

Computational Biology
(BIOSC 1540)
Mar 11, 2025
Lecture 09A
Structural Biology
Foundations
Announcements
Assignments
Quizzes
CBits
Supplementary material is available to read; not required, but recommended
After today, you should have a better understanding of
The definition and biological importance of structural biology
The atomic world of biology
At the foundation of biological processes lie atoms and their interactions
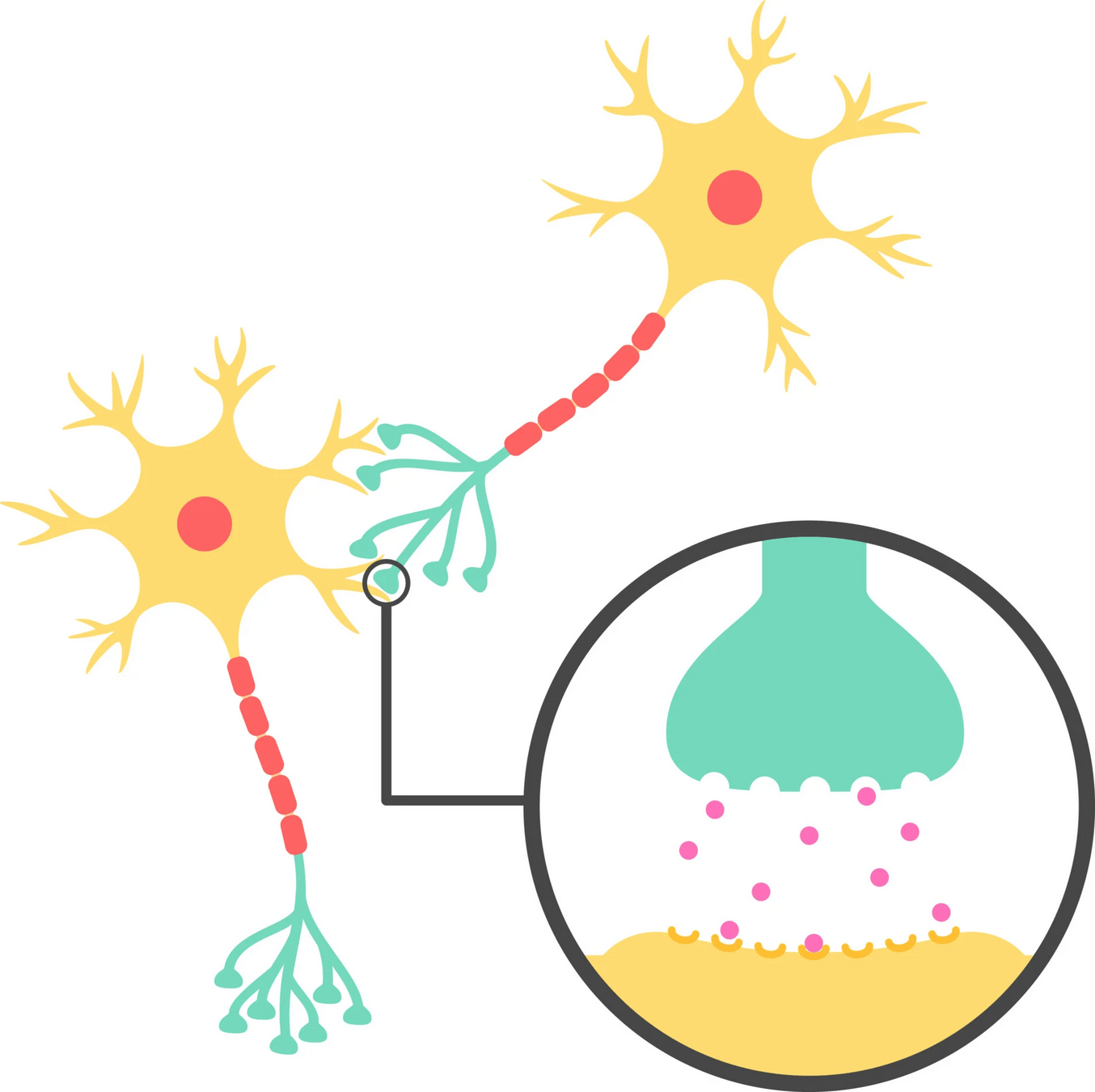
What is structural biology?
Structural biology studies the 3D shapes of biological macromolecules and how these shapes relate to function
Why study structure?
- Proteins and nucleic acids adopt specific shapes crucial for their biological roles.
- Example: The shape of an enzyme’s active site determines how it binds substrates and catalyzes reactions.
Primary Goal: To understand how molecular machines in cells work by deciphering their atomic arrangements.
Importance of structural biology: We cannot exploit what we do not understand
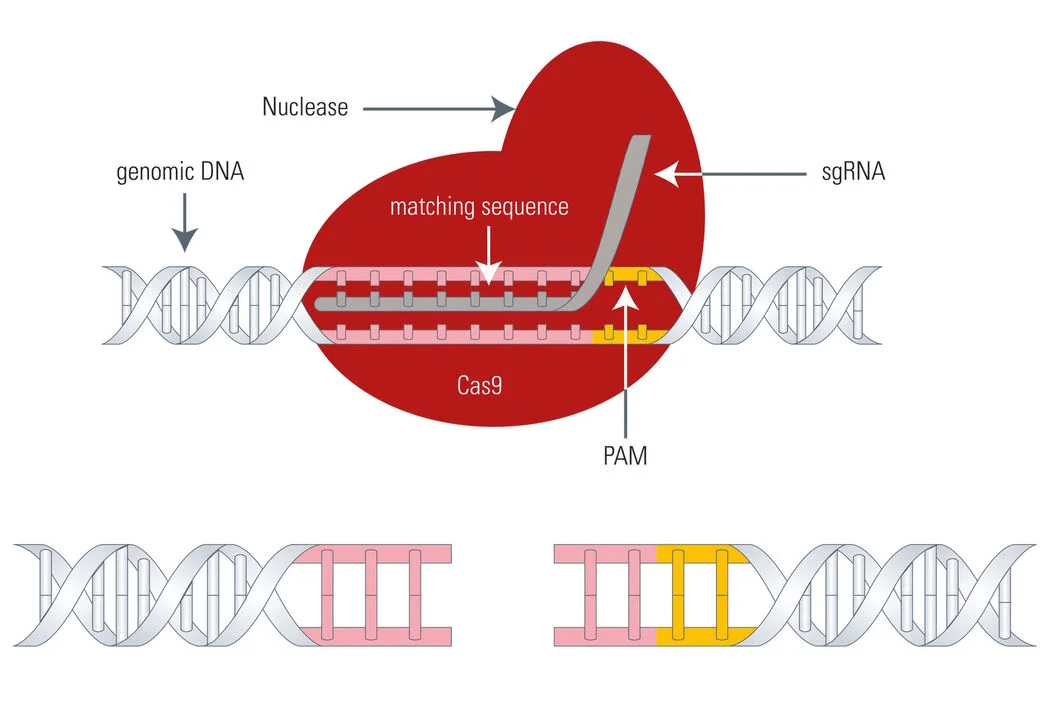
CRISPR-Cas9
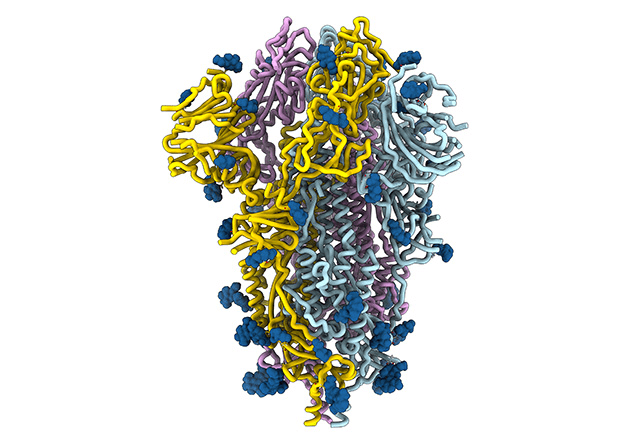
COVID-19 treatments
High-throughput sequencing
Innovation and biotechnology depend on molecular understanding
After today, you should have a better understanding of
The definition and biological importance of structural biology
Alex's research example: Engineering green fluorescent protein with Dr. Rosenbaum and Dr. Carlson
Green Fluorescent Protein (GFP) is a fluorescent protein from the jellyfish Aequorea victoria
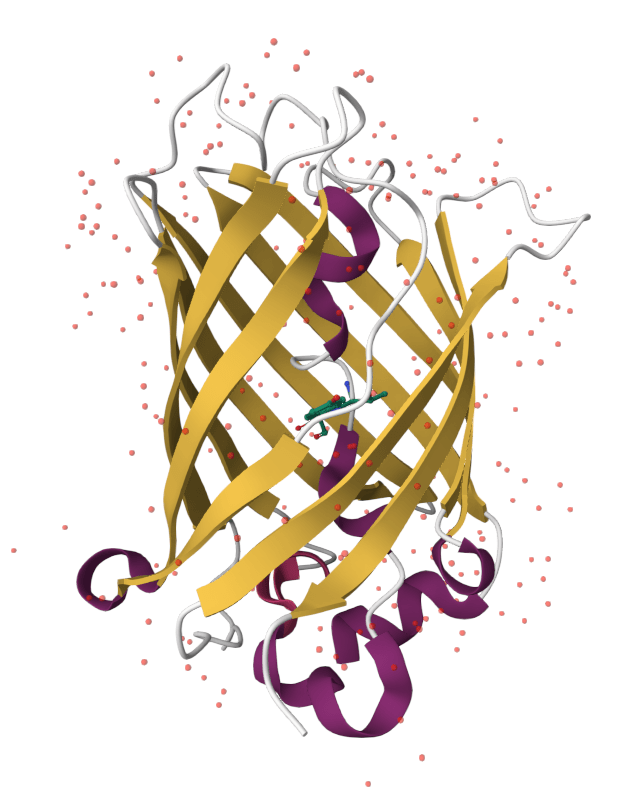
Enhanced GFP (eGFP) absorbs violet/blue light (400 - 490 nm) and emits green light ~507 nm
We can repurpose GFP to do many things!
Track molecules by adding it as a tag
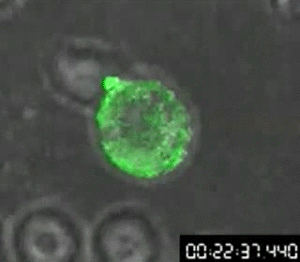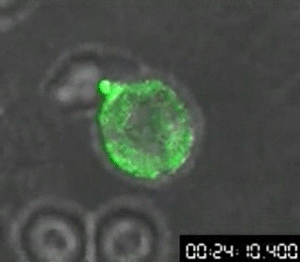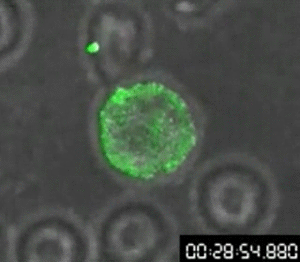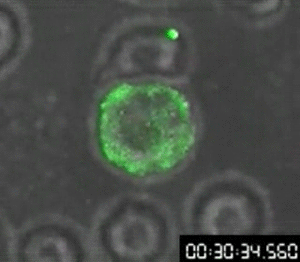
First video of cellular transfer of HIV
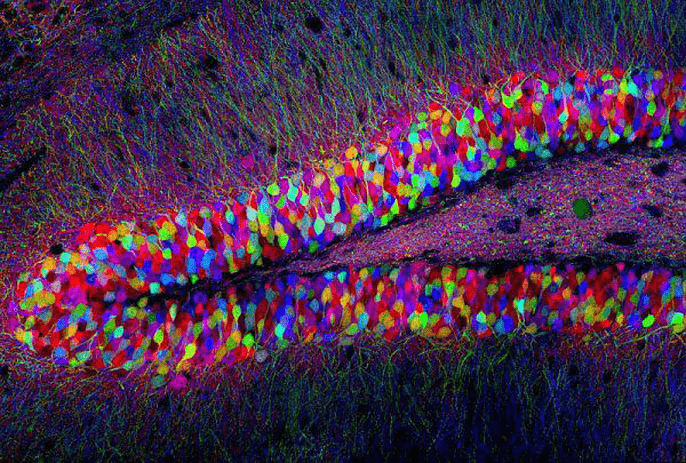
Differentiate cells with GFP variants
Multicolored GFPs used to map mouse brain
Redox reactions are a cornerstone of biology
Redox potentials indicate a solution's tendancy to gain or lose electrons
For example, mitochondria are highly reducing with a redox potential around -0.36 V

Reduction: NAD+ to NADH

Oxidation: NADH to NAD+
Remington and coworkers developed redox-sensitive GFPs
Hanson, G. T., et al. (2004). Journal of Biological Chemistry, 279(13), 13044-13053. DOI: 10.1074/jbc.M312846200

-0.310 V
-0.275 V
-0.240 V
Fluorescence ratio after 400/488 nm excitation correlated to redox potential of roGFP2 environment

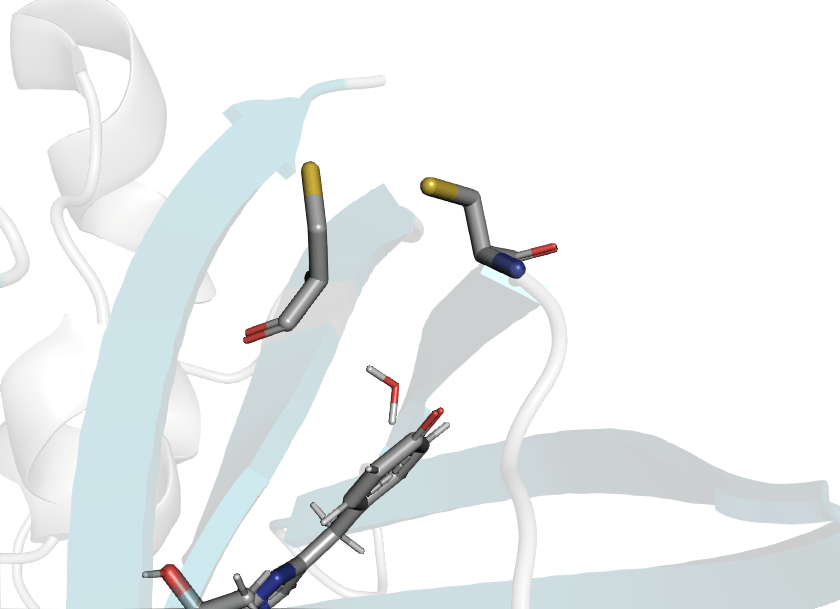
roGFP2 has S147C and Q204C mutations
Hanson, G. T., et al. (2004). Journal of Biological Chemistry, 279(13), 13044-13053. DOI: 10.1074/jbc.M312846200
PDB ID: 2Y0G
PDB ID: 1JC0
147
204
CRO
147
204
CRO
(Contains S65T "enhanced" mutation)
Wild type
roGFP2
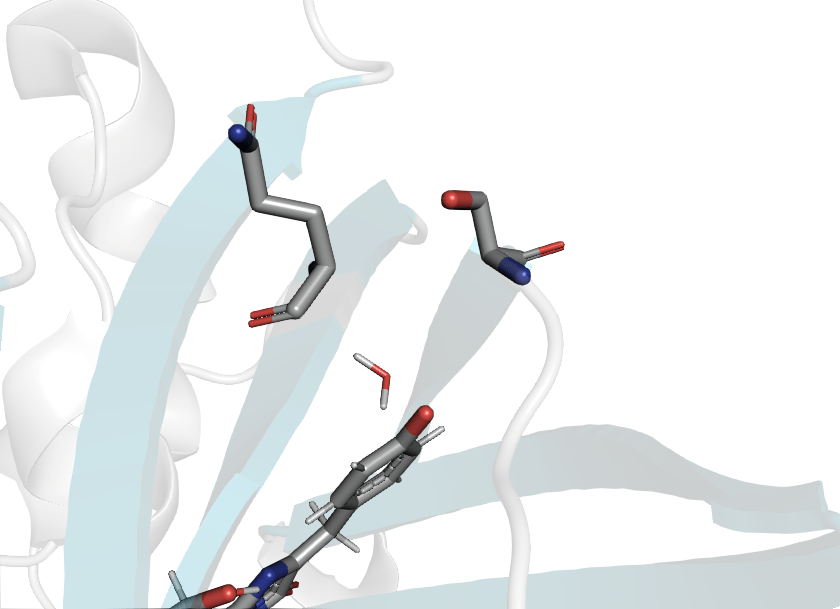
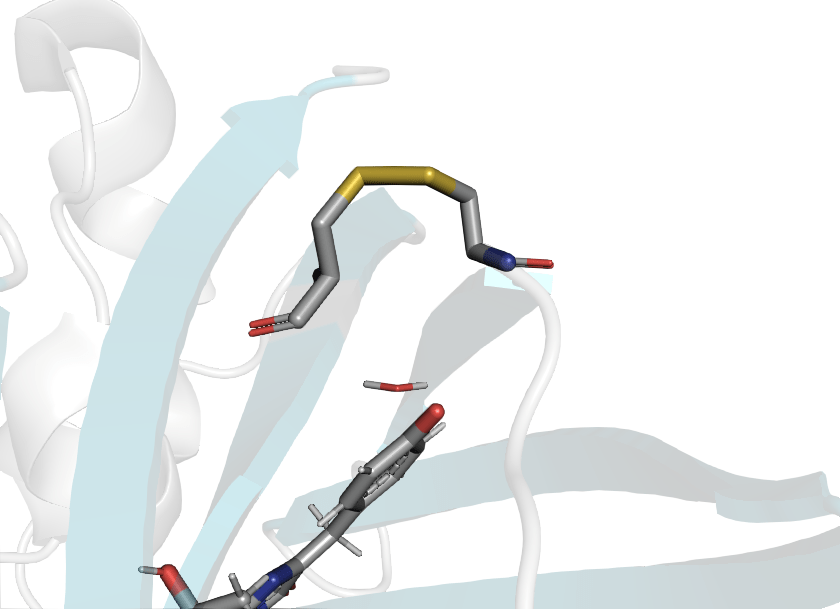
Disulfide bond formation is driven by redox potential

Hanson, G. T., et al. (2004). Journal of Biological Chemistry, 279(13), 13044-13053. DOI: 10.1074/jbc.M312846200
Reduced
Oxidized
PDB ID: 1JC0
PDB ID: 1JC1
(Contains S65T "enhanced" mutation.)
Cysteines can also bind metals
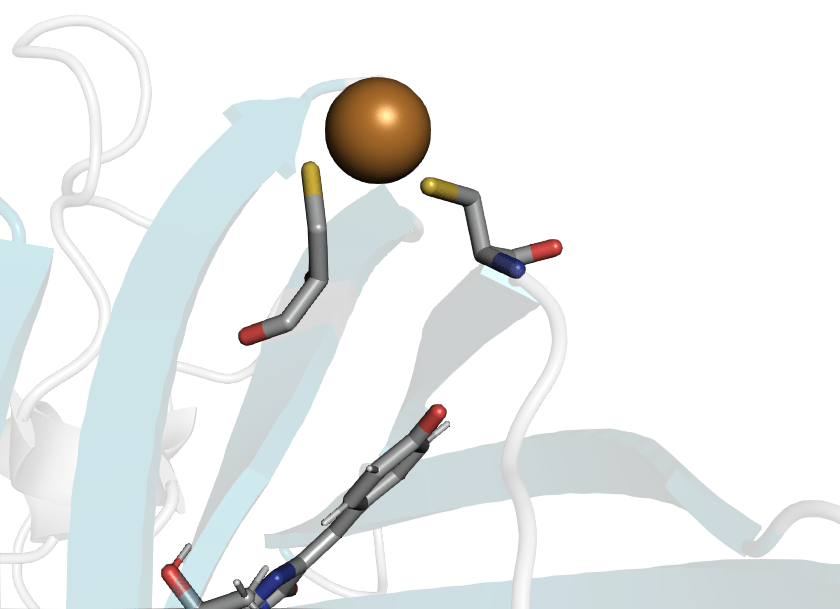
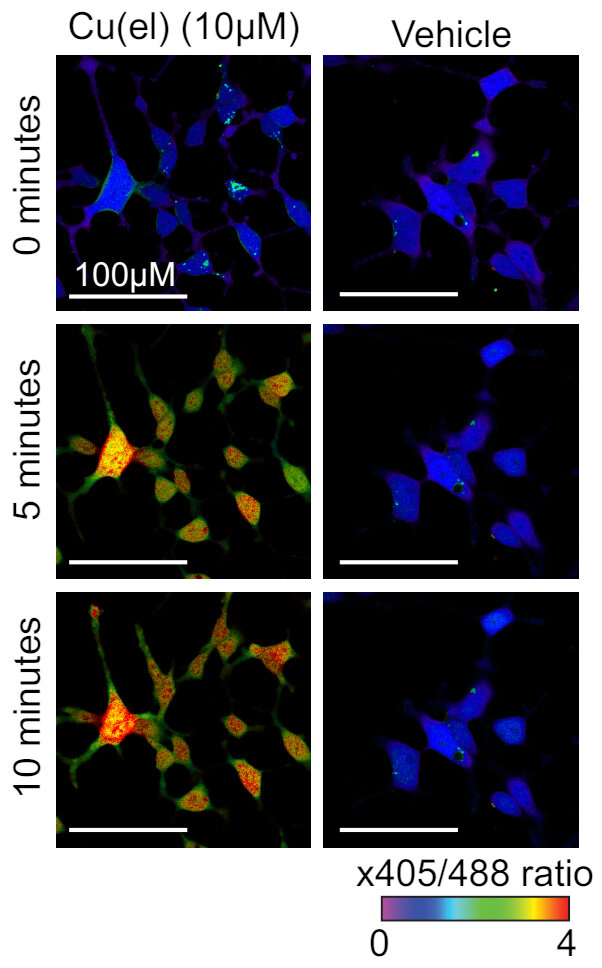
roGFP2 can bind Cu(I) to CYS147 and CYS204
Cu(I) sensing GFP
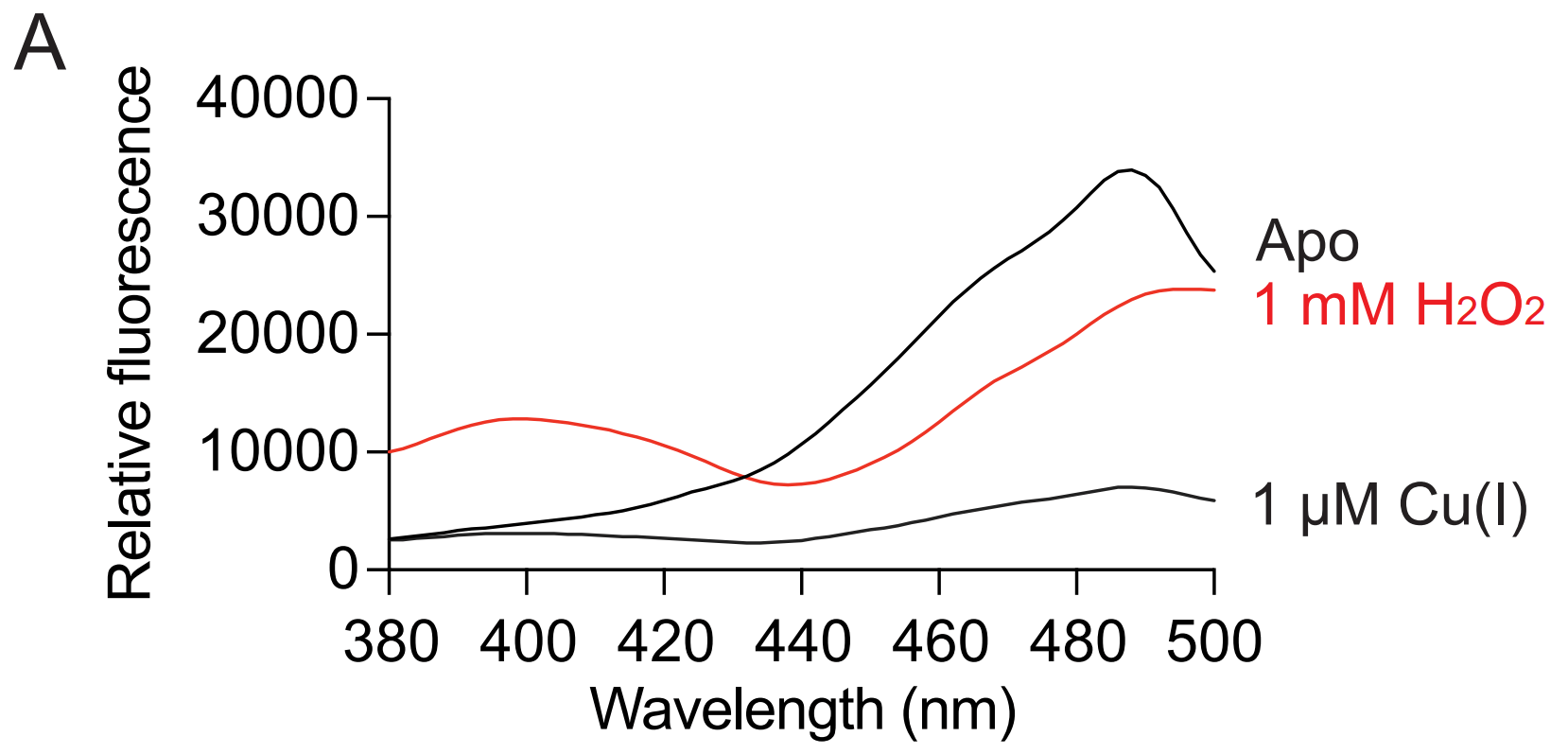
Computational question: How does Cu(I) binding quench roGPF2 florescence?
roGFP2 will also change fluorescence in a different way when copper is present
Cu(I) binding to Cys147 and Cys204 disrupts the chromophore's hydrogen bonding network
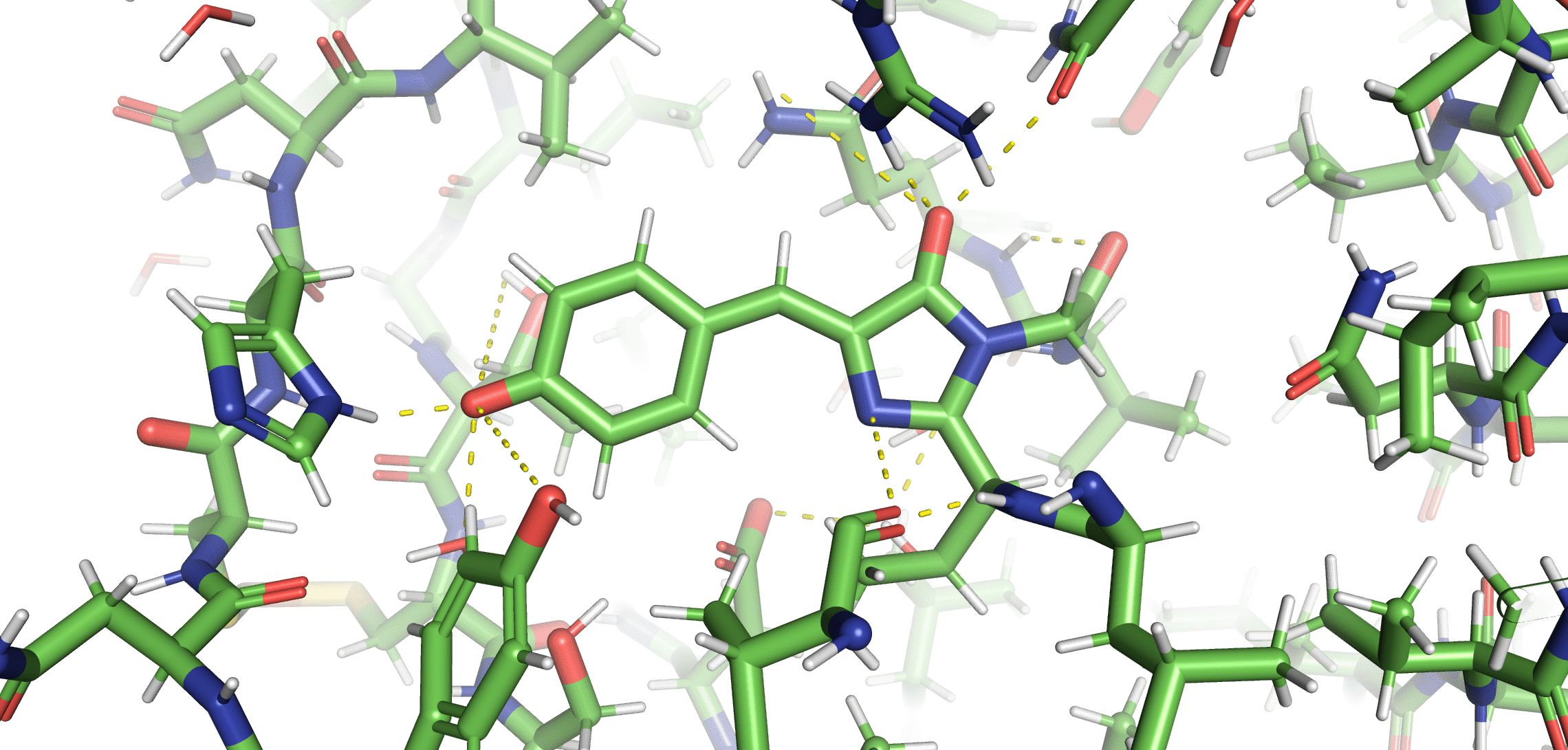

When the chromophore has increased flexability, it will de-excite through vibrations instead of emitting photons
Application: We can tailor the position of these cysteines to bind other heavy metals like lead, arsenic, etc.

After today, you should have a better understanding of
The definition and biological importance of structural biology
Alex's research example: Listeria monocytogenes with Dr. Cahoon
Dr. Cahoon is studying how Listeria monocytogenes (Lm) infects cells
Lm is a gram-positive bacteria responsible for listeriosis, a foodborne illness
Agbavor, C.; et al. DOI: 10.1128/mbio.00743-24
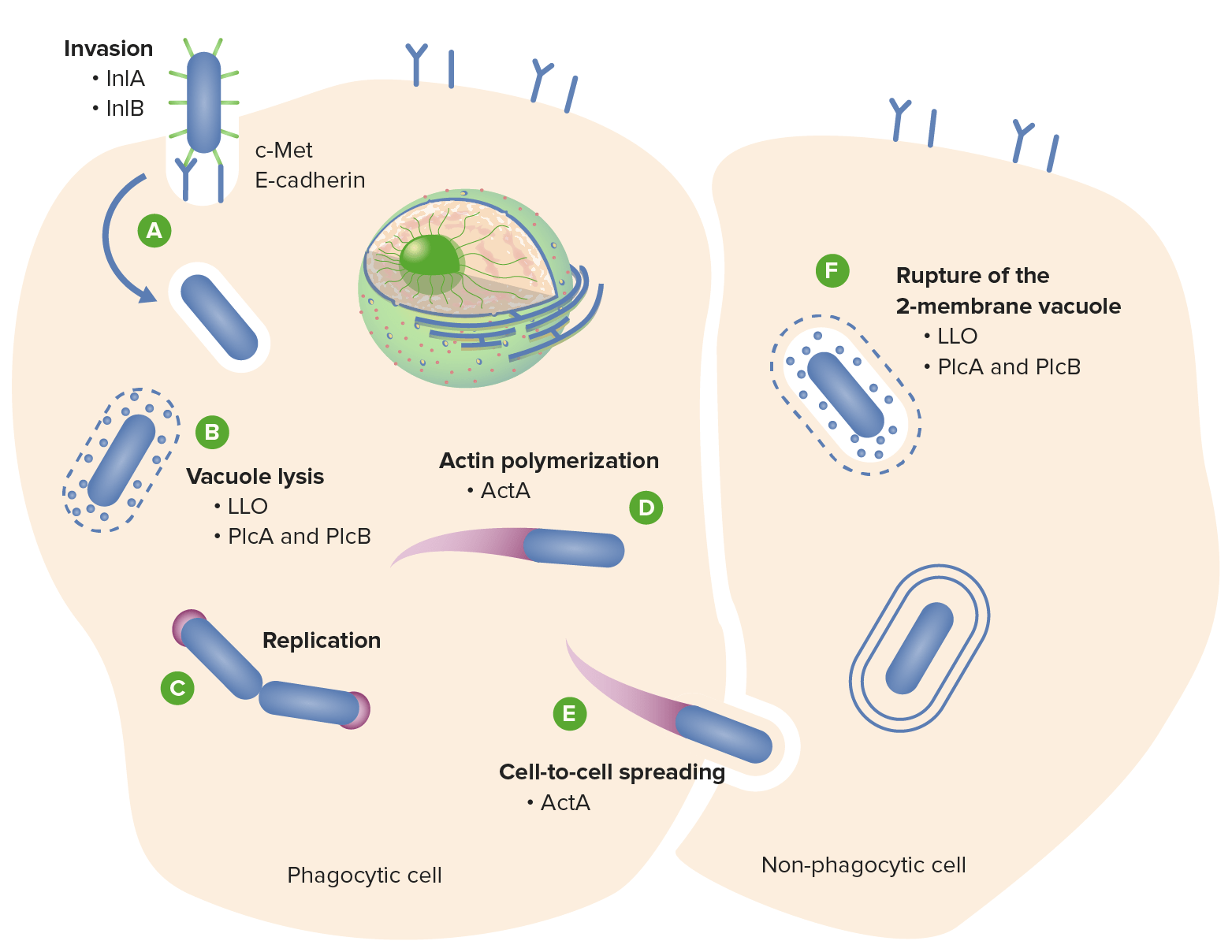
A key step in the Lm life cycle is escaping vacuoles and continue infecting
Lm secretes listeriolysin O (LLO) which forms pores in vacuoles allowing it to escape
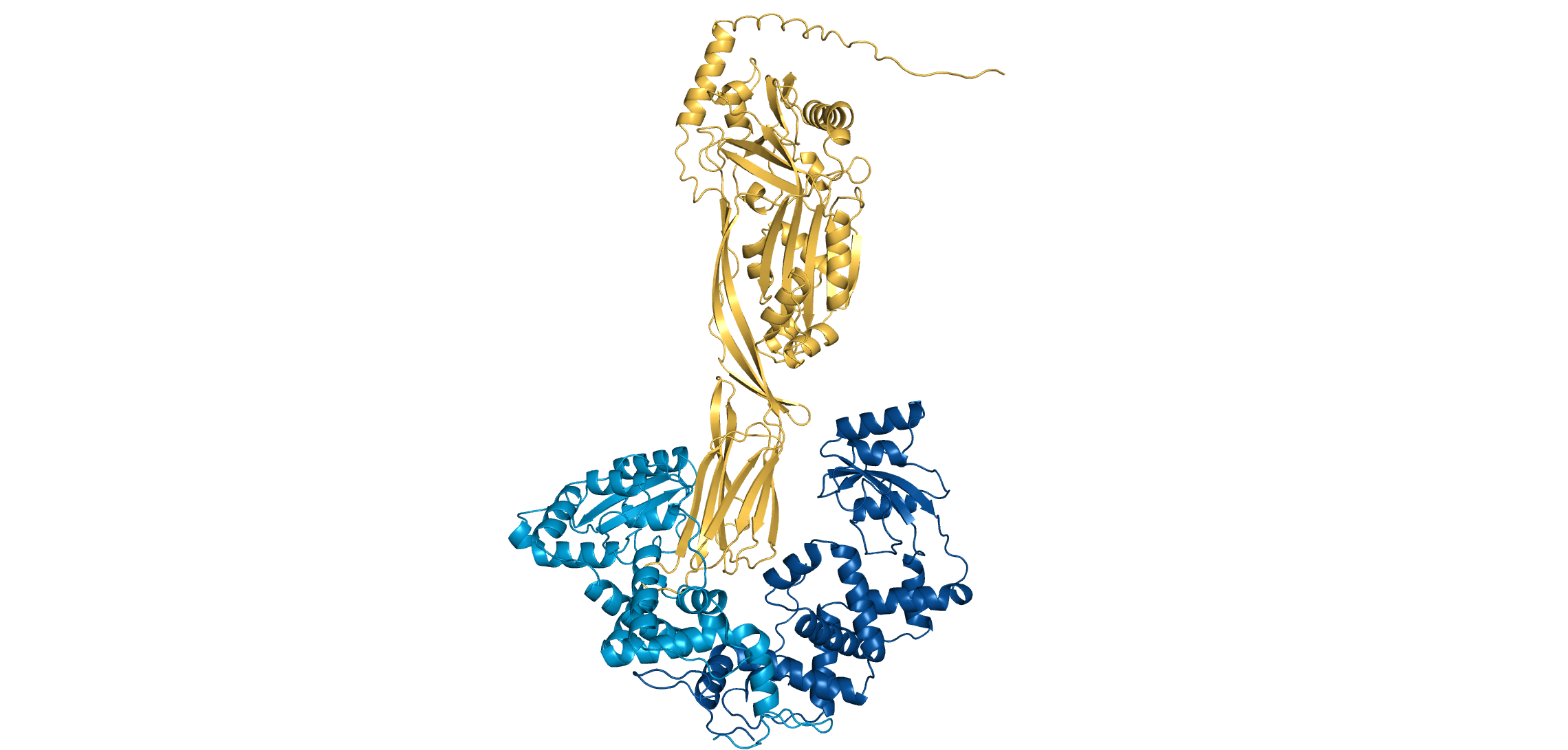
PrsA2
PrsA2
LLO
Lm secretes a pore-forming, cholesterol-dependent toxin called listeriolysin O (LLO) to escape vacuoles and infect cells
Agbavor, C.; et al. DOI: 10.1128/mbio.00743-24
The Cahoon lab (alongside several collaborators) demonstrated that PrsA2 (a chaperone) regulates LLO activity through a pH-dependent mechanism
At pH 7, PrsA2 remains bound to LLO, preventing it from forming pores. At pH 5, PrsA2 releases LLO to escape acidic vacuoles
This is a new project, so we do not know yet!
What is our computational question?
Are PrsA2-LLO interactions destabilized in acidic (i.e., pH 5) environments? If so, how?
Application: Once we understand this interaction, we can design a new antibiotic for gram positive bacteria
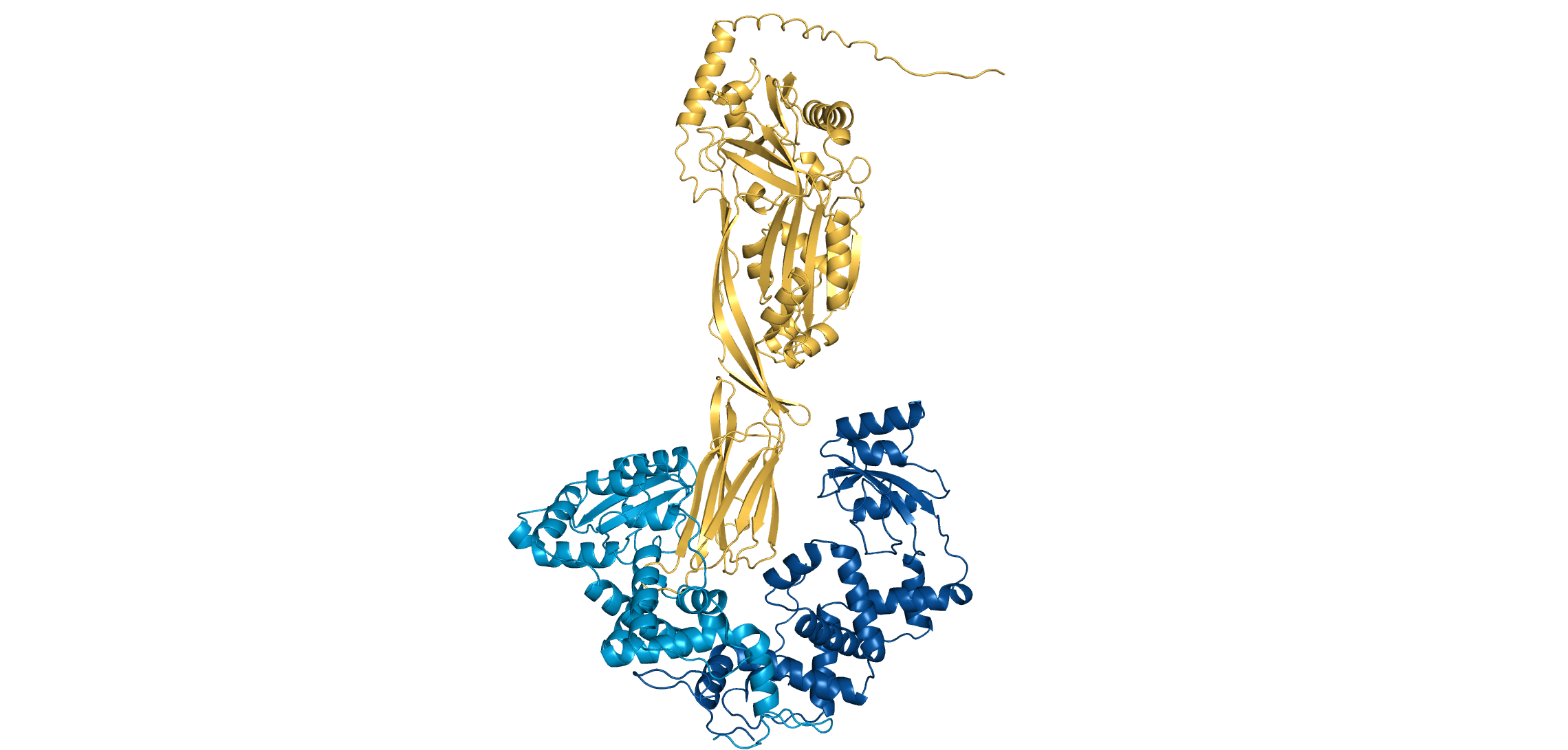
PrsA2
PrsA2
LLO
After today, you should have a better understanding of
Basic principles of protein structure
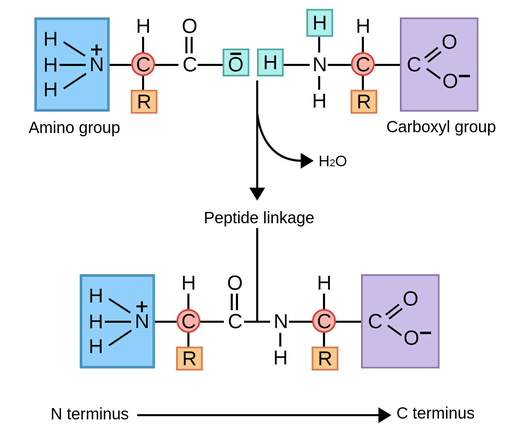
Amino acids are the fundamental building blocks of proteins
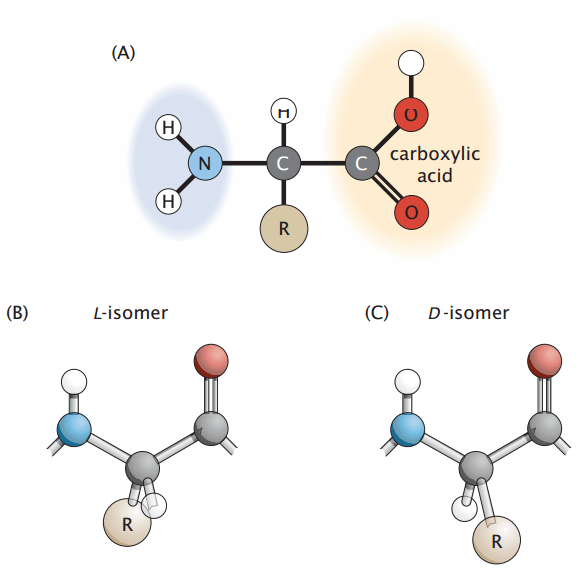
All proteins are composed of smaller molecules called amino acids, which are linked together in specific sequences.
Each amino acid contains a central carbon (alpha carbon) bonded to an amino group (NH₂), a carboxyl group (COOH), a hydrogen atom, and a variable side chain known as the R-group.
The primary structure of a protein is the linear sequence of amino acids held together by covalent peptide bonds
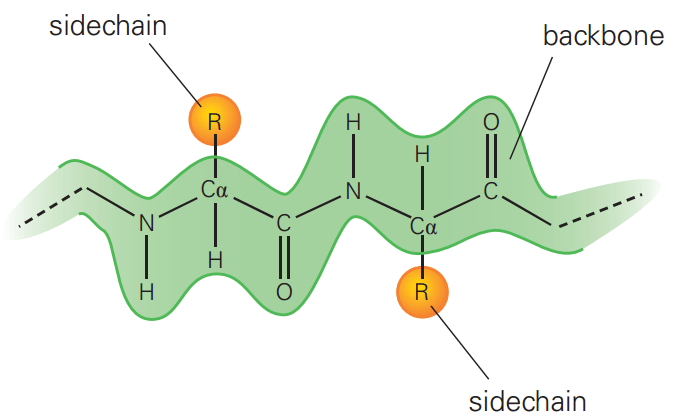
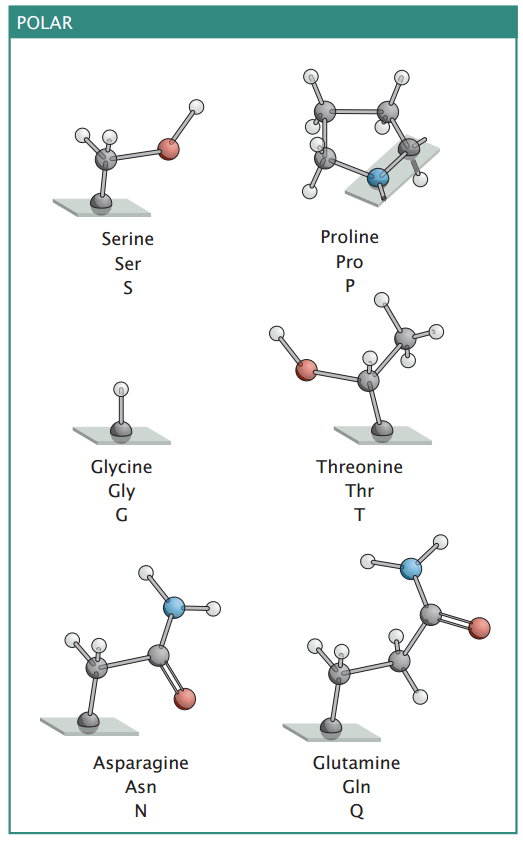
Polar amino acids enable interactions with water and other polar molecules
Polar amino acids have side chains that can form hydrogen bonds, making them hydrophilic
You will not be tested on your amino acid abbreviations
Polar amino acids contribute to protein solubility and help stabilize secondary and tertiary structures through hydrogen bonding.
Many polar amino acids are involved in enzymatic activity, facilitating catalytic reactions by stabilizing transition states or interacting with substrates.
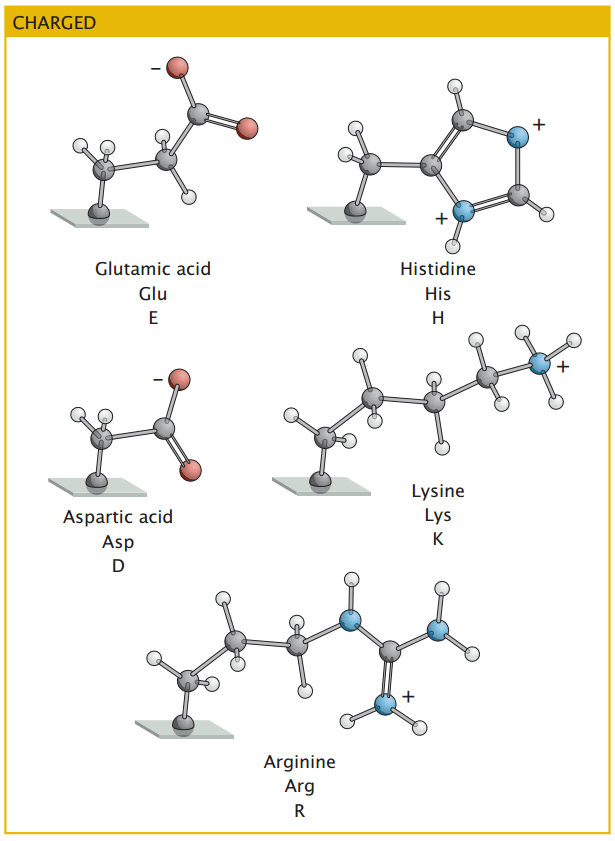
Charged amino acids play key roles in protein stability and interactions
Acidic amino acids carry negative charges and participate in ionic interactions that stabilize protein structures
Charged amino acids contribute to protein folding by forming salt bridges, which enhance stability.
The cellular environment's pH can influence these amino acids' charge state, affecting protein conformation and function.
Basic amino acids carry positive charges and frequently interact with negatively charged molecules like DNA and phospholipids.
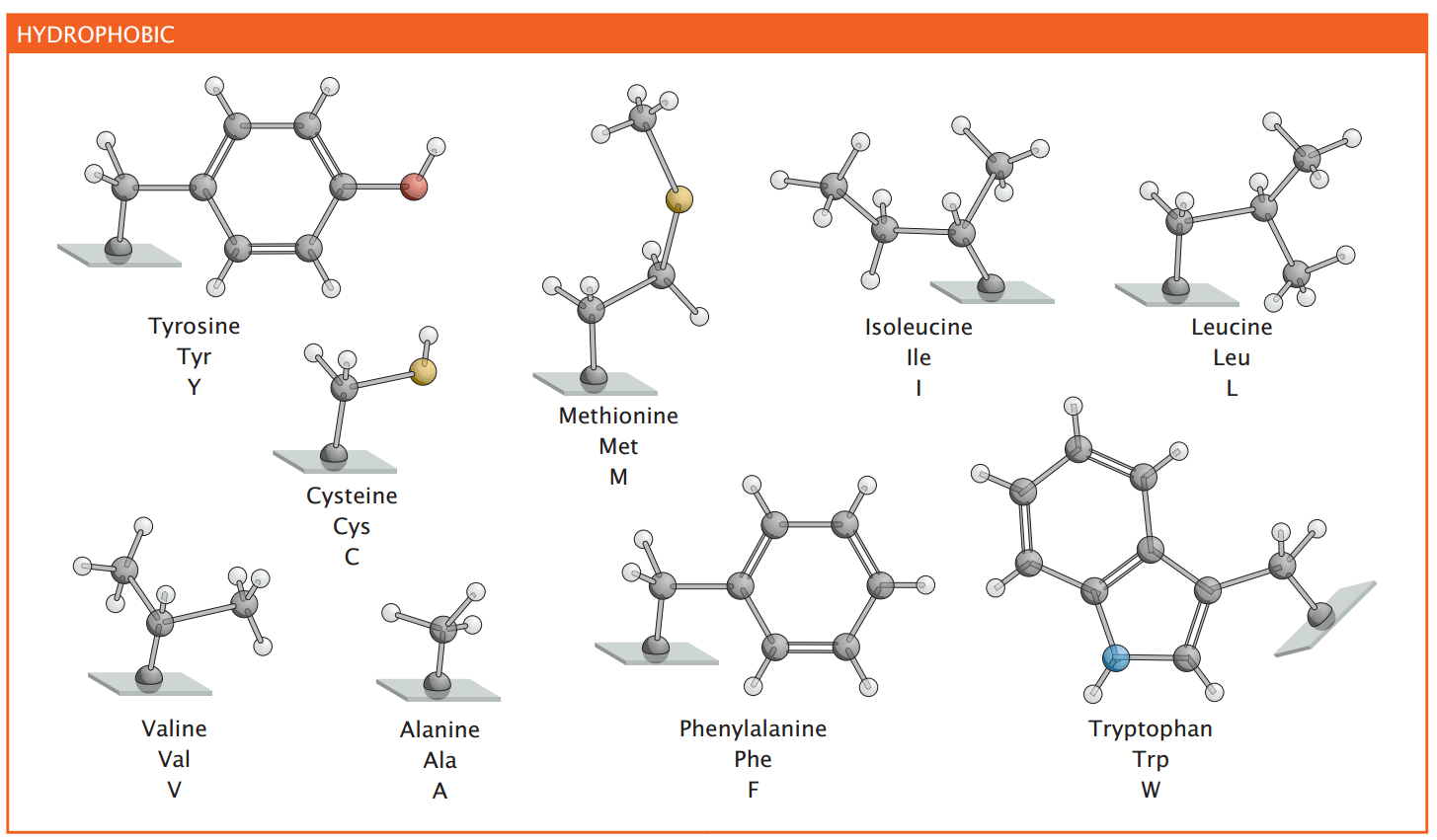
Nonpolar amino acids drive protein folding and membrane interactions
These amino acids are often found in the interior of globular proteins, stabilizing protein structure by minimizing exposure to water
Aromatic nonpolar amino acids participate in stacking interactions, influencing protein stability and ligand binding
The primary structure of a protein determines its final shape and function
The primary structure of a protein is the linear sequence of amino acids, held together by covalent peptide bonds
The primary structure alone does not reveal the protein's functional form or activity
While the primary sequence is critical, the folding process may also depend on cellular factors (e.g., chaperones)
After today, you should have a better understanding of
Basic principles of protein structure
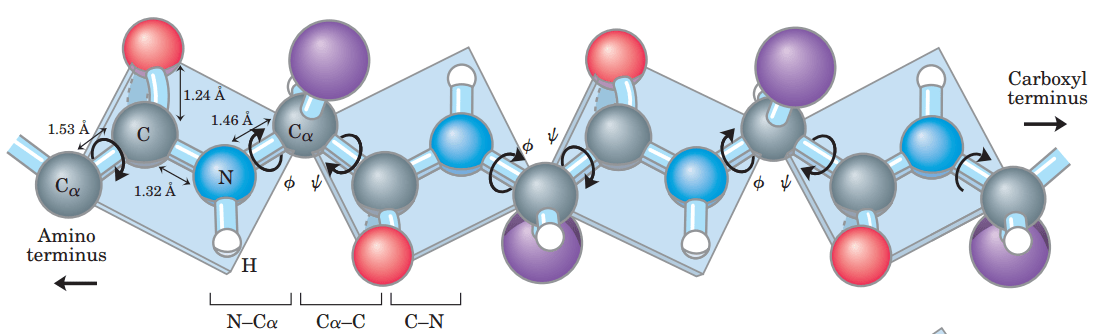
Phi (Φ) and Psi (Ψ) angles determine protein backbone flexibility and folding
Proteins are flexible due to rotation around specific backbone bonds: the phi (Φ) and psi (Ψ) angles.
Not all angle combinations are allowed due to steric hindrance—this is visualized in a Ramachandran plot, which maps permitted conformations.
Secondary structures provide local organization within proteins
Secondary structures refer to regularly repeating local conformations of the polypeptide backbone.
These structures help proteins achieve compact and stable folding while maintaining flexibility for function.
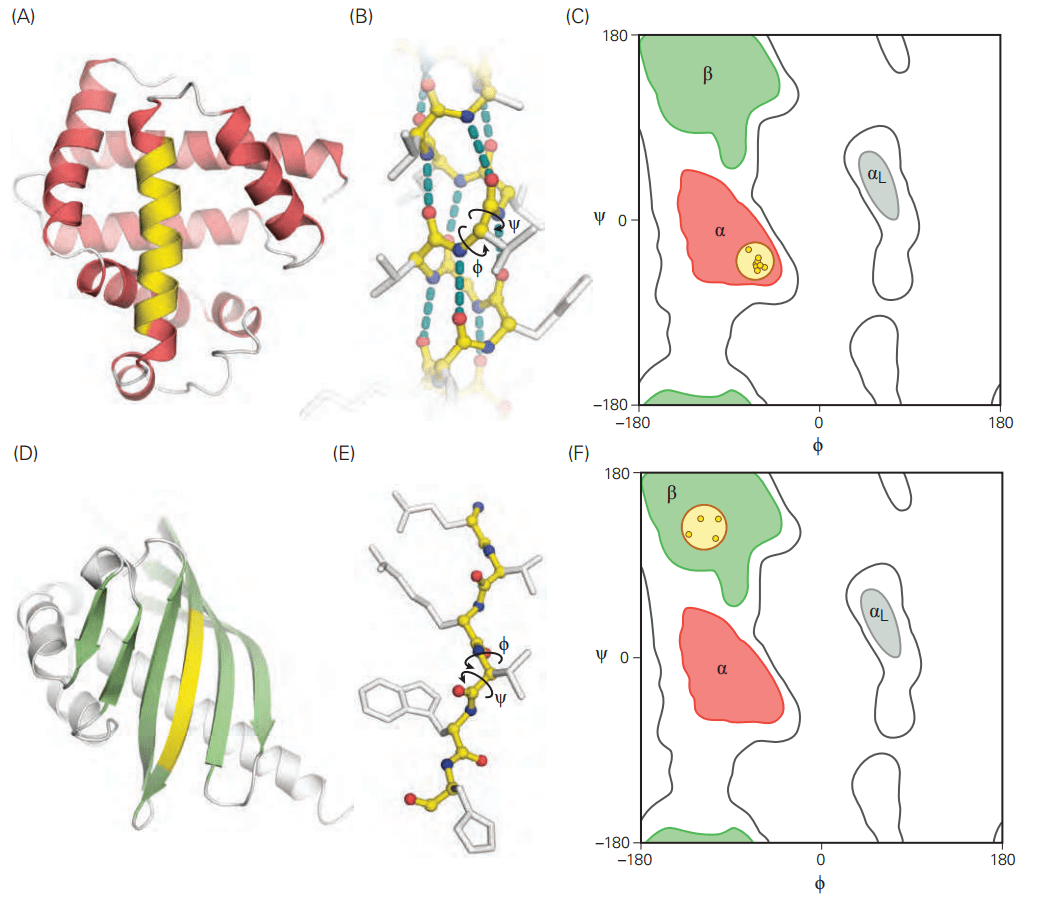
Alpha-helices are stabilized by hydrogen bonds and provide structural flexibility
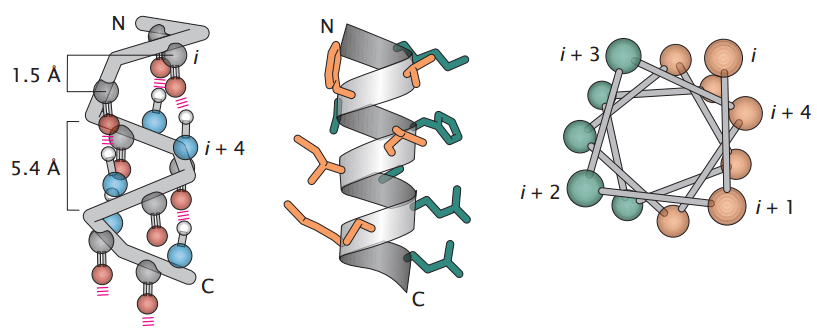
An alpha-helix is a right-handed coil with 3.6 amino acids per turn, stabilized by hydrogen bonds between the backbone carbonyl oxygen and the amide hydrogen of a residue four positions ahead.
Side chains project outward, allowing interactions with the surrounding environment.
Beta-sheets provide strength and stability to protein structures
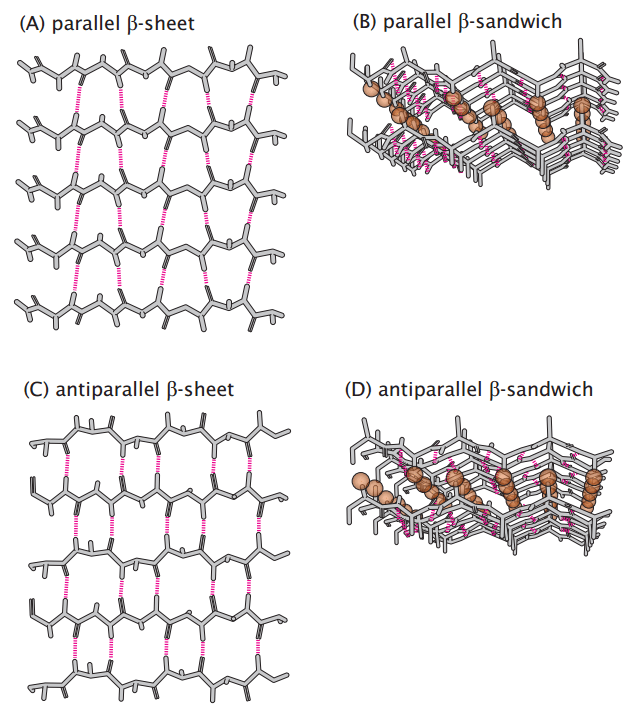

Beta-sheets consist of extended polypeptide strands aligned side by side, stabilized by hydrogen bonds between backbone atoms of adjacent strands.
Strands can be parallel (N-to-C direction aligned) or antiparallel (N-to-C in opposite directions), with antiparallel sheets being more stable.
Side chains alternate above and below the sheet, affecting interaction and stability.
After today, you should have a better understanding of
Basic principles of protein structure
Tertiary Structure
The tertiary structure refers to the complete 3D shape of a single polypeptide chain
Tertiary structures reveal active sites or binding pockets where catalysis or molecular interactions occur
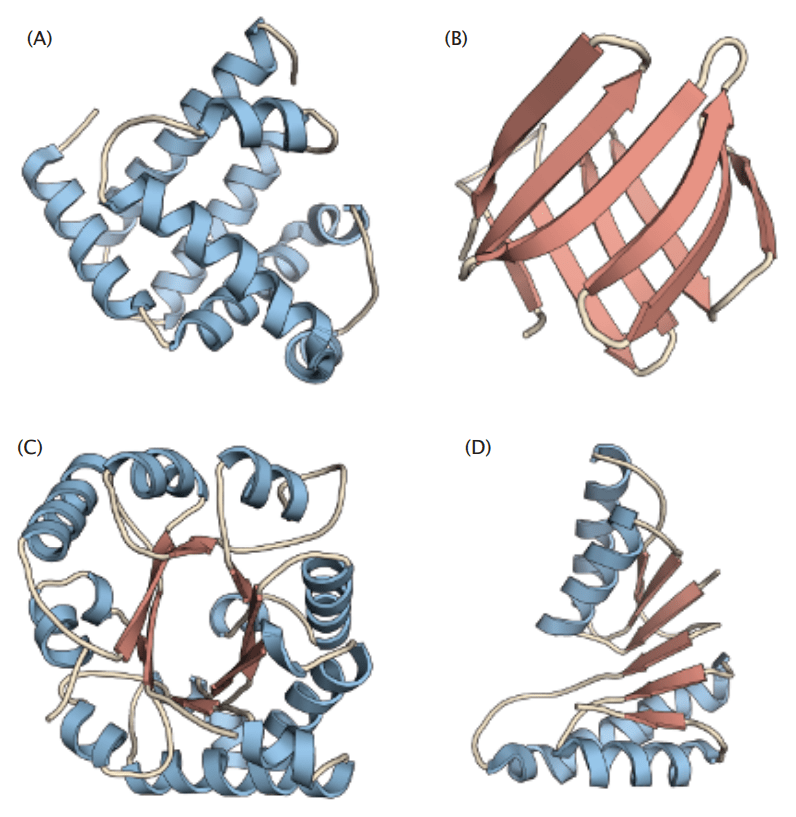
After today, you should have a better understanding of
X-ray crystallography and cryo-electron microscopy
How can we experimentally determine the 3D atomic structure of a protein?

Fundamentals of X-ray Crystallography
Basic Principle: Photons scatter when they interact with other particles
The scattered X-rays form a diffraction pattern unique to the crystal
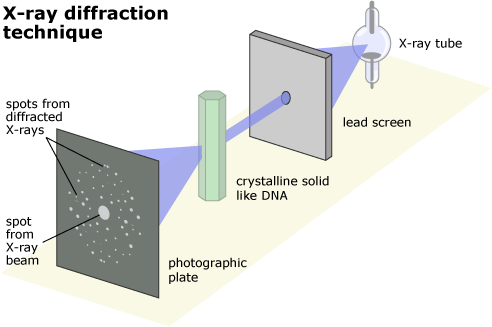
Probe: Photon (carrier of electromagnetic radiation)
X-rays undergo elastic scattering by electrons
- An incident photon induces an oscillating dipole by distorting the electron density (Rayleigh)
- An oscillating dipole acts as an electromagnetic source and re-emits photons at the same wavelength in all directions
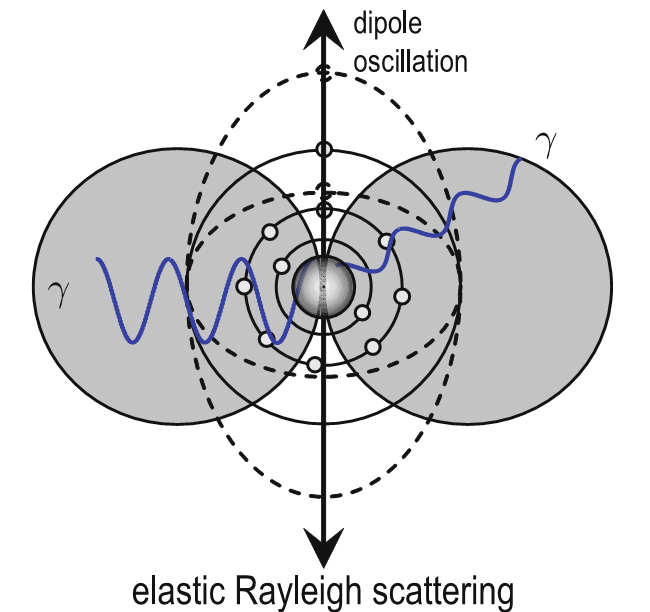
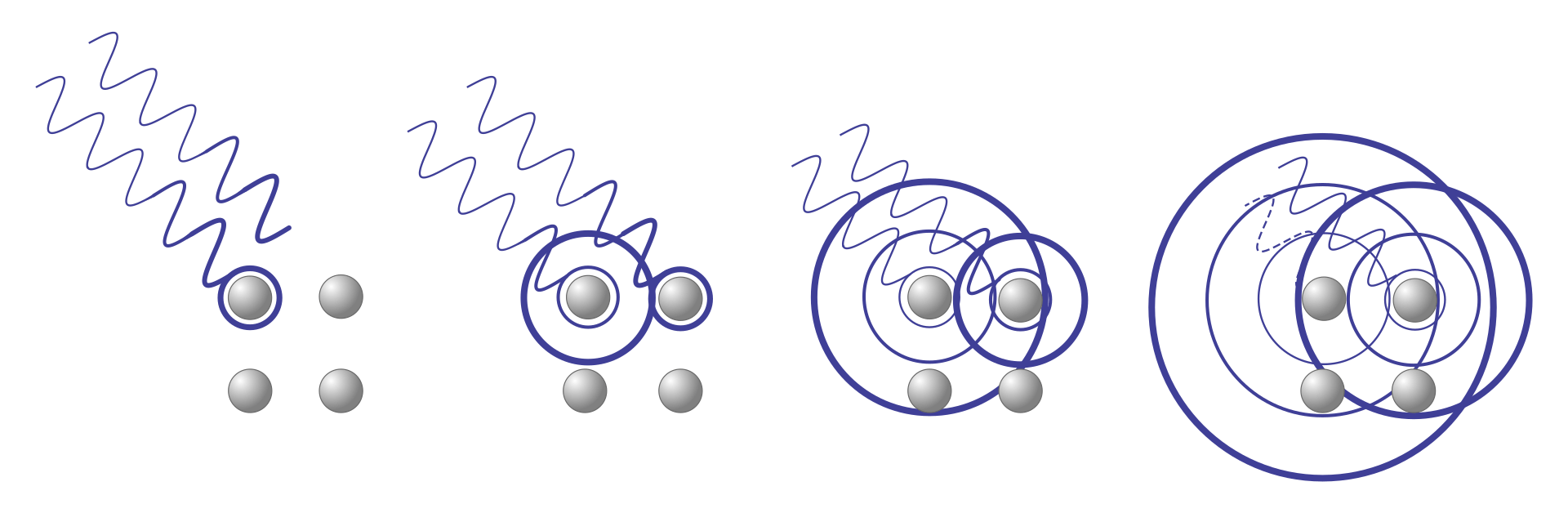
What happens when two waves overlap?
Constructive interference is needed to amplify signal for detectors
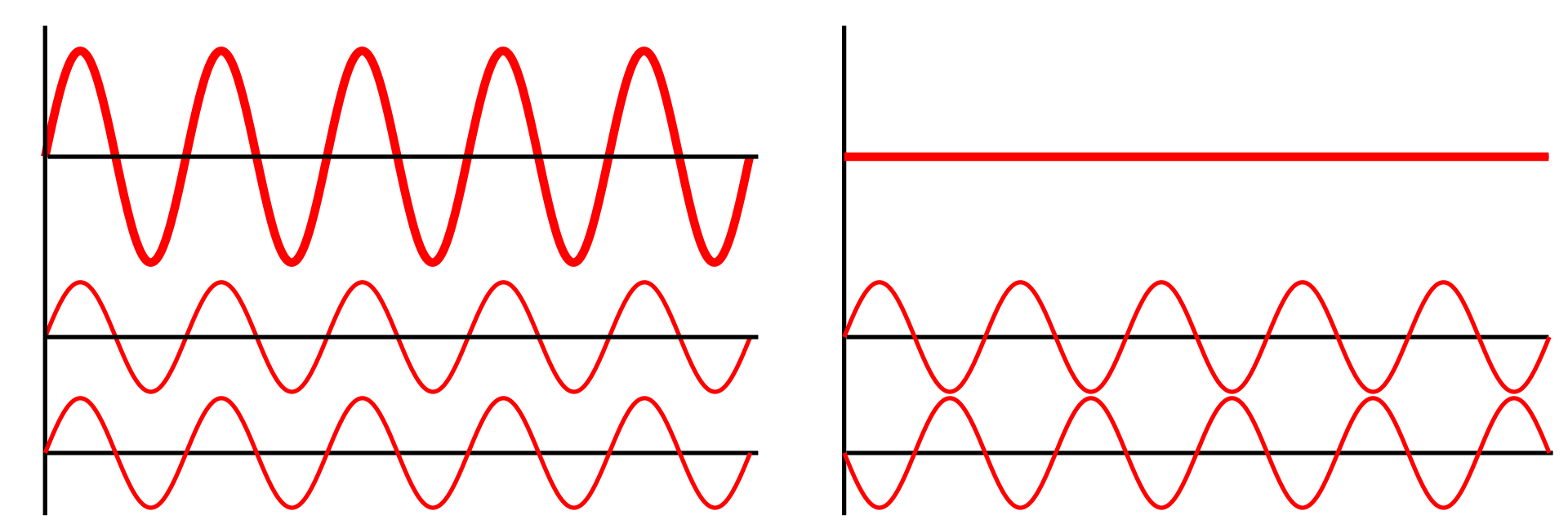



If wavelengths are similar and in phase, they constructively interfere
If waves are out of phase, they deconstructively interfere
Constructive interference leads to distinct patterns
If wavelengths are similar and in phase, they constructively interfere and form spots based on atom type and distance
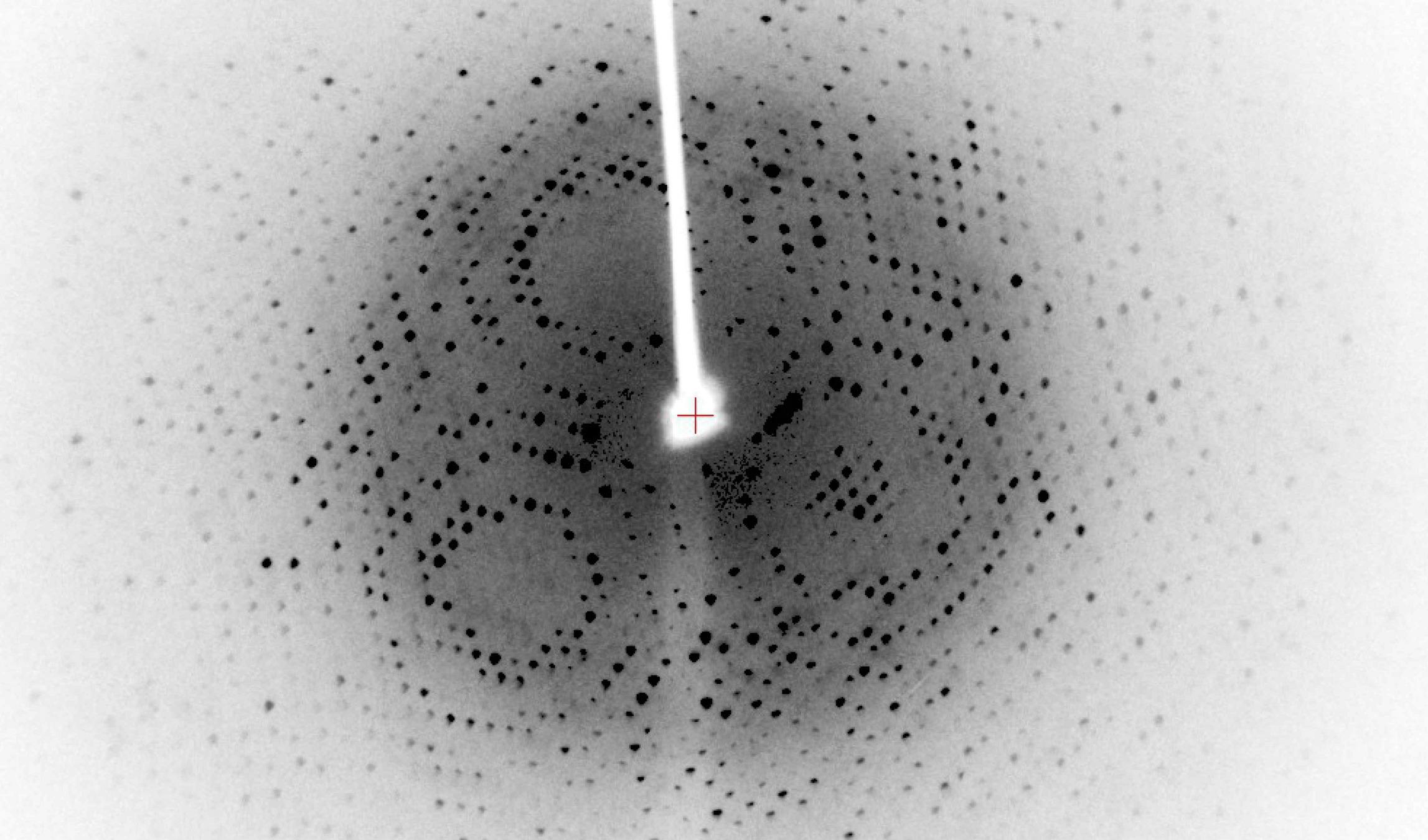
The diffraction pattern
The spots on the detector represent the reflections of the scattered X-rays
- Intensity of the spots reflects the electron density in the crystal
- Position and angle: The position of the spots corresponds to the geometry
The diffraction pattern does not directly show the atomic positions, but provides the data needed to infer the electron density

Building the electron density map
The 3D electron density map reveals the distribution of electrons in the crystal, indicating where atoms are located
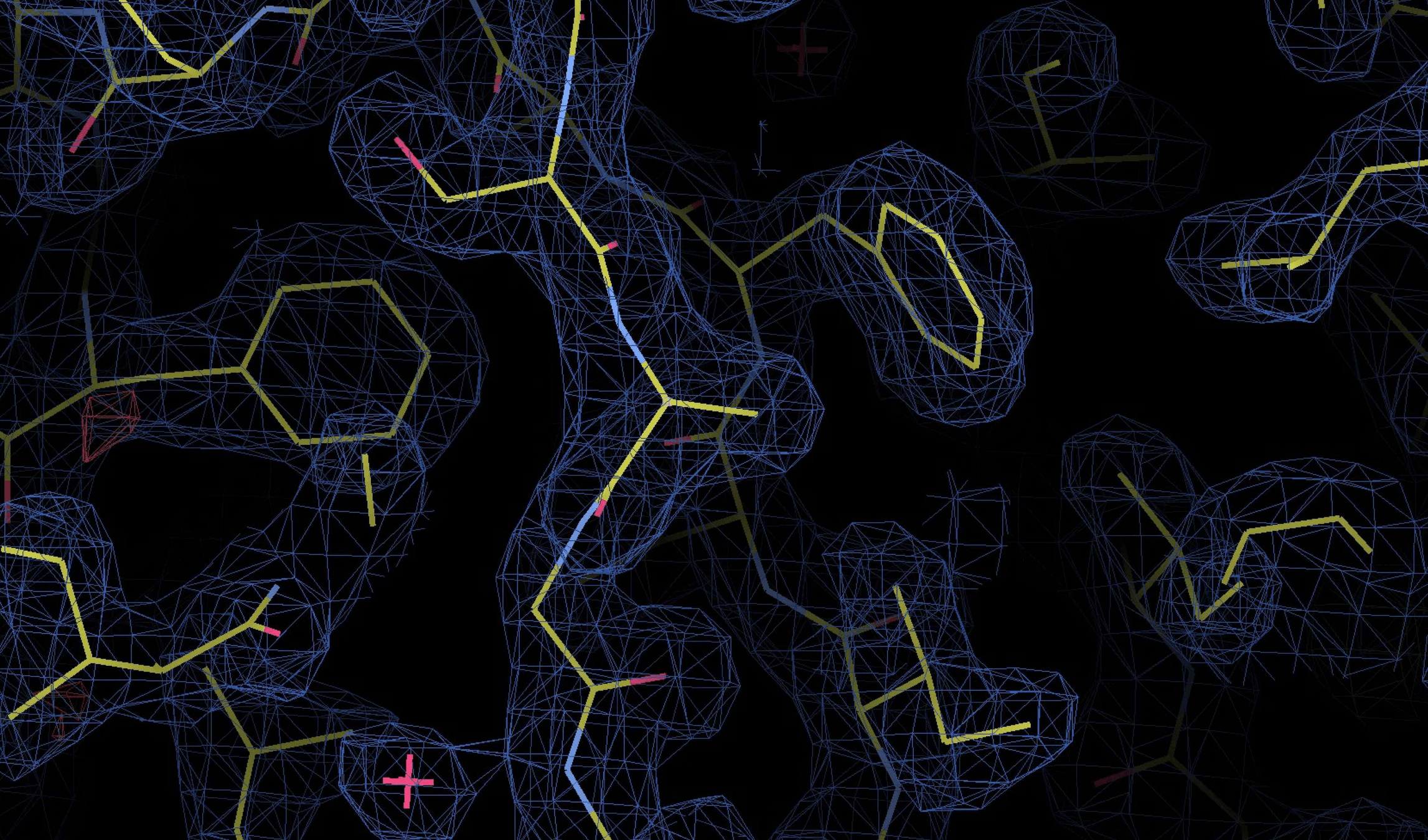
The electron density map is interpreted by fitting atomic models (e.g., amino acids for proteins) into the density
Low-resolution data make it difficult to assign atomic positions precisely, leading to uncertainty in the model
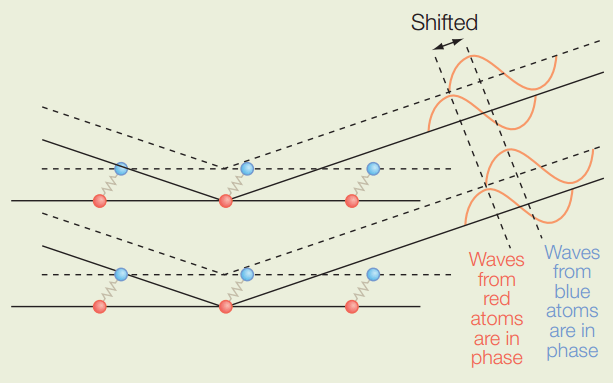
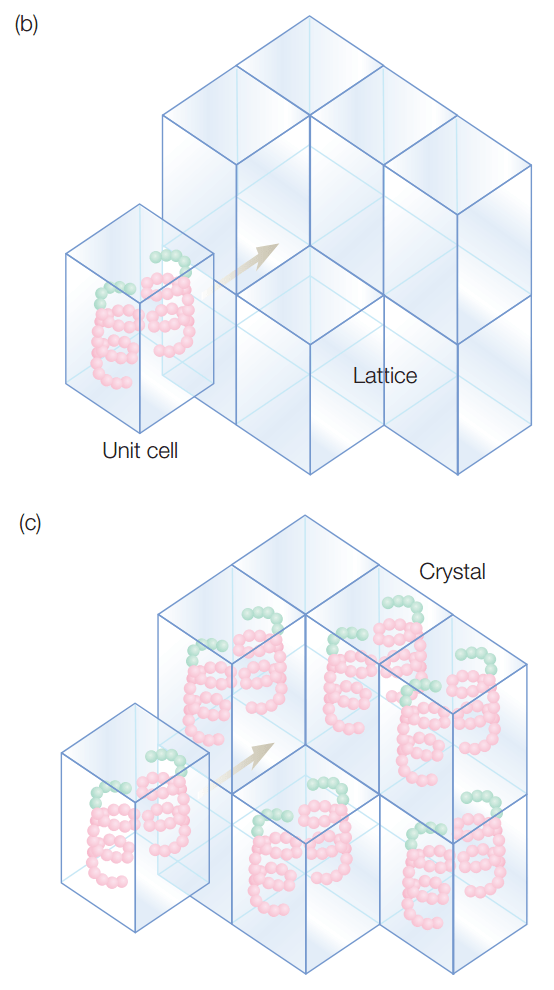
Why do we need crystals?
Crystals have the same repeating unit cell, which amplifies our signals
If in solution, particles would be
- Too sparse to diffract
- Moving and diffraction pattern would constantly change

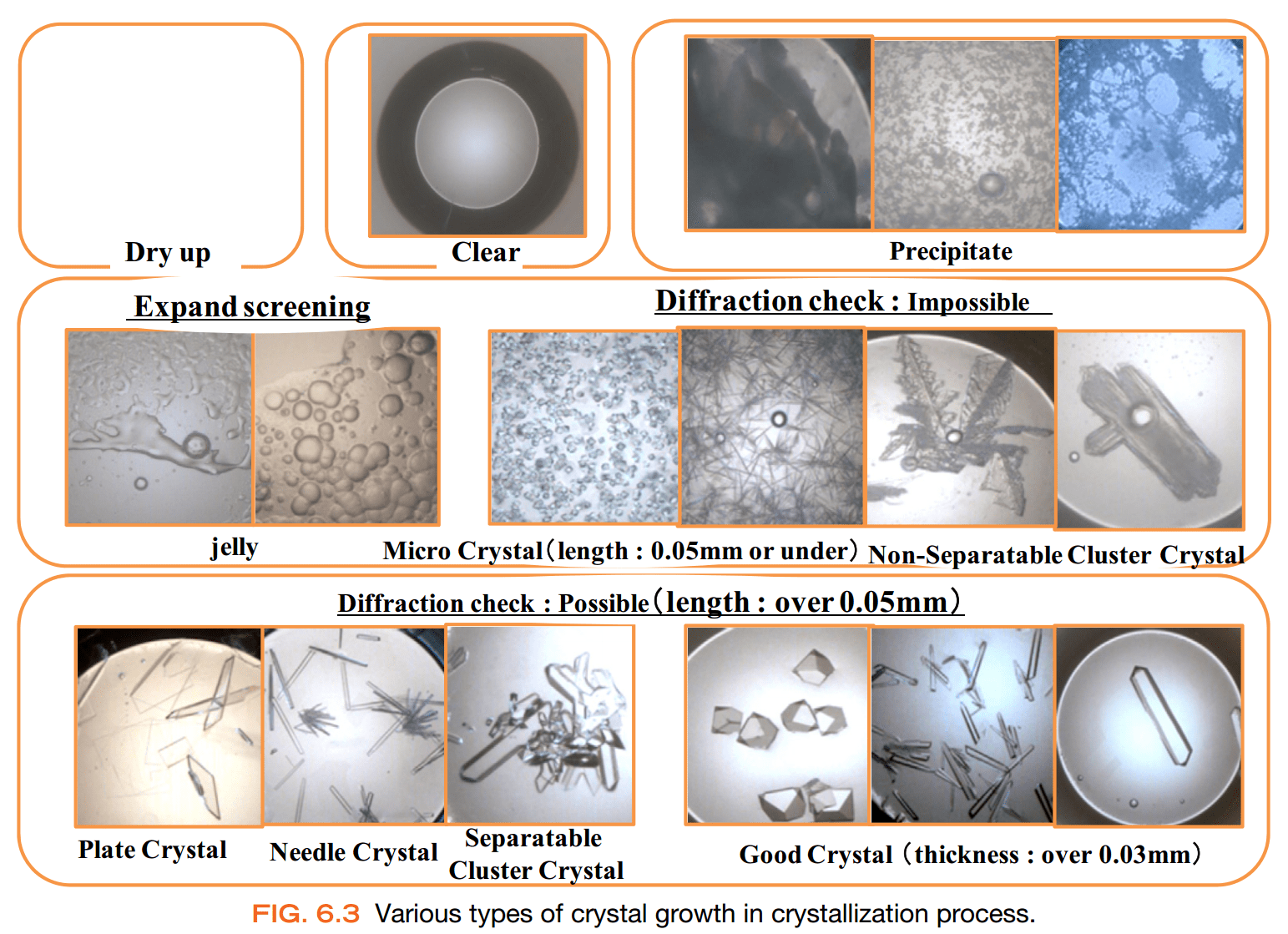
Crystal quality
After today, you should have a better understanding of
X-ray crystallography and cryo-electron microscopy
Why Cryo-EM?
In Cryo-EM, a beam of high-energy electrons is used instead of photons
Why Electrons?
- Electrons have a much shorter wavelength (~0.02 Å at 300 keV) than photons
- Light elements scatter electrons more effectively than X-rays
No crystals: The sample is rapidly frozen in vitreous ice to preserve its native structure
- By freezing the sample, biological molecules are imaged in their native hydrated state
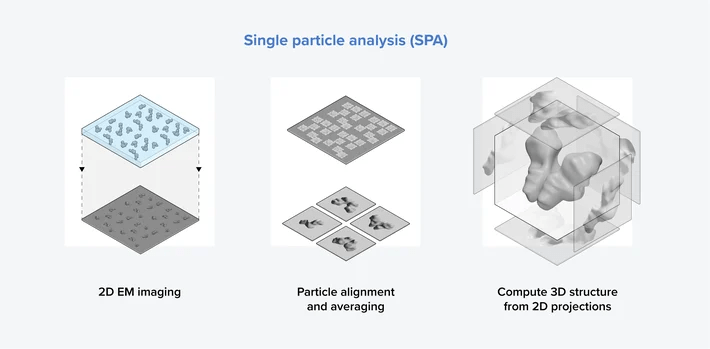
Single Particle Analysis (SPA)
Single Particle Analysis is the main Cryo-EM technique used to determine the 3D structures of individual macromolecules
- Millions of images of individual particles are collected from a thin layer
- Particles are computationally aligned and classified into different orientations


After today, you should have a better understanding of
The challenge of protein disorder
Challenge of flexibility and disorder in biomolecules
Molecules are not static
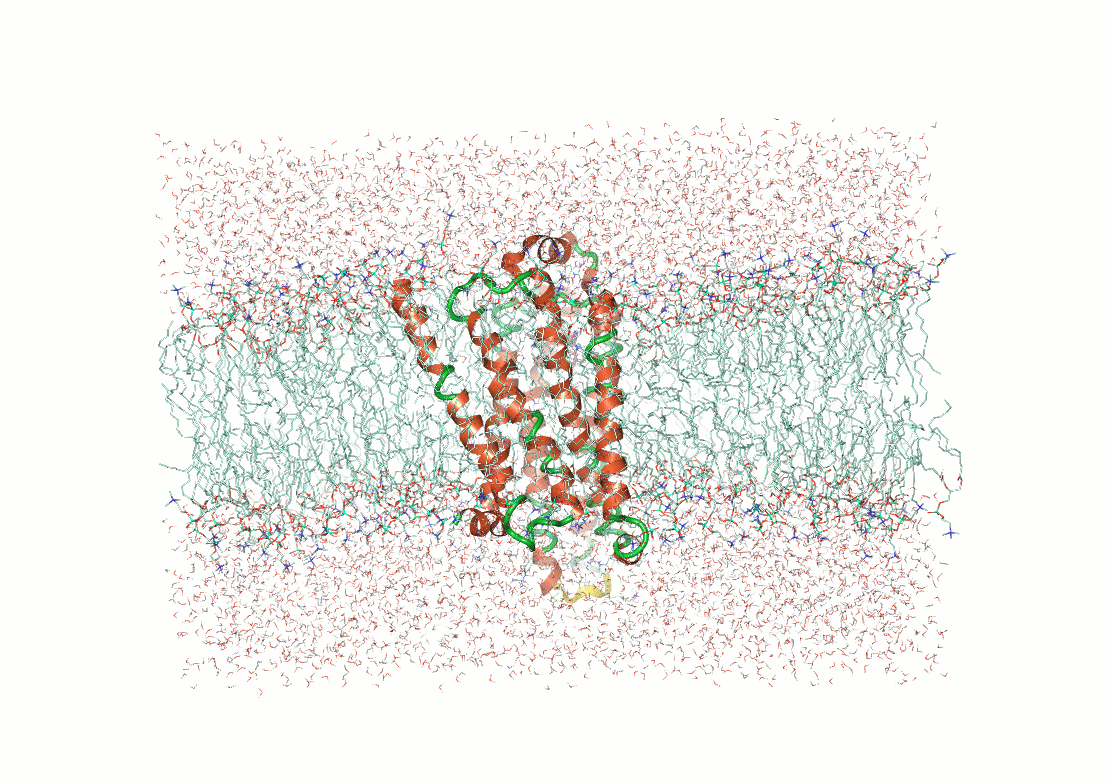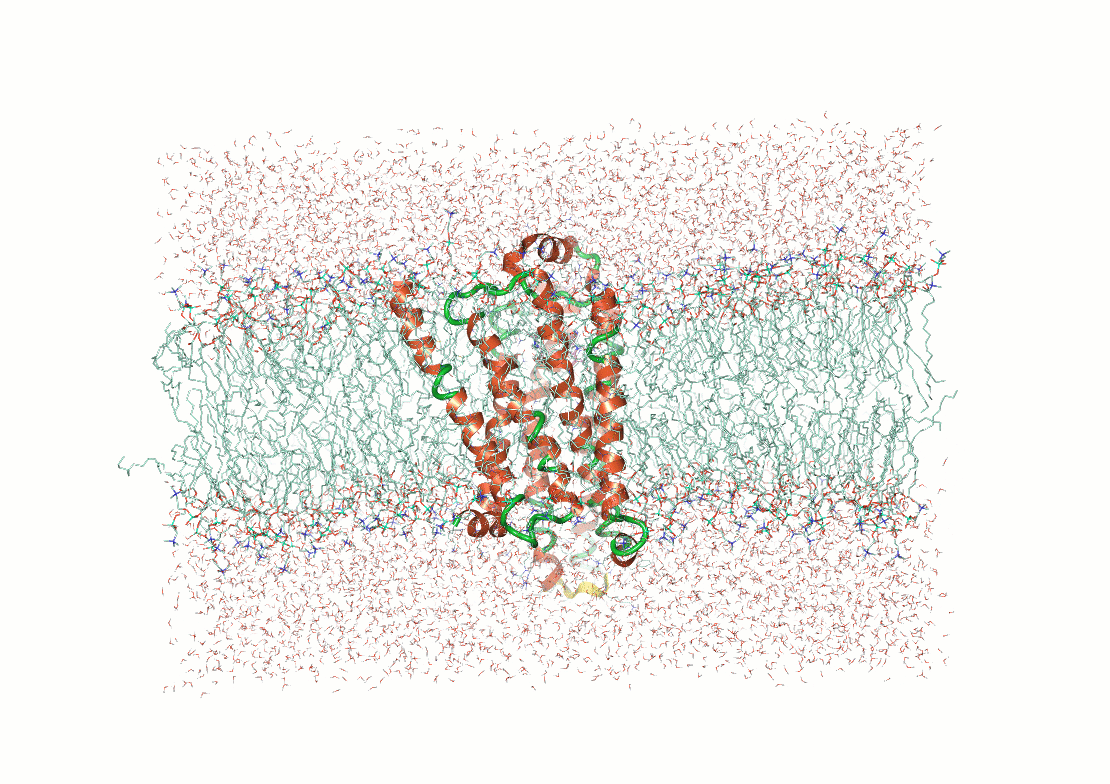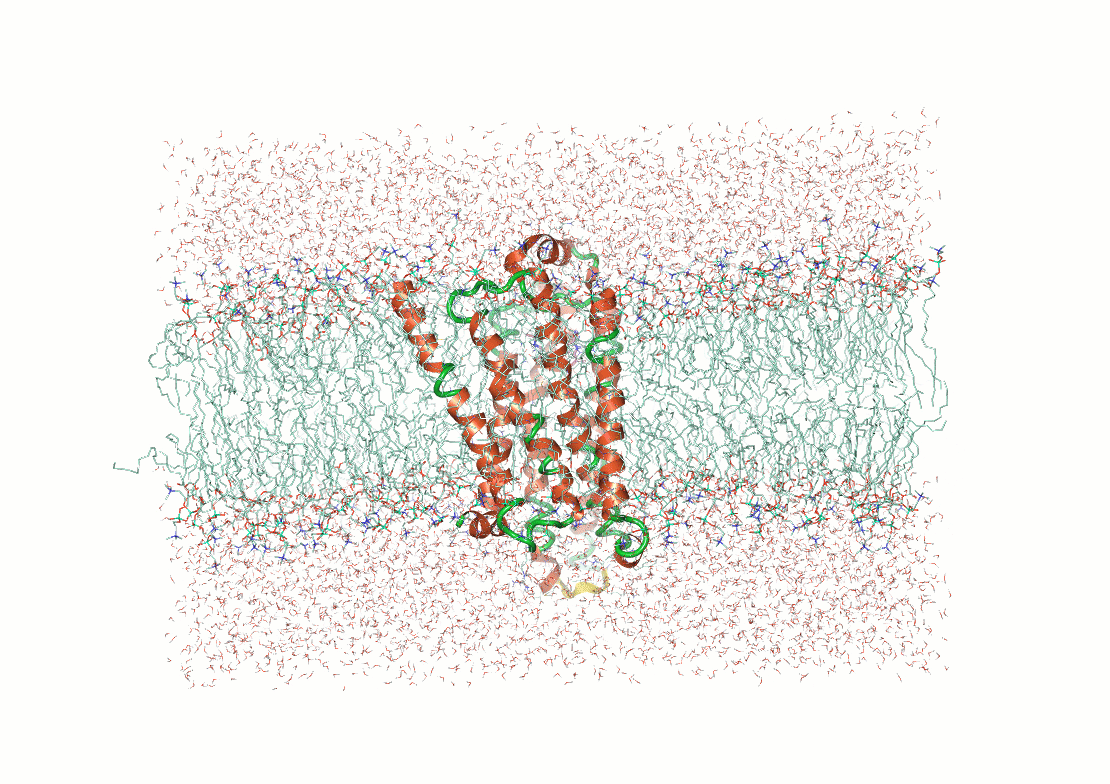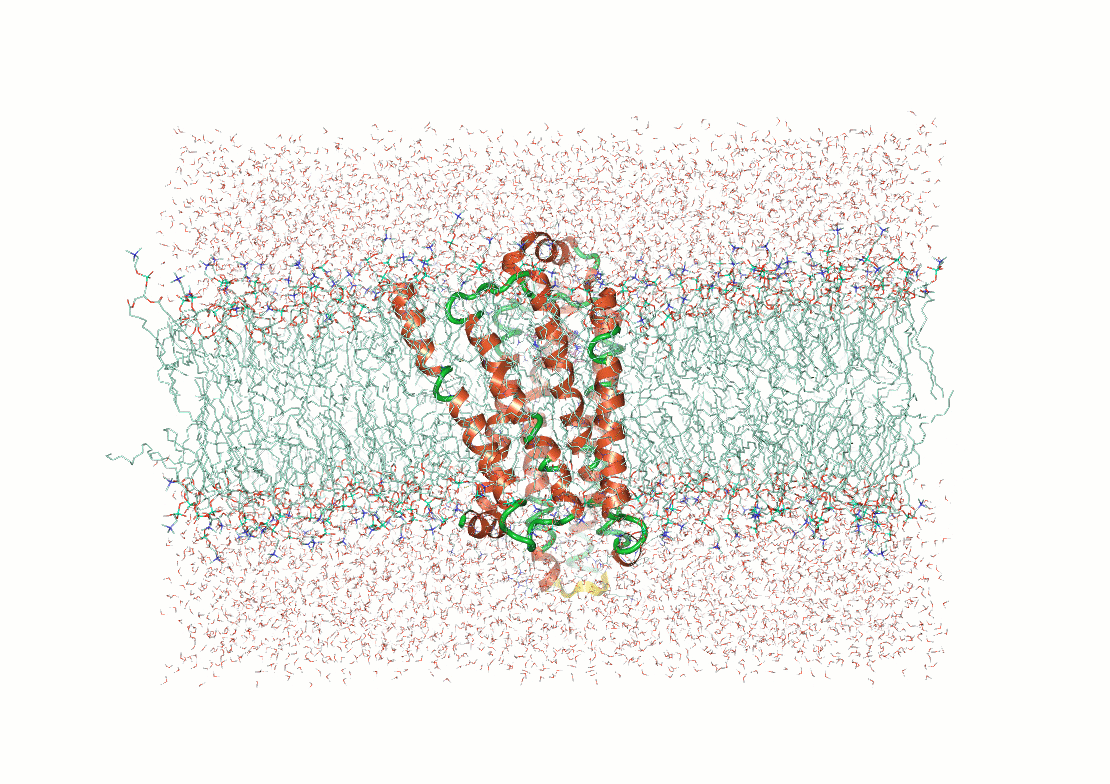
Example: The p53 tumor suppressor protein has flexible regions critical for its regulation and binding interactions
Proteins often exhibit flexibility, disordered regions, and multiple conformations
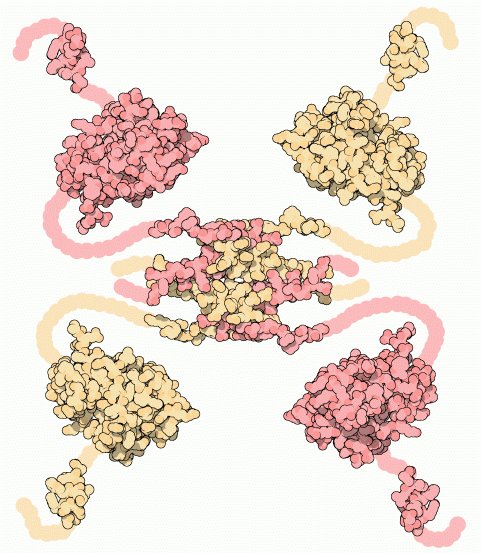
Why It Matters: Structural techniques often require ordered or stable configurations
Challenges in X-ray Crystallography
- Flexible or disordered regions do not pack into crystals well, often leading to failure in obtaining high-quality crystals.
- Even in cases where crystallization is successful, flexible or disordered regions often do not show up clearly in the electron density map.
- Crystals capture a single molecule conformation, often ignoring the flexibility or dynamic range.

Cryo-EM and Conformational Flexibility
One strength of Cryo-EM is its ability to capture multiple conformational states of a molecule, providing insights into flexibility and structural heterogeneity.
Challenge: A major issue in Cryo-EM is that highly flexible or disordered molecules may appear as fuzzy or low-resolution regions in the final structure
Advanced computational techniques are required to sort out different conformations present in the Cryo-EM data
Intrinsically Disordered Proteins (IDPs)
Intrinsically disordered proteins (IDPs) or regions lack a stable 3D structure under physiological conditions but are still functional, often gaining structure upon binding to partners
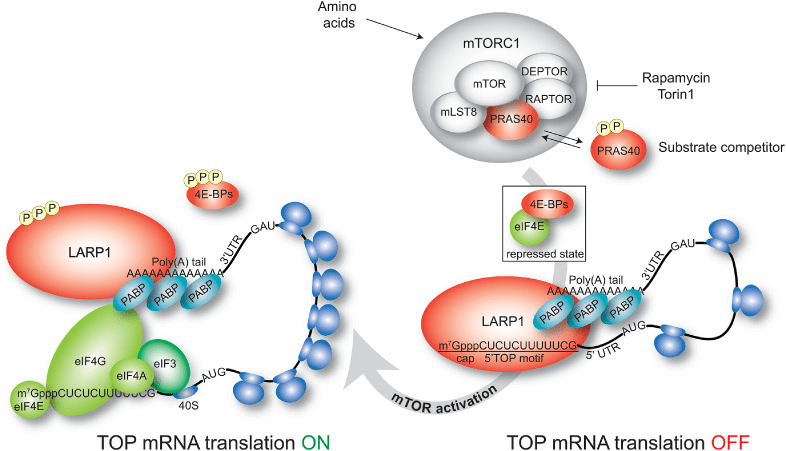
Before the next class, you should
Lecture 09B:
Structural Biology -
Methodology
Lecture 09A:
Structural Biology -
Foundations
Today
Thursday
Atomistic structure determines behavior of biomolecules
Why is Green Fluorescent Protein (GFP) fluorescent, but not the chromophore in solution?
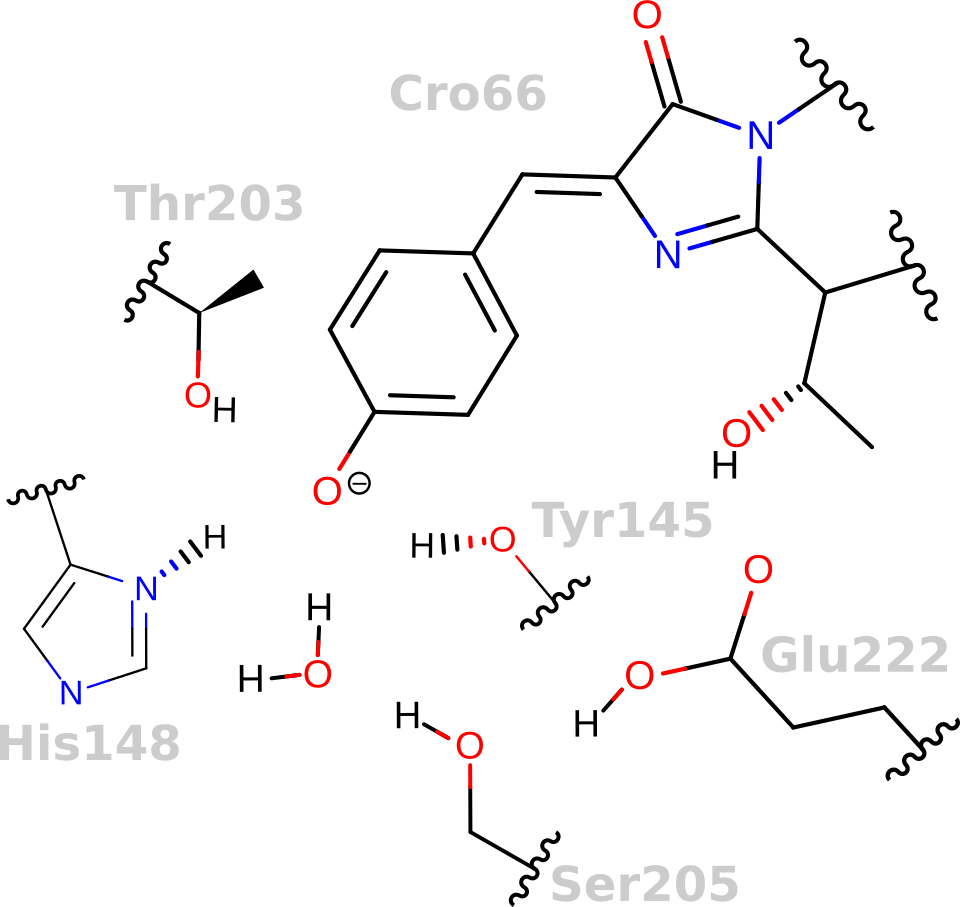

GFP keeps the chromophore planar and facilitates an excited-state proton transfer
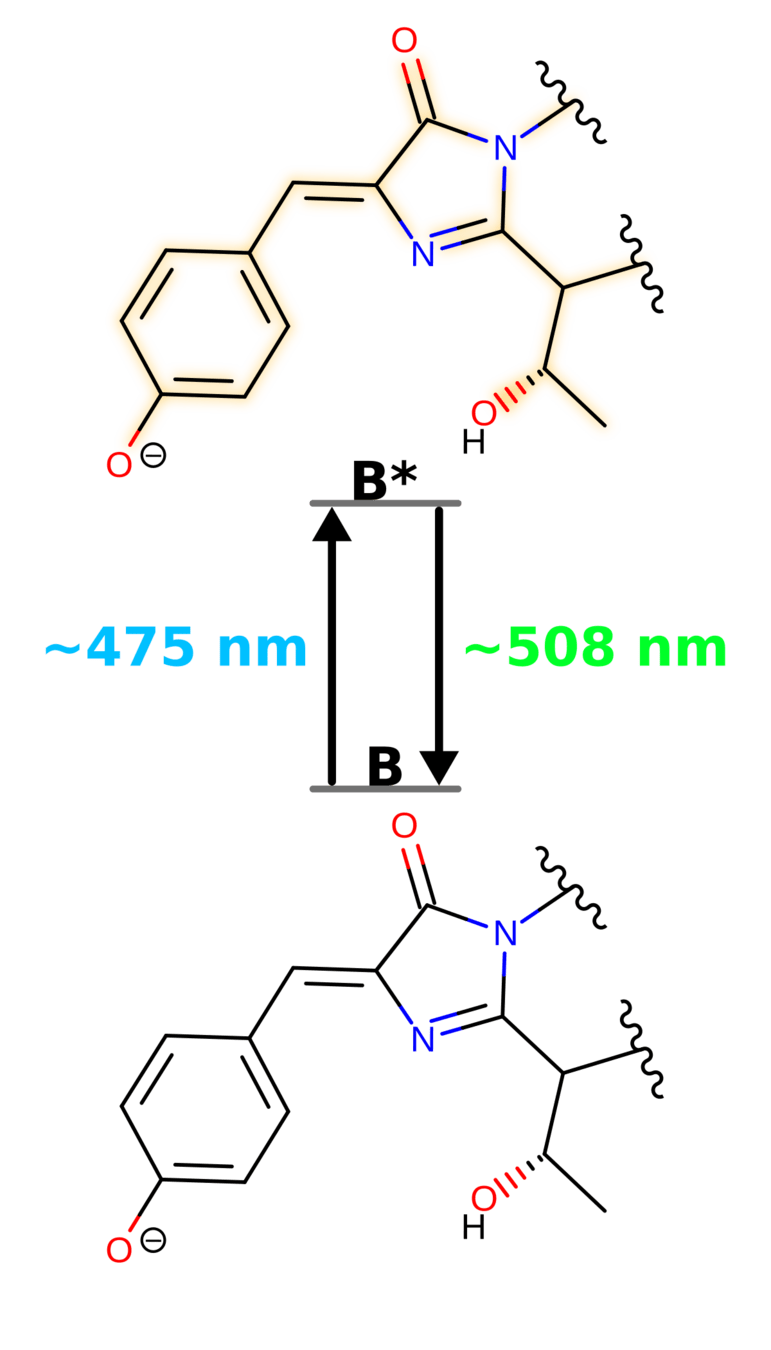
Fluorescent
Not Fluorescent
Particle behaviors are determined by quantum numbers
Principle quantum number
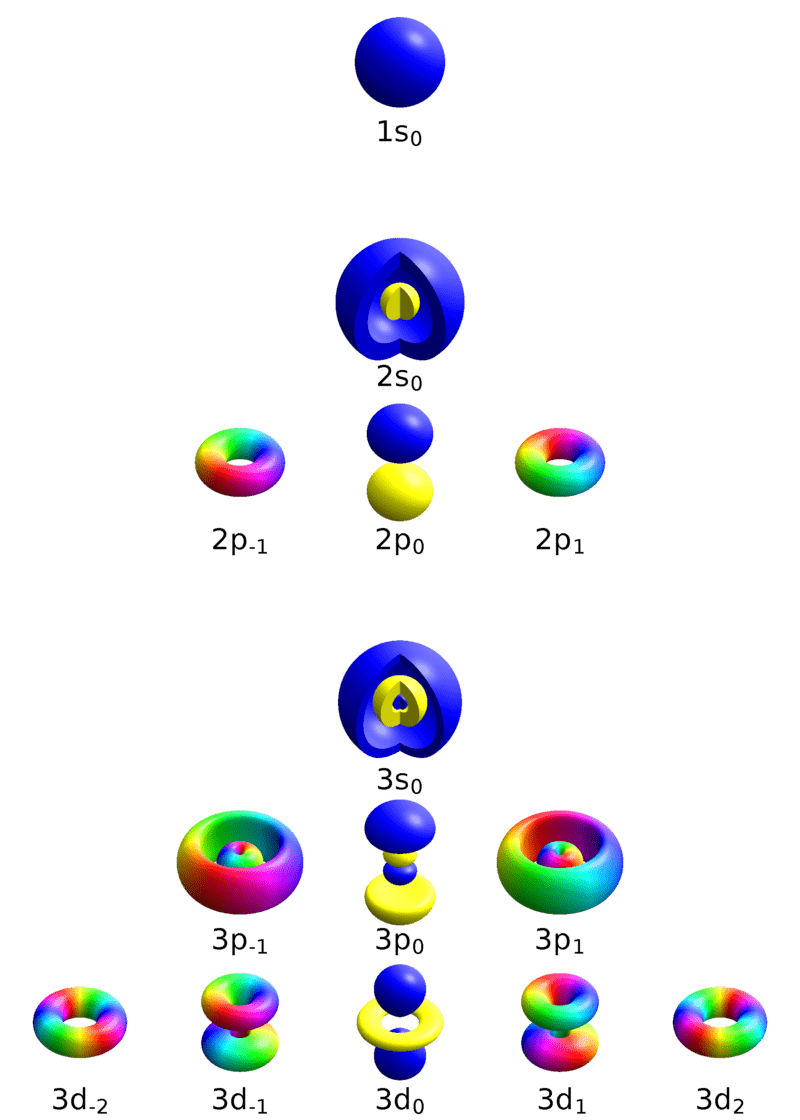
1
2
3
Orbital quantum number
0
-1
0
1
-1
0
1
-2
2
Magnetic quantum number
1
1
2
3
2
1
(You don't need to know what these mean)
An electron at (n, l, m) will have a specific energy level and characteristics
Each atom contributes electrons to the molecule
Benzene has . . .
Six carbon atoms with 1s2 2s2 2p2
Six hydrogen atoms with 1s
located at the center of each atom's position












Electrons "mix" into molecular orbitals to a specific energy level
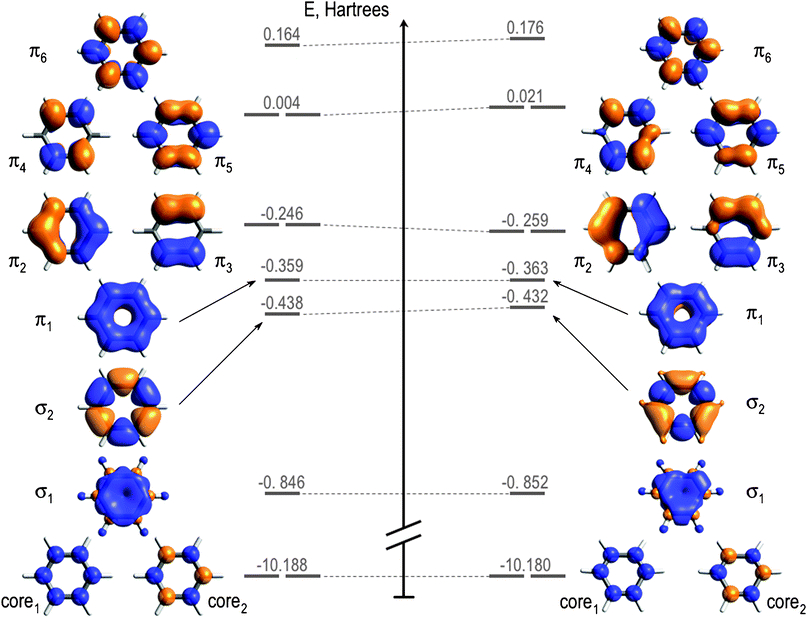
Particles (e.g., electrons and photons) can interact with these molecular orbials
D6h structure
D3h structure
These molecular orbitals determine behavior
Changing the positions (or symmetry) change molecular orbitals
Result: An electron density distribution unique to that structure
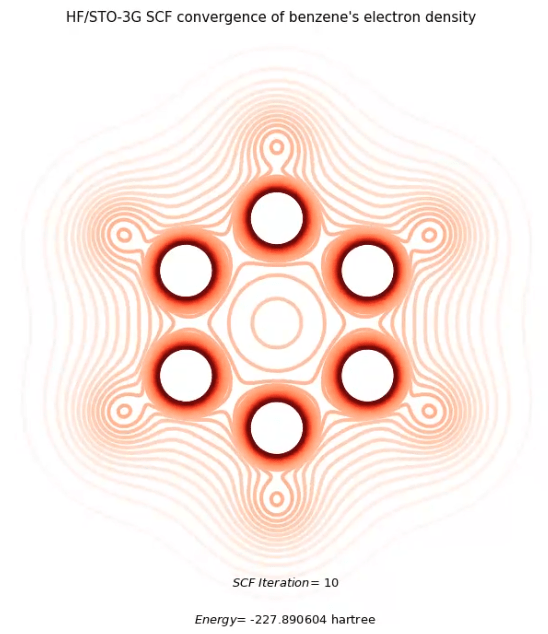
All experimental techniques are based on probes interacting with molecule's electron density to reveal structural information
- X-ray Crystallography: How a crystal of molecules diffracts X-rays
- NMR Spectroscopy: How atomic nuclei interact with magnetic fields and radiofrequency pulses
- Cryo-Electron Microscopy: How molecules scatter electron beams
Electron density of benzene
After today, you should have a better understanding of
Basic principles of X-ray crystallography and cryo-electron microscopy
UniProt is a protein information database
Let's find information about our project's drug target: Dihydrofolate reductase
UniProt is a comprehensive database to access curated data about protein structures, functions, sequences, and annotations.
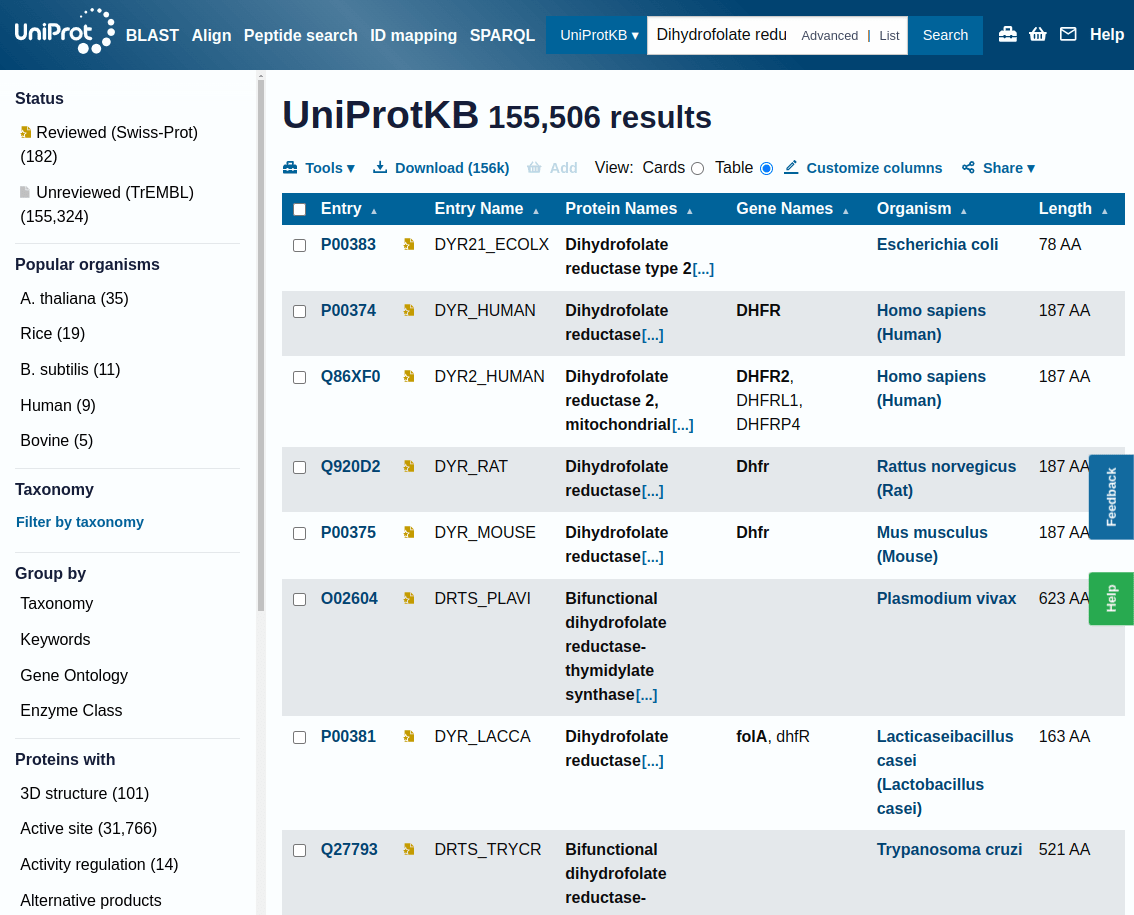
This page shows the results of a search in UniProtKB for a specific protein, in this case, "Dihydrofolate reductase"
On the left side, you have multiple filters to narrow your search results:
-
Reviewed (Swiss-Prot): Experts manually curated and verified these entries, ensuring high accuracy
-
Unreviewed (TrEMBL): These entries are automatically generated and have not been manually reviewed
Each row in the table represents a different protein entry
Entry ID: A unique identifier for the protein (e.g., P00383). You can click on this ID for detailed information about the protein
Protein Data Bank contains structures
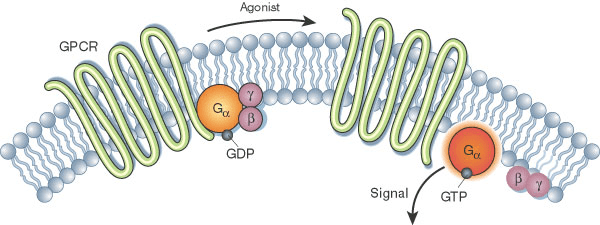
Conformational Heterogeneity and Biological Function
Many proteins function by switching between different conformations, which is essential for their activity (e.g., enzymes, transporters, and receptors).
- Example: G-protein coupled receptors (GPCRs) adopt different conformations when bound to different ligands, triggering different cellular responses.
After today, you should have a better understanding of
Types and biological significance of atomic interactions
Covalent
Two types of atomistic interactions
Covalent
Noncovalent
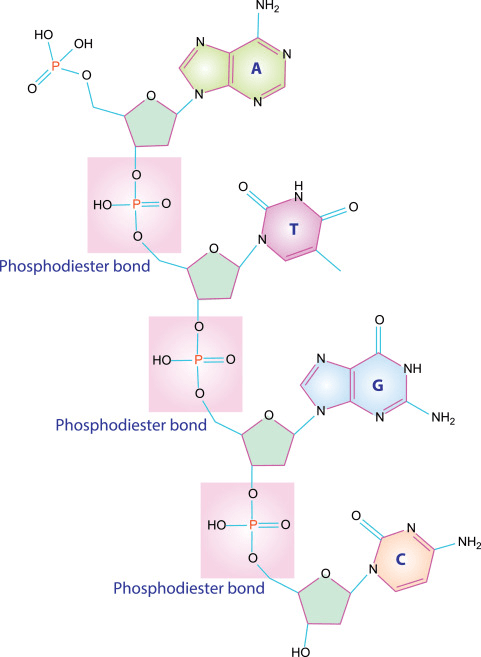
Covalent bonds: The framework of biomolecules
- Peptide bonds covalently link amino acids into polypeptide chains
- Phosphodiester bonds form the sugar-phosphate backbone of DNA and RNA
- Glycosidic bonds join monosaccharides to form complex sugars
Covalent bonds are formed when atoms share pairs of electrons that holds molecules together

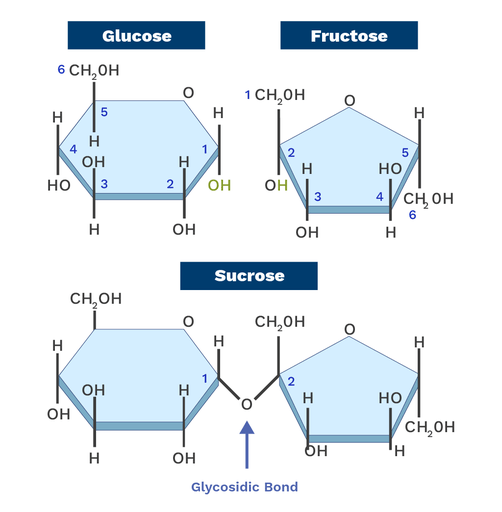
Relevant characteristics of covalent bonds
- Single Bonds: Allow rotation, contributing to molecular flexibility
- Double/Triple Bonds: Restrict rotation, affecting the rigidity and function of molecules
Strength and stability: Covalent bonds provide the necessary stability for complex biological structures
Directionality: Covalent bonds limit the specific angles and orientations leading to the 3D shapes of biomolecules
After today, you should have a better understanding of
Types and biological significance of atomic interactions
Noncovalent
Two types of atomistic interactions
Covalent
Noncovalent

Noncovalent Forces: The Dynamic Glue
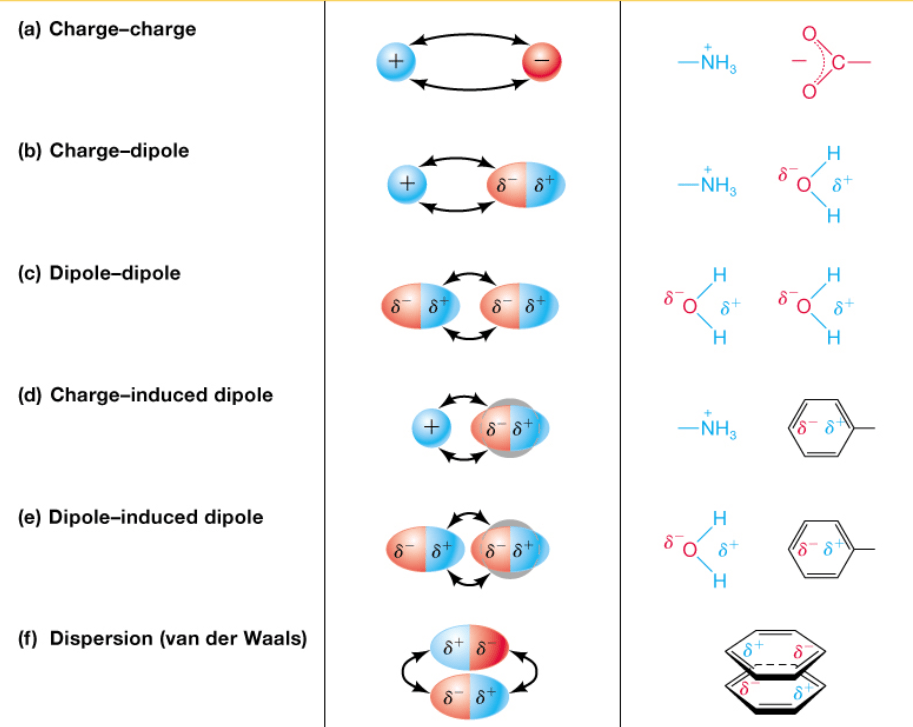

Noncovalent interactions are weaker than covalent bonds and involve electrostatics
We will cover this in L10
Noncovalent interactions drive most of biology
Macromolecular structure
-
Membrane Formation
-
Protein-Protein Interactions
- Base pairing in DNA and RNA
- Protein folding
Molecular recognition
- Enzyme-Substrate Binding
- Antigen-Antibody Interactions
BIOSC 1540: L09A (Structural biology)
By aalexmmaldonado
BIOSC 1540: L09A (Structural biology)
- 225



