Computational Biology
(BIOSC 1540)
Apr 1, 2025
Lecture 12A
Docking
Foundations
Announcements
Assignments
Quizzes
Final exam
OMETs
- If the response rate is 80% or higher, I will drop your lowest assignment
Drug targets
After today, you should have a better understanding of
Proteins
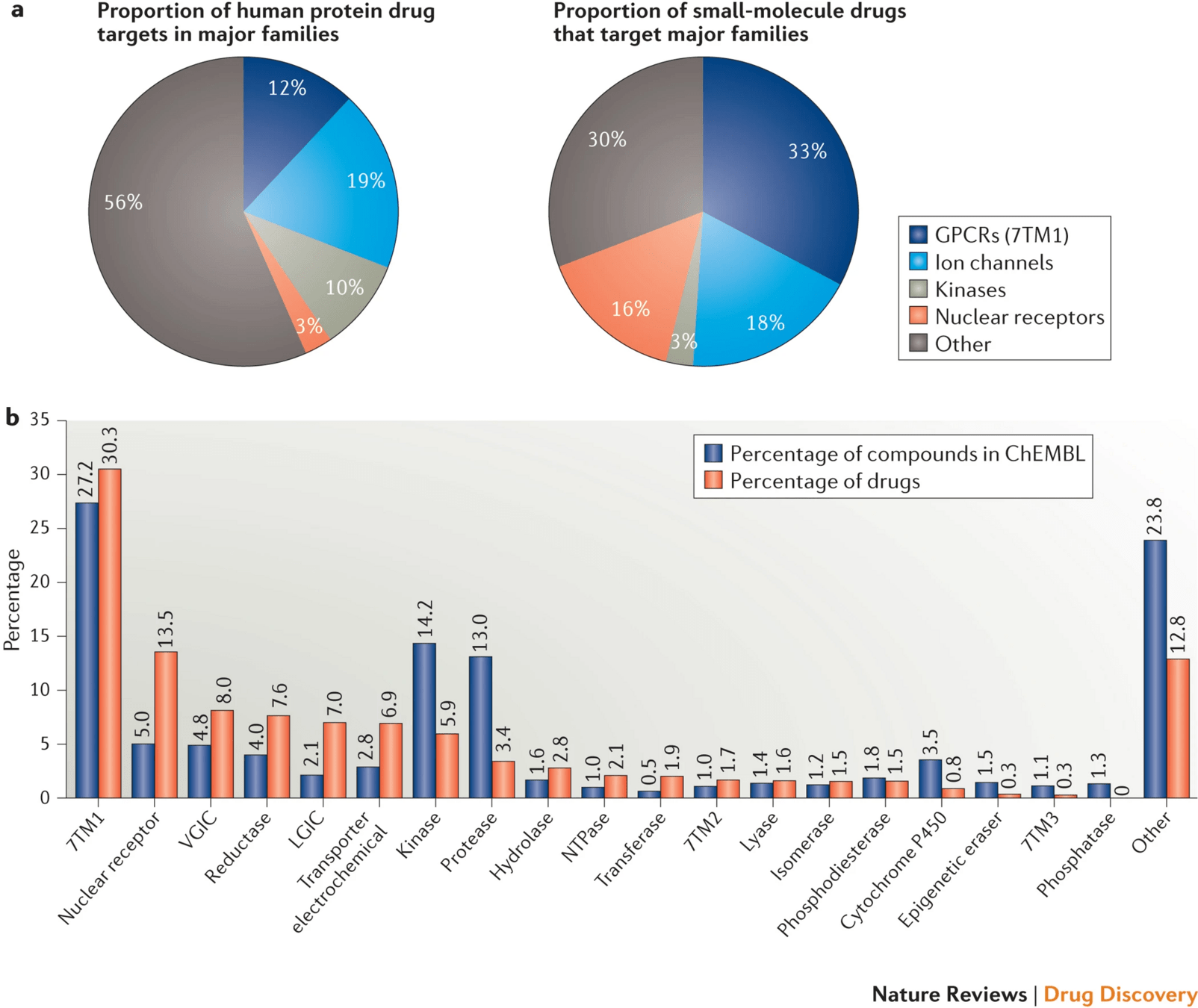
Proteins are the primary targets of most small-molecule drugs
Over 90% of FDA-approved drugs act on proteins—enzymes, receptors, ion channels, and transporters.
These proteins play key roles in signaling, metabolism, immune response, and other vital functions.
Modulating protein activity with small molecules allows us to influence biological pathways precisely.
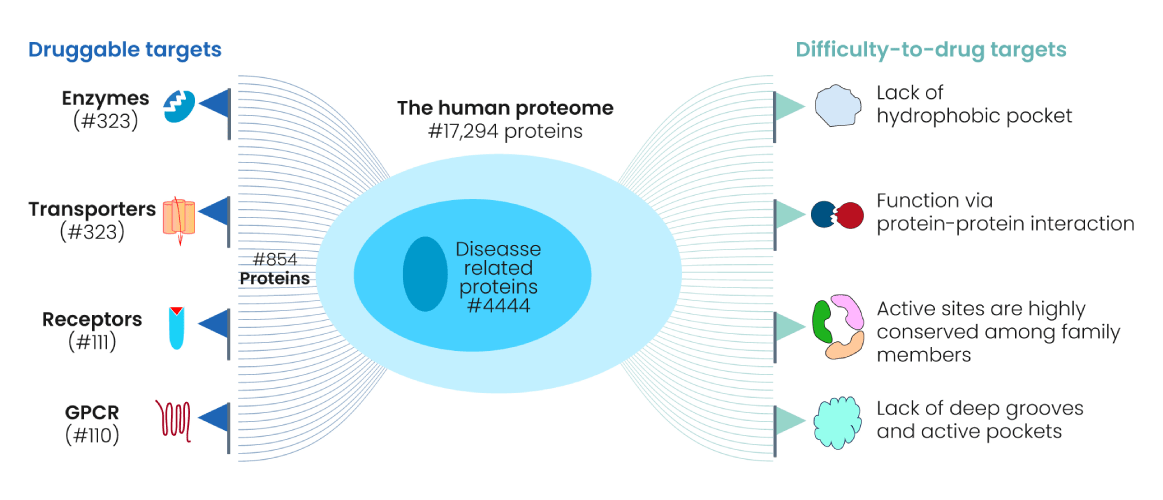
A protein is a good drug target when it is causally linked to disease
Not every protein is "druggable"—target selection must be biologically and chemically justified.
Genomics, proteomics, and phenotypic screens help identify candidate targets.
Criteria for Selecting a Protein Target
- Disease Relevance: The protein plays a critical role in the disease mechanism.
- Druggability: The target has a structure that allows it to bind with drug-like molecules.
- Specificity: Targeting the protein minimizes effects on healthy cells, reducing side effects.
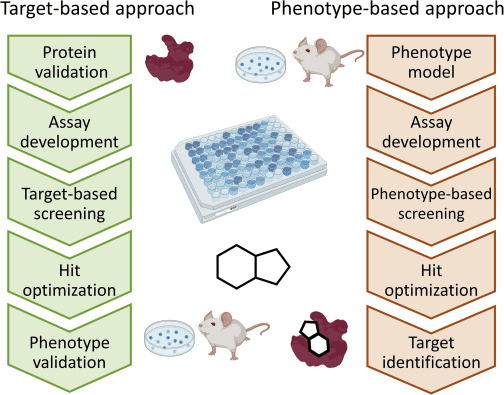
Target-centric drug discovery begins with the biology, not the molecule
In target-centric approaches, researchers start with a well-understood protein and search for molecules that modulate its activity.
This contrasts with phenotypic screening, which starts with observed effects and works backward to find the target.
Target-centric methods allow rational design, structure-based modeling, and docking campaigns.
Drug targets
After today, you should have a better understanding of
Bruton’s tyrosine kinase (BTK)
Identifying the right protein target is crucial for developing effective and safe drugs
Proteins regulate nearly all cellular processes and drugs can inhibit or activate proteins to correct disease states
Example: Bruton’s tyrosine kinase (BTK) is a critical signaling enzyme that causes XLA, a genetic disorder marked by a severe lack of mature B cells that leads to immunodeficiency.

Family-based genotyping of affected males revealed tight linkage between the disease and genetic markers.
Positional and sequence-based analyses identified BTK as the XLA gene
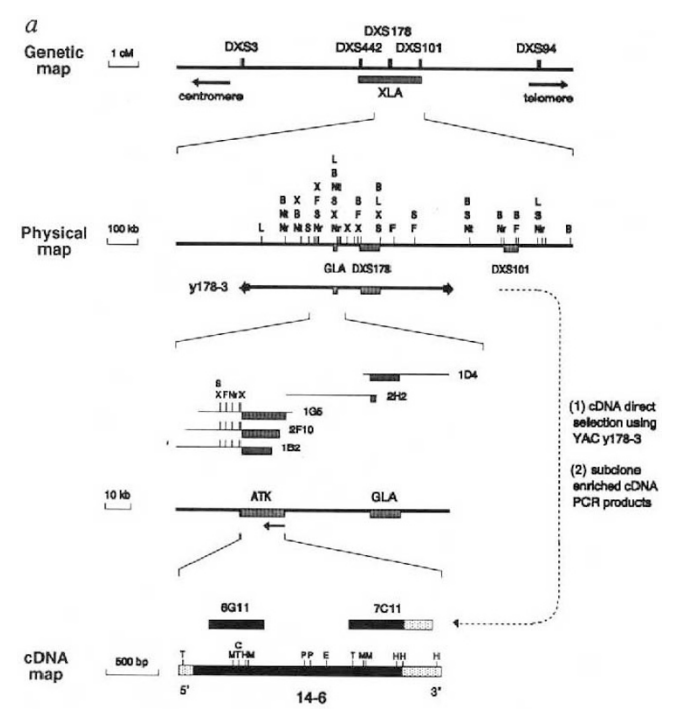
Bioinformatic comparison of the novel gene’s sequence to known kinases identified conserved domains with non-receptor tyrosine kinases.
In silico analysis of XLA patient sequences showed missense mutations affecting critical residues (e.g., Lys430 in the ATP-binding site).
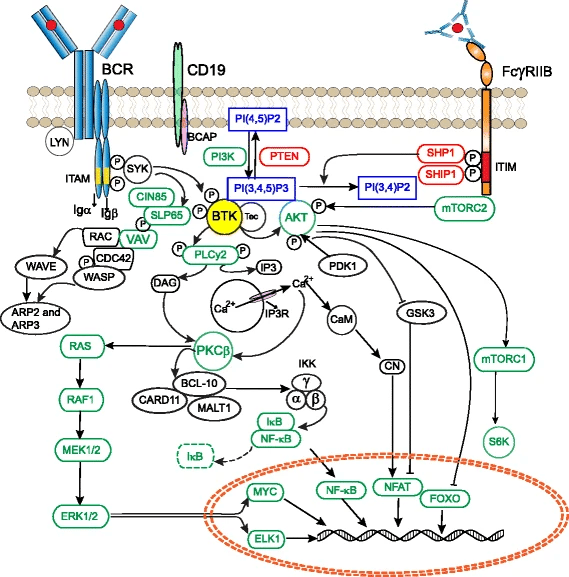
Tyrosine phosphorylation datasets and known B-cell signaling proteins placed BTK at a convergence point of multiple BCR-related pathways
Network-based analyses revealed BTK as a central hub in BCR signaling
Comparative genomics shows high conservation of BTK’s domains across vertebrates that are overrepresented among central signaling hubs.
Dihydrofolate Reductase (DHFR)
Drug targets
After today, you should have a better understanding of
THF production is crucial for cellular growth
THF is needed for
- Producing red blood cells,
- Synthesizing purines,
- Interconverting amino acids,
- Methylating tRNA,
- Generating and using formate.
Disrupting THF production has a cascading effect on essential cellular processes, primarily affecting DNA and RNA synthesis and amino acid metabolism
This is a useful process for drug design
DHFR is responsible for synthesizing THF
Dihydrofolate reductase (DHFR) is a crucial enzyme that produces THF from dihydrofolate (DHF)
DHF + NADPH
THF + NADP(+)
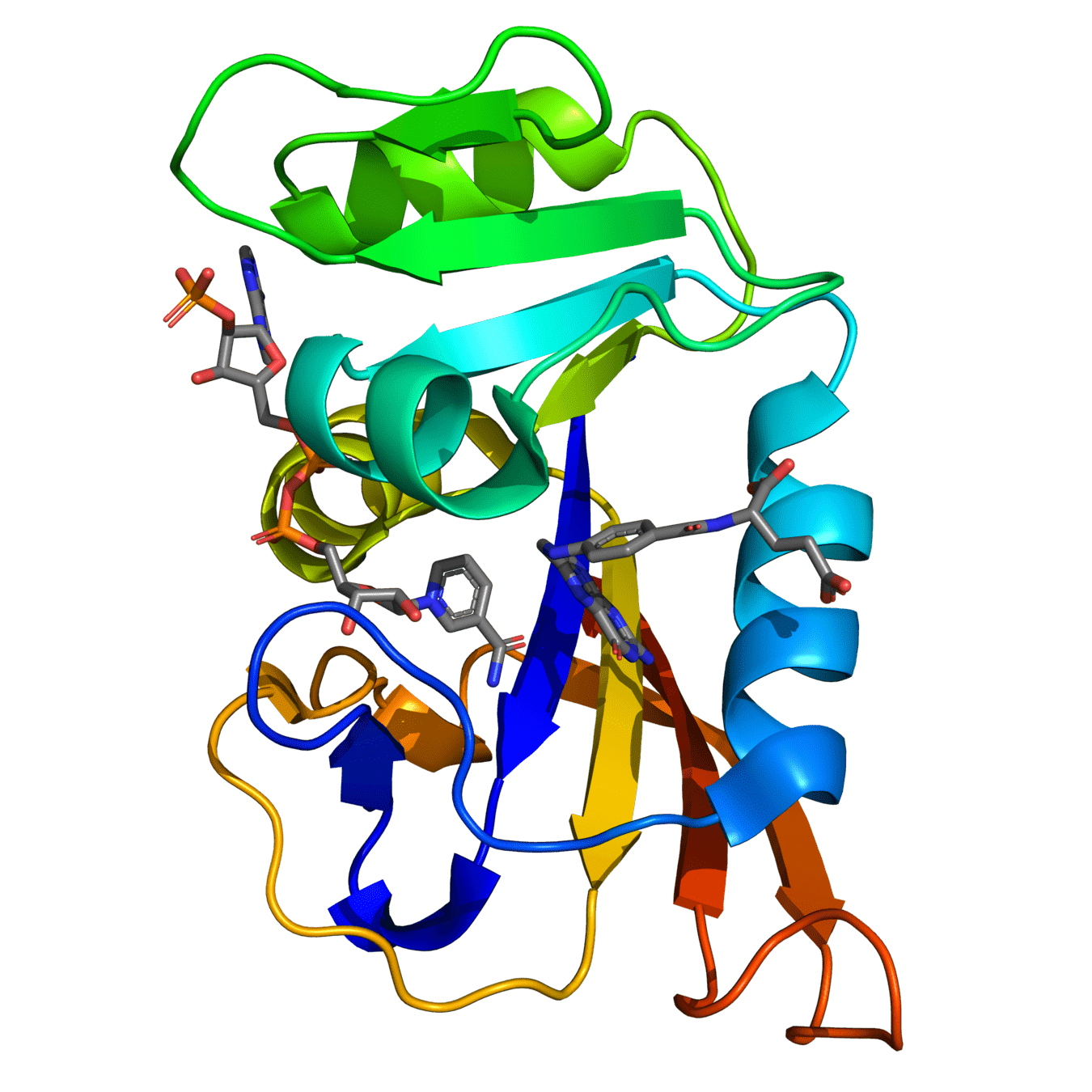
DHF
NADPH
(We will use this protein for our project)
DHFR has been extensively studied as an antibiotic (e.g., trimethoprim) and cancer (e.g., methotrexate) target
DHFR conservation complicates drug design
Patient could have deleterious side effects
What would happen if a patient with a bacterial infection is prescribed a drug loosely targeting DHFR?
Both proteins have high structural similarity, even around the active site
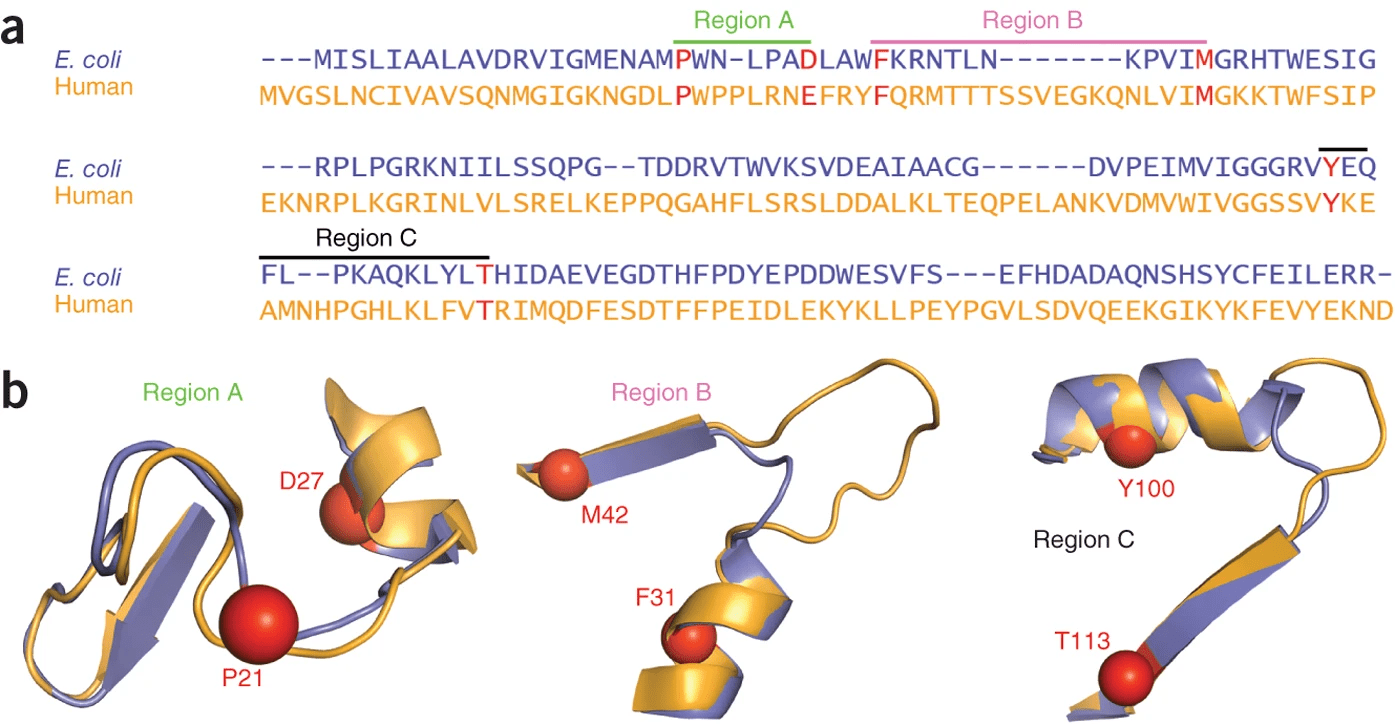
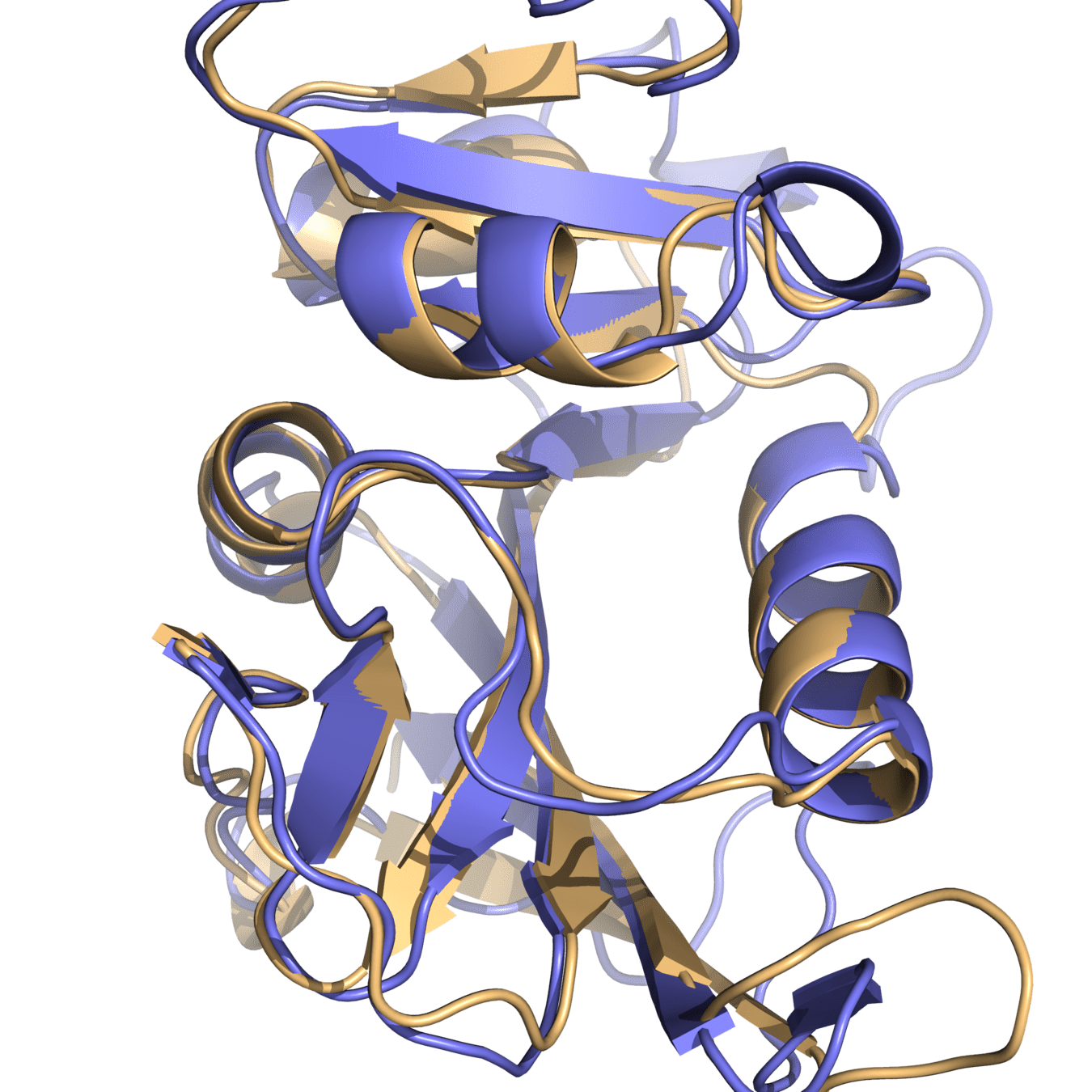
DHFR conservation complicates drug design

Bacteria and humans have similar structures, but their dynamics are different

Outcome: We need to ensure drugs only bind to bacterial proteins by exploiting dynamic insights
Structure-based drug design
After today, you should have a better understanding of
We can modulate protein's function by binding small molecules to specific sites
A protein’s activity can be altered by binding a small molecule—often called a ligand—to a functional site on its surface.

This interaction can inhibit, activate, or subtly reshape the protein’s behavior.
These small molecules act like “molecular switches” that control protein action without altering the underlying gene.
Binding affinity and specificity determine a molecule’s effectiveness as a modulator
Not every small molecule that binds a protein is useful—effective modulation depends on how tightly and selectively it binds.
High-affinity binding ensures that a drug is effective at low doses, while specificity minimizes off-target effects.
These properties can often be optimized through structure-based design and screening campaigns.
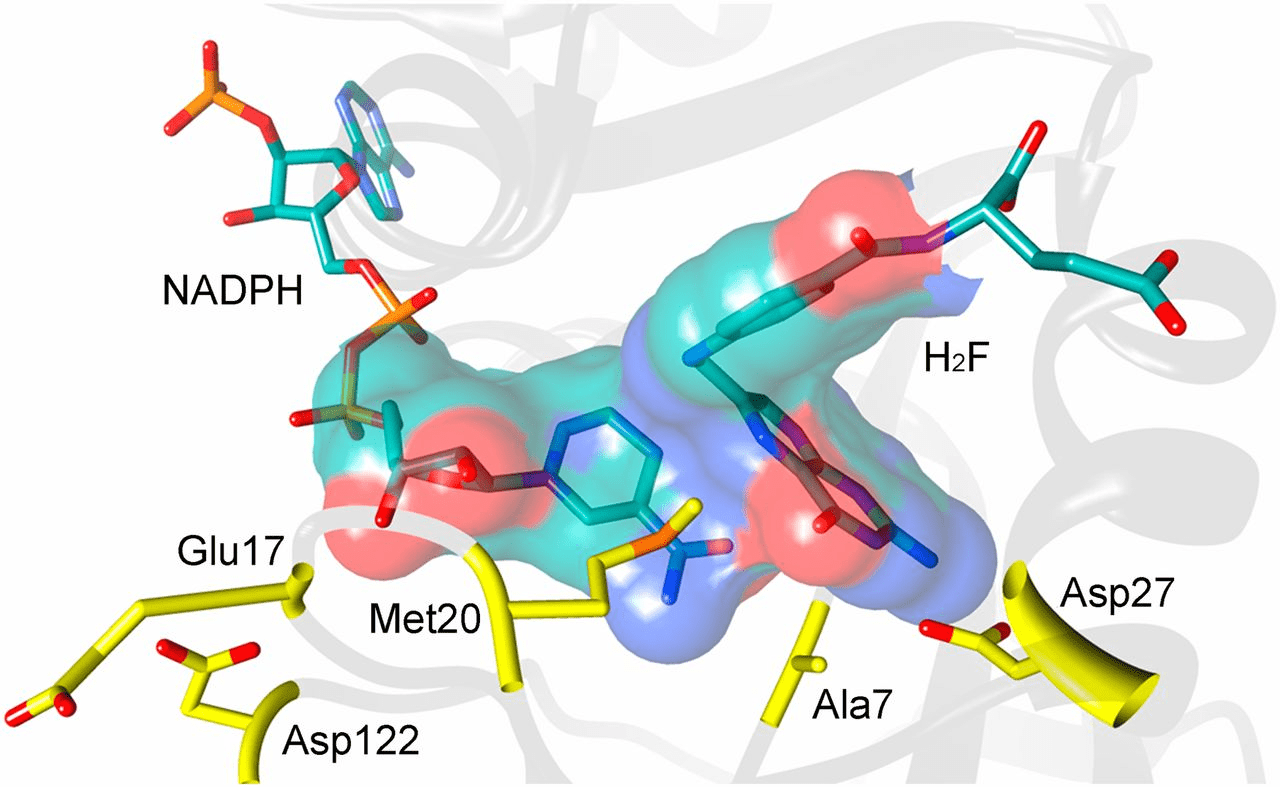
Critical ligand interactions in DHFR
Chemical space contains an astronomical number of possible compounds to explore
Estimated to be between 1060 to 10200 possible small organic molecules
We need methods to navigate chemical space and identify promising leads accurately and efficiently
High-throughput screening (HTS) allows testing of thousands of compounds against the target protein
- Library Preparation: Collection of diverse compounds
- Assay Development: Design of biological assays to measure compound activity against the target
- Screening: Compounds are tested in miniaturized assays
- Data Analysis: Identification of "hits" that show desired activity
Virtual screening evaluates vast libraries to identify potential leads efficiently
Experimental assays are still expensive, and limited to commercially available compounds
Instead, we can use computational methods to predict which compounds we should experimental validate
Can screen millions to billions of compounds in silico, thereby dramatically expanding our search space

Thermodynamics of binding
After today, you should have a better understanding of
Selective binding to a protein is governed by thermodynamics (and kinetics)
Binding occurs when a compound/ligand interacts specifically with a protein
Protein
Ligand
Binding
Protein-
ligand
We can model this as a reversible protein-ligand binding
Gibbs free energy combines enthalpy and entropy
Entropy
Enthalpy
Accounts for energetic interactions
How much conformational flexibility changes
By predicting the free energy of binding, we can identify small molecules with high affinity to our drug target
Accurate and efficient binding predictions are essential
Objective: Directly predict binding affinity from protein and ligand structures with high accuracy and minimal computational resources.
We can carefully simplify our modeling to improve speed with (hopefully) minimal impact to accuracy
Avoid sampling all microstates and determine one "optimal" protein-ligand structure
Using this bound structure, predict a "score" that is correlated to binding affinity
This is called docking
Docking simplifies the binding free energy prediction problem to enhance speed
Identifying relevant protein conformations
After today, you should have a better understanding of
Accurate, reproducible docking requires a relevant protein conformation
Significance of Protein Conformation in Docking
- Protein-ligand interactions are highly dependent on the protein's 3D structure.
- Using an rare protein conformation can lead to inaccurate docking results.
Docking still considers the protein structure, but we only select one
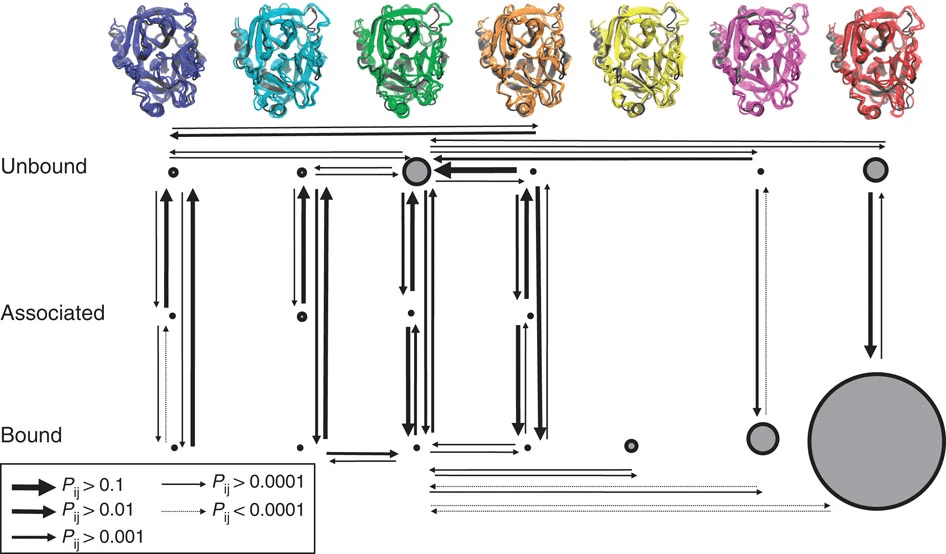
Sources of Protein Conformational Data
Experimental Methods
- Molecular Dynamics (MD) Simulations: Explore the conformational space over time.
- Normal Mode Analysis (NMA): Identifies collective motions in proteins.
- Ensemble Generation Methods: Generate multiple protein conformations for docking.
X-ray Crystallography: Provides high-resolution structures but may miss dynamic conformations.
Computational Techniques
NMR Spectroscopy: Captures ensembles of conformations but is limited to smaller proteins.
Will discuss these in L14
Experimental Structure Selection Criteria
-
Resolution and Quality
- Prefer structures with higher resolution (e.g., <2.5 Å).
- Assess reliability using R-factors and validation reports.
-
Ligand-Bound vs. Apo Structures
-
Ligand-Bound (Holo) Structures
- Provide direct insight into binding site conformation.
-
Apo Structures
- May reveal binding site flexibility in the absence of ligands.
-
Ligand-Bound (Holo) Structures
-
Relevance to Target Ligand
- Choose structures co-crystallized with ligands similar to those of interest.
Importance of Water Molecules
Role in Binding: Structured water molecules can mediate interactions between the protein and ligand.
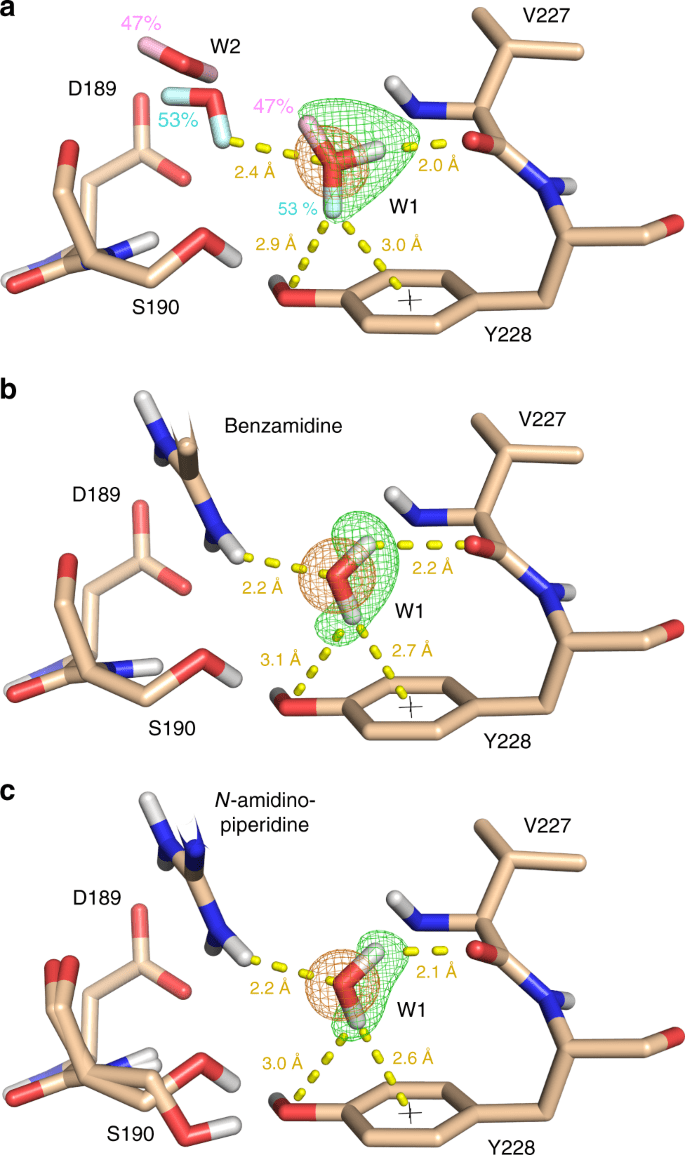
Handling Water in Docking
- Some docking programs allow explicit water molecules in the binding site.
- Alternatively, consider their effect implicitly in scoring functions.
Inclusion Criteria: Retain water molecules that are conserved across multiple crystal structures.
Binding pockets
After today, you should have a better understanding of
Types
Accurate Binding Pocket Detection is Crucial for Docking
The binding pocket is the specific region where a ligand interacts with a protein
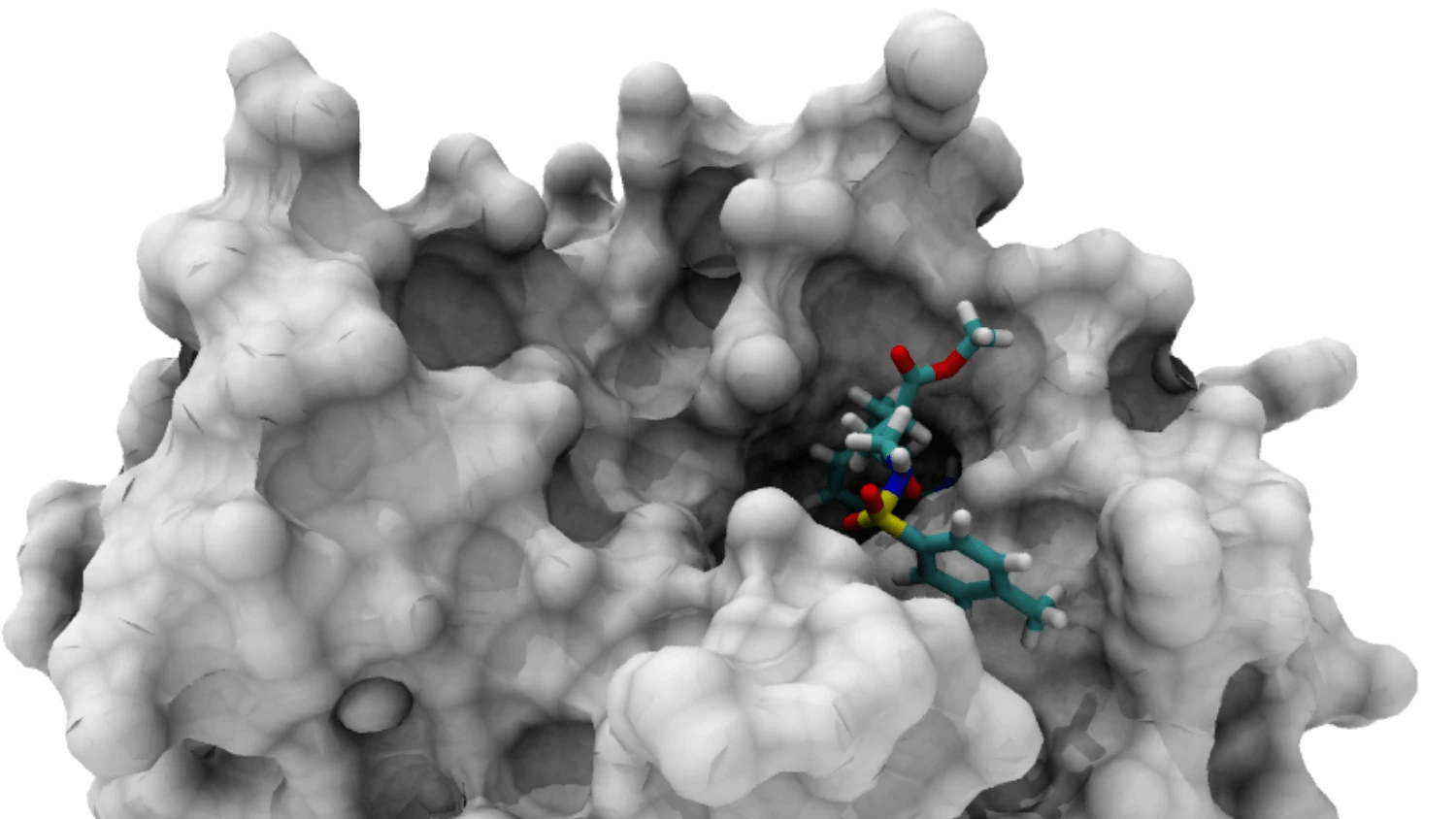
Accurate identification of binding pockets is essential for successful docking and virtual screening.
Understanding Protein Surface Topography
Active Site: The functional region where biochemical reactions occur (often a binding pocket in enzymes).
Protein Surface Characteristics
- Convex Regions: Typically inaccessible to ligands.
- Concave Regions (Cavities): Potential binding sites.
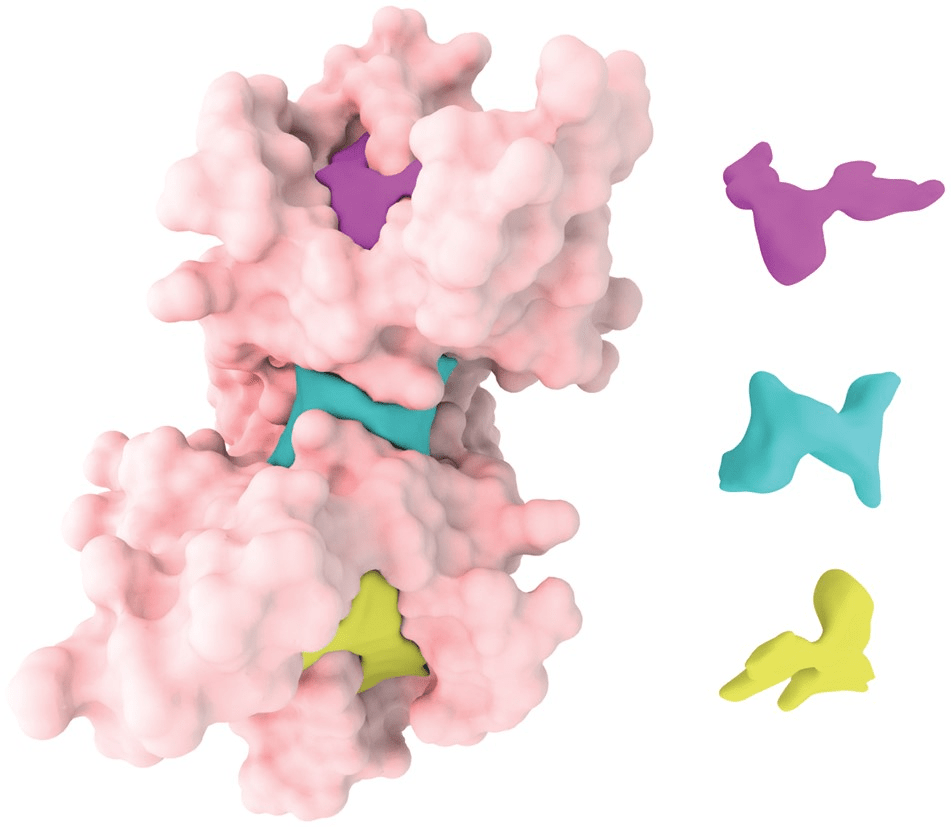
Binding Pocket: A cavity that can accommodate a ligand.
Binding pockets are classified by location, accessibility, and regulatory function
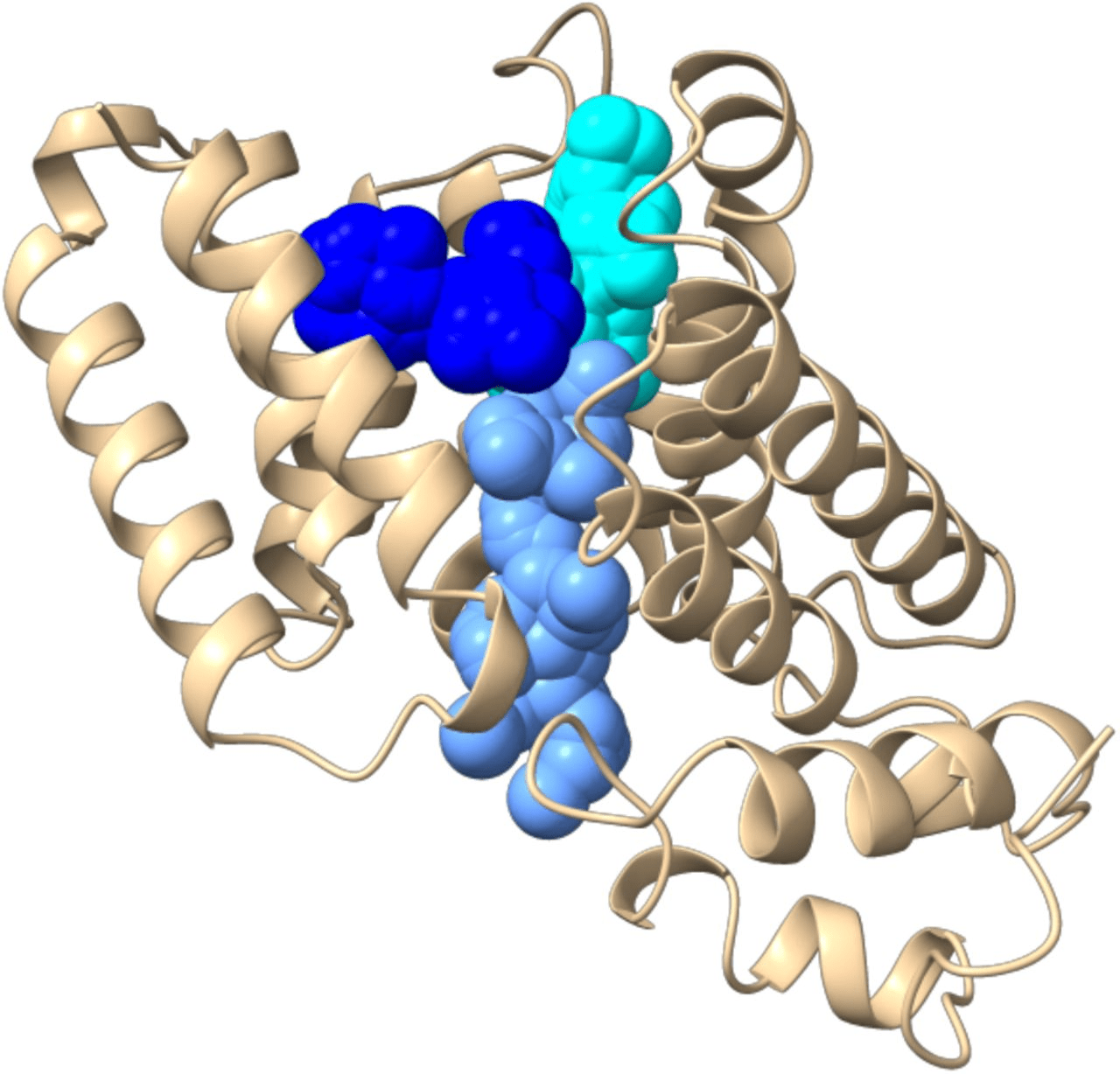
Orthosteric sites are the natural binding sites of endogenous ligands or substrates
Cryptic Sites: Binding pockets not apparent in the unbound protein structure but form upon ligand binding or conformational change.
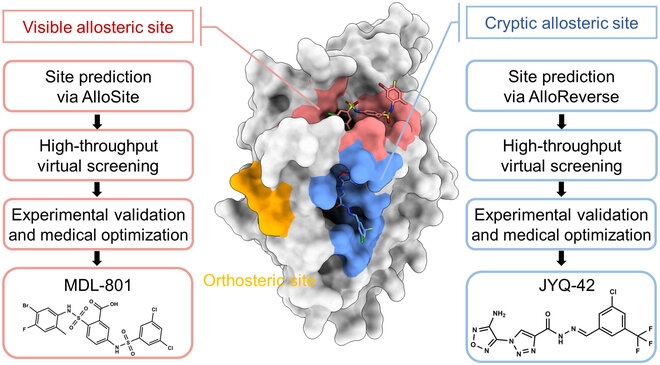
Allosteric sites are spatially distinct from the orthosteric site and modulate protein activity indirectly.
Binding pockets
After today, you should have a better understanding of
Detection
Alpha shapes detect pockets by reconstructing molecular surface topology
Alpha shapes extend the idea of a convex hull to capture the "shape" of a protein surface with cavities and tunnels.
Think of shrinking a sphere around atom centers—small alpha values allow more detailed surface features to be resolved.
Cavities enclosed by the alpha shape can be interpreted as binding pockets.
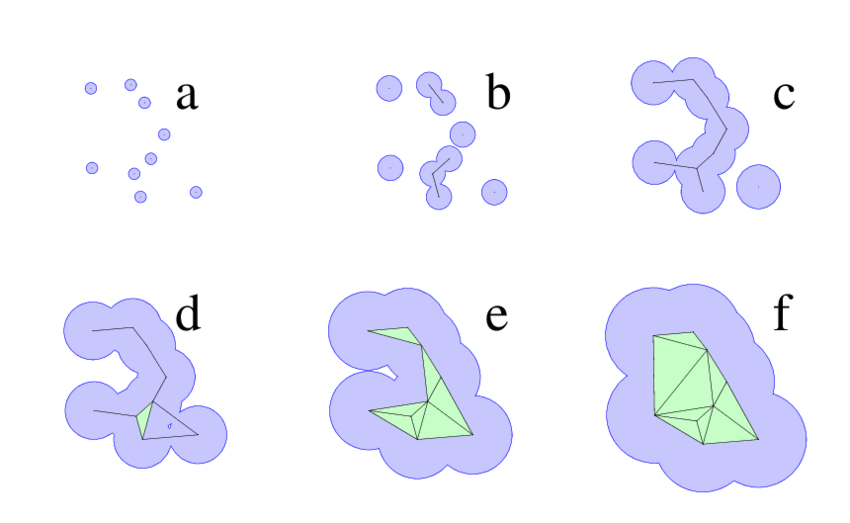
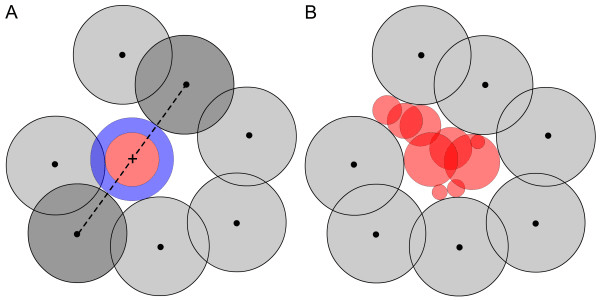
Alpha shape–based methods reveal pockets as topological voids
Proteins are modeled as a union of spheres (atoms), and the alpha shape filters through those to reveal cavities.
The algorithm identifies pocket volume, enclosure, and surface accessibility—critical for ligand fit.
These pockets are purely geometry-based, independent of electrostatics or residue type.
Grid-based methods map the protein onto a 3D lattice to identify cavities
The protein is embedded in a 3D grid, and each voxel is labeled as protein, solvent, or cavity.
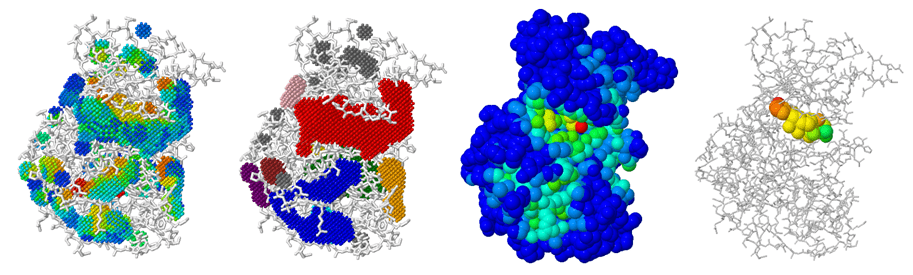
Grid points near the surface but not occupied by protein atoms are clustered into potential pockets.
Detecting Cryptic Binding Sites
Cryptic sites are hidden in the unbound structure and require conformational changes to become apparent.
Strategies
- Use enhanced sampling MD methods like metadynamics.
- Apply pocket detection to multiple conformations.
Case Study: Identification of allosteric sites in Hsp90
Scoring functions
After today, you should have a better understanding of
Scoring functions estimate how well a ligand binds to a protein
Scoring functions assign a numerical value to a protein-ligand pose, reflecting its favorability.
Scoring functions attempt to approximate this balance using simplified models.
The best pose maximizes favorable interactions (e.g., hydrogen bonds, hydrophobic packing) and minimizes clashes or strain.
A good scoring function may rank correctly even if absolute energies are wrong.
Force-field-based scoring calculates interactions from first principles
They consider van der Waals attraction/repulsion, electrostatics, and sometimes torsional strain.

Advances in scoring functions are driven by data and AI
Machine learning models (e.g., RF-Score, DeepDock) are trained directly on binding data.
These models learn nonlinear relationships and use structural and chemical features.
Require large, high-quality datasets and careful validation.
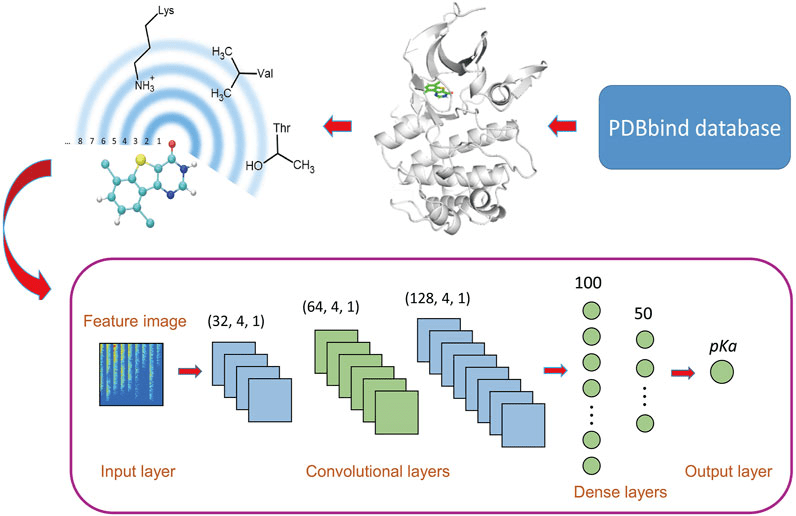
Ligand pose optimization
After today, you should have a better understanding of
Accurate Docking Depends on Optimized Ligand Poses

Docking needs to generate diverse conformations
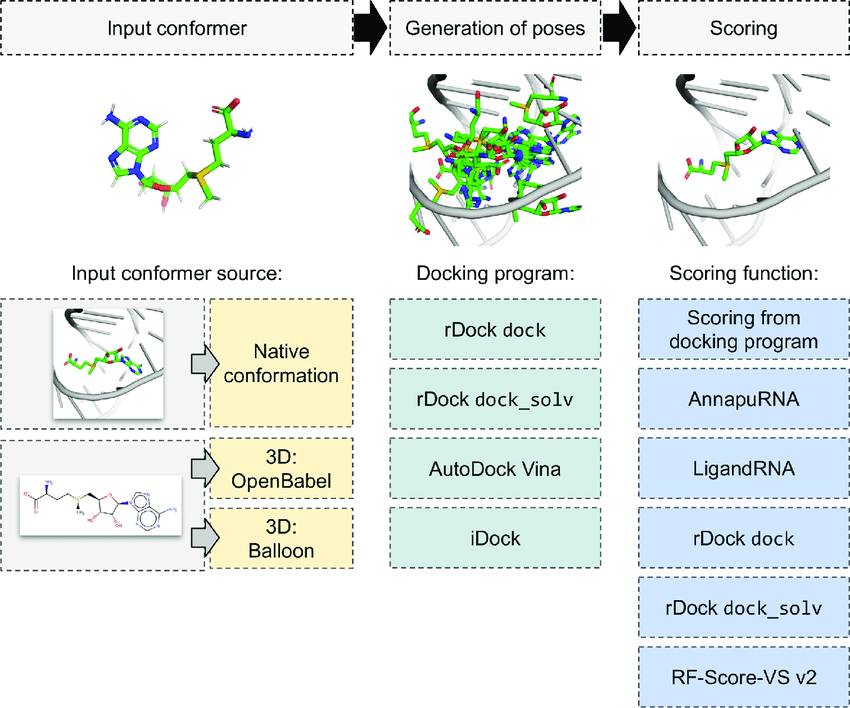
Search strategies
- Systematic
- Stochastic
- Empirical
- Machine learning
Systematic searches numerically iterate over all possible conformations

Identify important degrees of freedom
- Angles
- Dihedrals
Scan along each angle with a step size of a N degrees
Remove structures with high strain
Systematic searches are only possible for very small molecules
How many different conformations would we have in this molecule if we scanned only dihedrals every 45 degrees?

Systematic searches are only possible for very small molecules
8 dihedrals

1
2
3
4
5
6
7
8
8 angles
8 × 8 × 8 × 8 × 8 × 8 × 8 × 8 = 16,777,216
That's a lot of structures, and many of them will clash!
We almost never do a systematic search in practice without some precautions to combinatorics
Stochastic algorithms provide better balance of sampling and cost
Monte Carlo
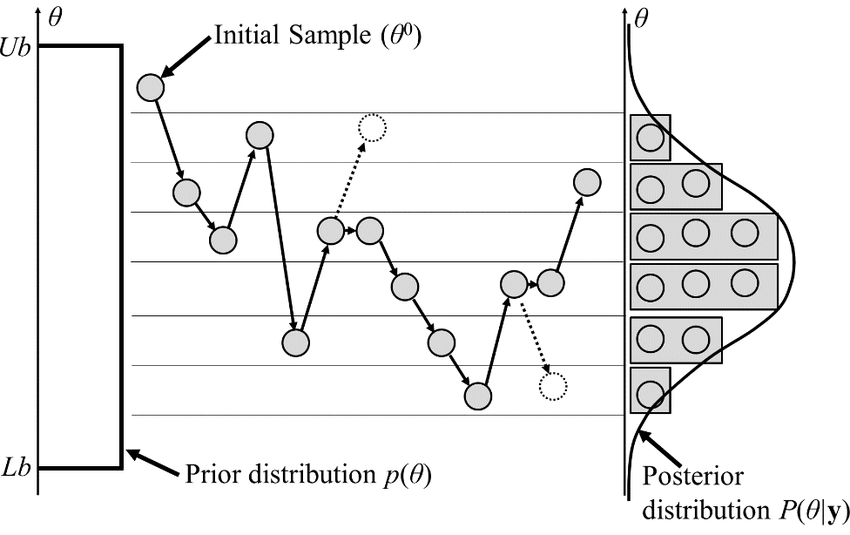
Steps:
- Generate conformation
- Compute energy change
- If energy change less than a random sample: make move
- Repeat
Allows us to sample efficiently
We also have conformer libraries
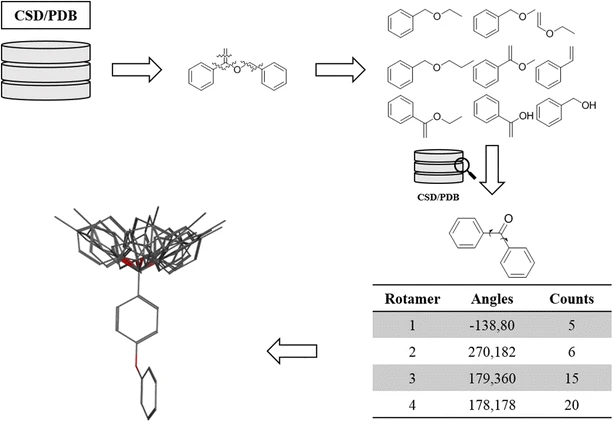
Use pre-generated libraries
Before the next class, you should
Lecture 12B:
Docking -
Methodology
Lecture 12A:
Docking -
Foundations
Today
Thursday
BIOSC 1540: L12A (Docking)
By aalexmmaldonado
BIOSC 1540: L12A (Docking)
- 183