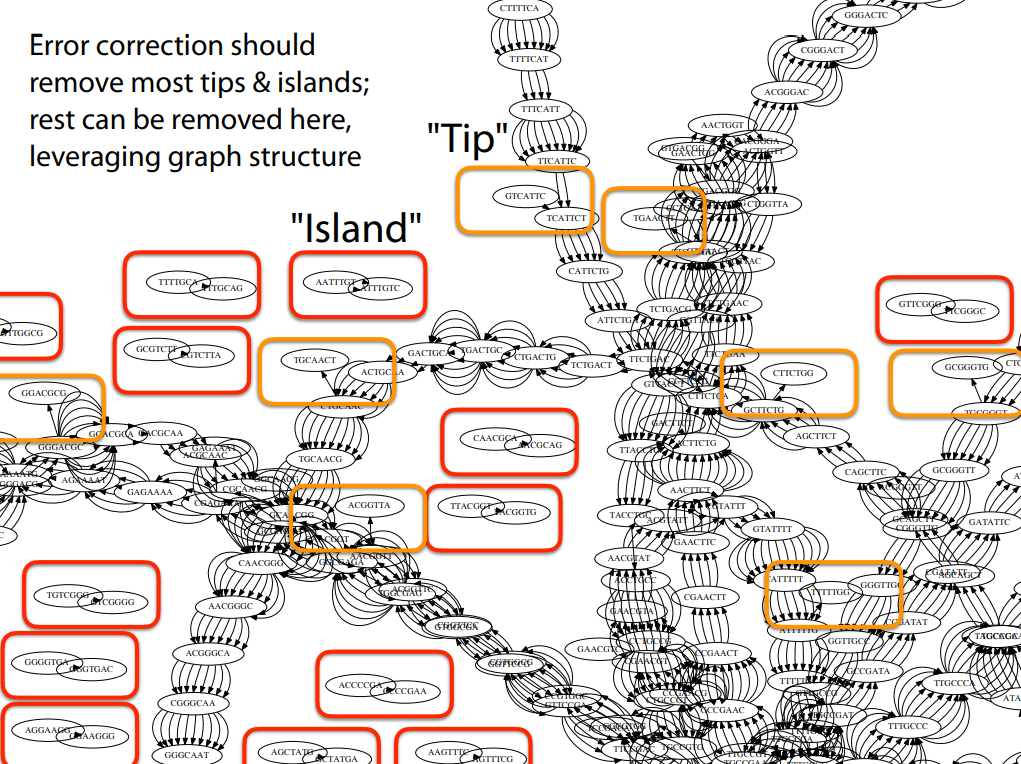
Islands are reads we couldn't merge
After today, you should have a better understanding of
Graph traversal methods for extracting contigs
De Bruijn graphs are traversed to extract contiguous genome sequences
Traversal is the process of finding contigs (continuous DNA sequences) by walking through the De Bruijn graph
TACC
ACCG
CCGG
GATT
ATTA
TTAC
TACA
ACAG
CAGA
AGAT
GGAT
Edges: Represent k-mer overlaps between nodes.
Nodes: Represent k-mers derived from sequencing reads.
Standard traversal methods, such as breadth-first search (BFS) and depth-first search (DFS), are building blocks for more advanced assembly techniques.
After today, you should have a better understanding of
Graph traversal methods for extracting contigs
DFS explores as far as possible along each branch before backtracking
Imagine exploring a maze with this strategy:
- Keep walking forward until you hit a dead end
- Backtrack only when necessary
- Take the first unexplored path you see
A
/ \
B C
/ / \
D E FDFS Traversal from A (one possible order):
- A → B → D (follow first path to end)
- Backtrack to A
- A → C → E
- Backtrack to C
- C → F
BFS explores all neighbors of a node before moving deeper into the graph
Imagine you're dropping a pebble in a pond:
- First, you see ripples reach nearby points
- Then, they spread outward in circles
- Each "wave" represents a level of exploration
A
/ \
B C
/ / \
D E FDFS Traversal from A (one possible order):
- A → B → D (follow first path to end)
- Backtrack to A
- A → C → E
- Backtrack to C
- C → F
Standard traversal methods struggle with genome assembly challenges
Specialized traversal methods, like Eulerian and Hamiltonian paths, address these challenges.
Before the next class, you should
Lecture 04A:
Genome annotation -
Foundations
Lecture 03B:
Genome assembly -
Methodology
Today
Tuesday
Quiz 01
After today, you should have a better understanding of
Graph traversal methods for extracting contigs
Eulerian paths traverse every edge exactly once, aligning with k-mer overlaps
- Definition: A path that visits every edge exactly once in a graph.
-
Relevance to De Bruijn Graphs:
- Nodes = k-1-mers, Edges = k-mer overlaps.
- Finding an Eulerian path reconstructs the genome sequence.
-
Key Properties:
- All nodes must have equal in-degree and out-degree, except at most two nodes (semi-balanced).
- Efficient traversal in graphs where overlaps are abundant.
- Example:
AAT → ATG → TGG → GGCforms a single path when traversing overlaps.
An Eulerian path resolves overlaps in sequencing reads
-
Graph Example:
- Nodes: AAT, ATG, TGG, GGC, GCG, CGT.
- Edges: AAT → ATG, ATG → TGG, TGG → GGC, GGC → GCG, GCG → CGT.
-
Traversal: Follow the edges to reconstruct the genome:
AATGGCGT. - Activity: Ask students to find the Eulerian path for a similar graph with cycles.
Eulerian paths are fundamental to extracting contigs from De Bruijn graphs
- Eulerian Path: A path through a graph that visits every edge exactly once.
-
Why Eulerian Paths?
- De Bruijn graphs use k-mer overlaps to simplify assembly into an edge-traversal problem.
- Each edge represents an overlap, making the Eulerian path equivalent to contig assembly.
-
Key Properties for Eulerian Paths:
- All nodes except two must have equal in-degree and out-degree.
- At most two nodes can be semi-balanced (differing in degree by one).
Hamiltonian paths visit every node exactly once, aligning with reads
- Definition: A path that visits every node exactly once in a graph.
-
Relevance to Overlap Graphs:
- Nodes = Reads, Edges = Overlaps between reads.
- A Hamiltonian path assembles the genome by arranging reads in sequence.
-
Limitations:
- Computationally intractable for large datasets due to the complexity of visiting every node.
- Less practical than Eulerian paths in modern genome assembly.
Suppose we want the shortest superstring
This is a valid superstring, but why would we want the shortest?
BAAAABBBAABAABBBBBAAABAB

Talk with your neighbors
Overlap maximization
- Reduces redundancy
- Maximizes confidence with highest overlaps
Repeat resolution
Resolves repeats by favoring collapsed arrangements
Evolutionary pressure
Most genomes have selective pressure to be efficient
Let's formulate our problem
Suppose we have a collection of strings (i.e., reads)
(In CS, we call a sequence of characters a string)
BAA
AAB
BBA
ABA
ABB
BBB
AAA
BAB
We want to assemble these strings into a single, continuous string (i.e., contig)
What's the easiest way?
Concatenate
BAAAABBBAABAABBBBBAAABAB
Done!
Well, no.
Right?
This is called a "superstring"
Simplifying De Bruijn graphs resolves ambiguities for effective traversal
- Tip Removal: Remove dead-end branches caused by errors or low coverage.
-
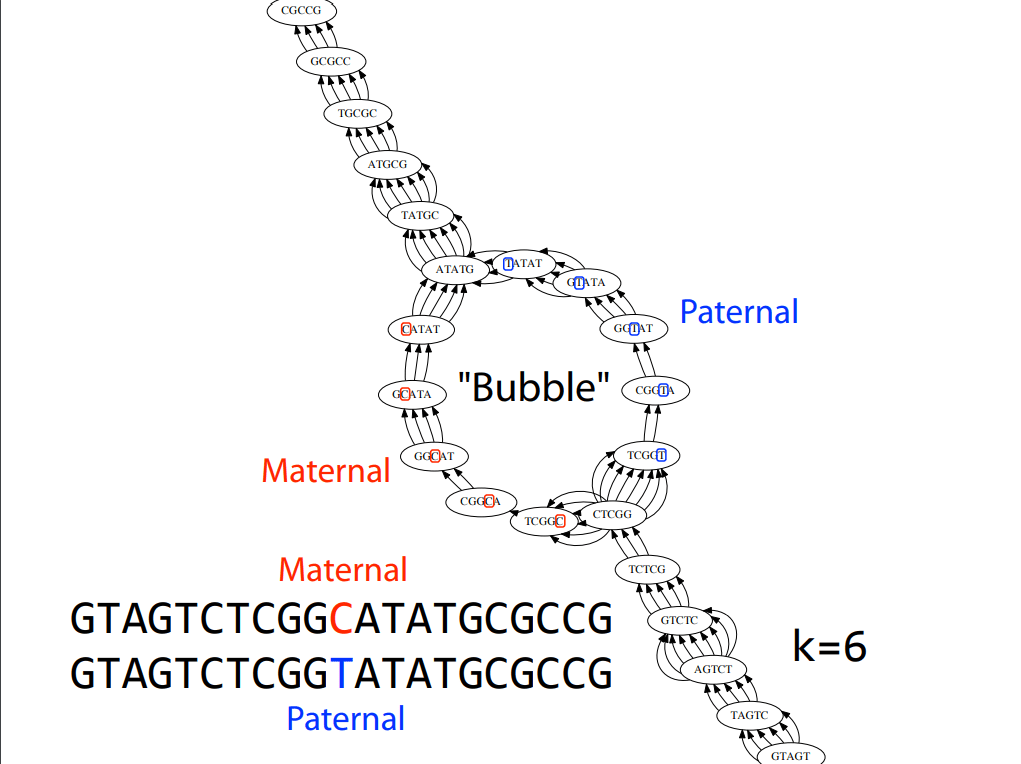
Bubble Removal:
- Bubbles are alternative paths between nodes due to sequencing errors or variants.
- Identify and merge the correct path using coverage information.
-
Compression:
- Linear paths (chains of nodes with degree 1) are collapsed into single edges to reduce graph size.
- Result: A simplified graph enables easier and more accurate traversal.
After today, you should have a better understanding of
De Bruijn graphs and their role in assembly
Error correction
Errors dramatically increase the number of edges and unconnected graphs
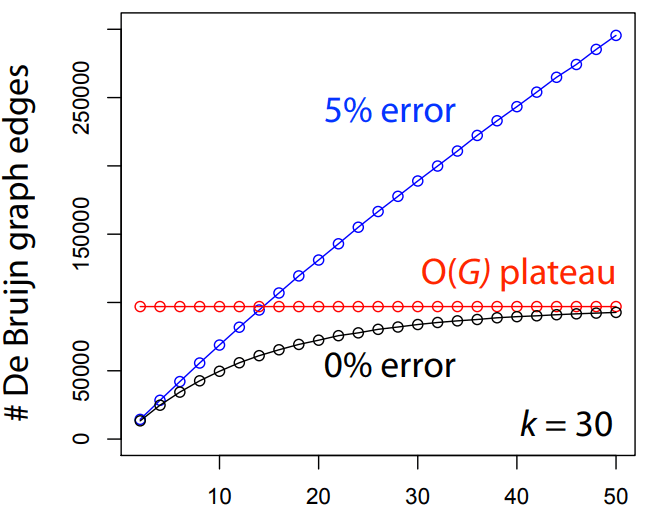
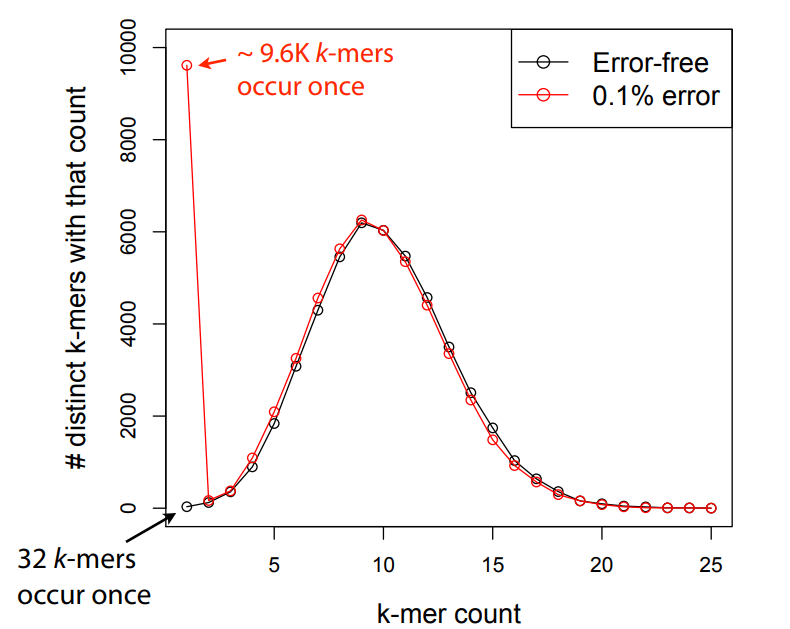
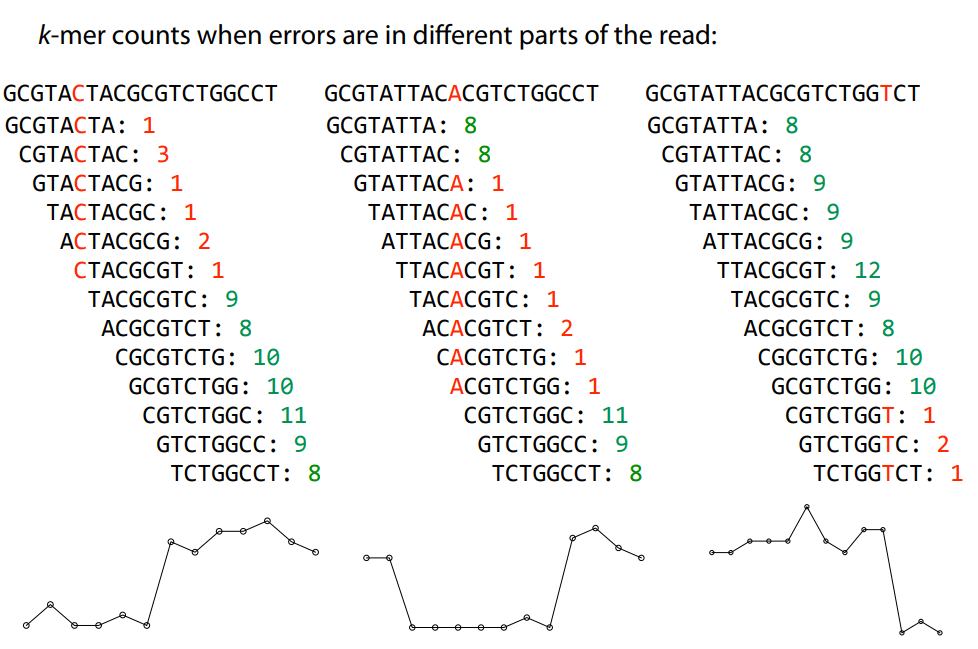
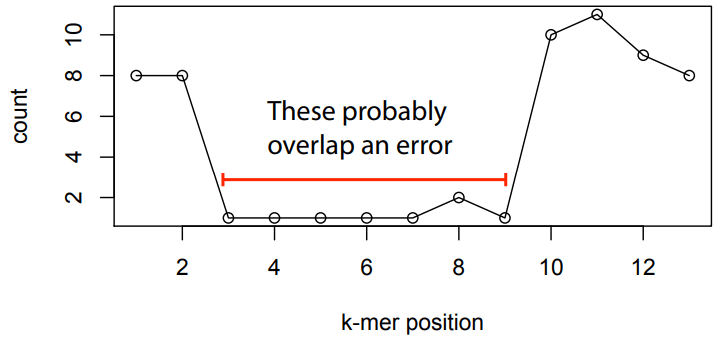
Errors affect k-mer counts


Error correction
BIOSC 1540: L03B (Genome assembly)
By aalexmmaldonado
BIOSC 1540: L03B (Genome assembly)
- 197



