Predictive Medicine Using Interpretable Recurrent Neural Networks

IST Master's Thesis
Student:
André Ferreira
Supervisors:
Alexandra Carvalho
Susana Vinga
22/10/2020
Predictive Medicine Using Interpretable Recurrent Neural Networks

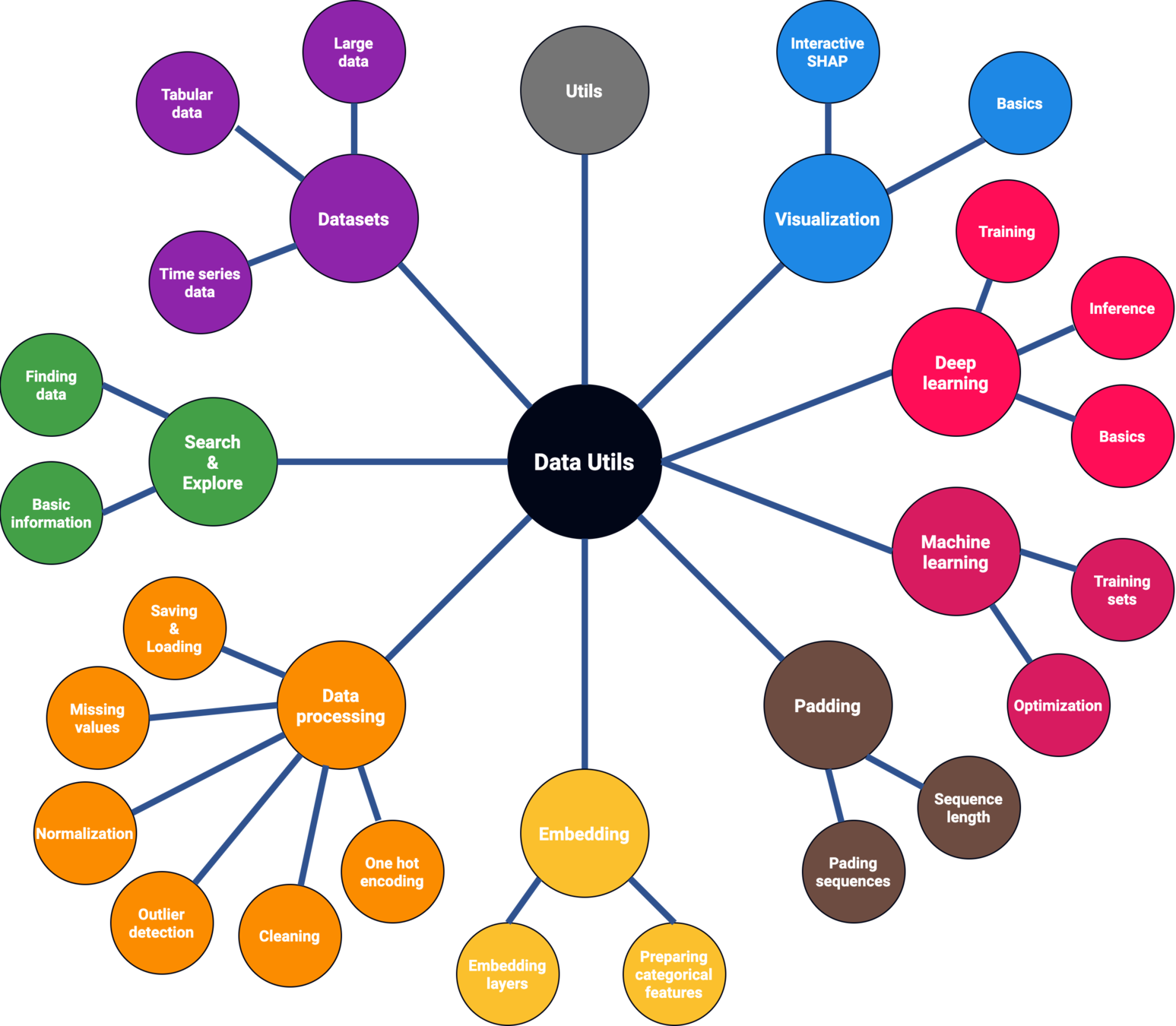
Motivation
Deep learning has disrupted several fields with state-of-the-art results and new possibilities.
The performance and interpretability tradeoff


Motivation
Deep learning has disrupted several fields with state-of-the-art results and new possibilities.
The performance and interpretability tradeoff
However, these models' complexity makes them harder to interpret.
Lack of trust in AI in critical use cases, including healthcare.
Predictive Medicine Using Interpretable Recurrent Neural Networks

Motivation
Predictive Medicine Using Interpretable Recurrent Neural Networks

Future of healthcare
Healthcare could benefit from AI.
9.9 million
Forecasted global shortage of physicians, nurses and midwives in 2030.
Data

Dataset Report
Diagnostic:
ALS (Amyotrophic lateral sclerosis).
Label:
Use of non-invasive ventilation (NIV) over the next 90 days.
#Patients:
840
#Features:
46
Information:
Demographics
Medical history
Onset evaluation
Genetic
Functional scores
Neurophysiological tests
How can we facilitate AI in healthcare?
Already taken care of


How can we facilitate AI in healthcare?
Already taken care of

The core pillars

Already taken care of

The core pillars
How can we facilitate AI in healthcare?
Already taken care of

The core pillars
But first, a bit of context...
But first, a bit of context...
Related work
📄
Background
Recurrent Neural Networks (RNN) are particularly good at handling sequential data.
In each timestamp , a cell:
Recurrent Neural Networks
But first, a bit of context...
Related work
📄
Background
Recurrent Neural Networks (RNN) are particularly good at handling sequential data.
Recurrent Neural Networks
— Updates the memory with the current input;
— Calculates an output ;
— Sends the memory to the next identical cell.


In each timestamp , a cell:
But first, a bit of context...
Related work
📄
Background
Recurrent Neural Networks
However, RNNs have some problems, specially in long sequences, when we backpropagate the error.
But first, a bit of context...
Related work
📄
Background
Recurrent Neural Networks
If
The gradient vanishes to 0.
But first, a bit of context...
Related work
📄
Background
Recurrent Neural Networks
Else, if
The gradient explodes to infinity.
But first, a bit of context...
Related work
📄
Background
LSTM

Long Short-Term Memory (LSTM) reduce the risks of gradient problems, by adding a "gradient highway".
Pointwise operations, instead of vector multiplications, avoid repeated scaling.
But first, a bit of context...
Related work
📄
Background
Model variations
LSTMs and other RNN-based models can have variations.
For example, a model can be bidirectional.
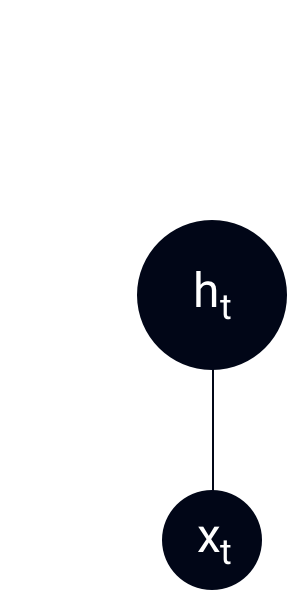
Model
ht
ht
But first, a bit of context...
Related work
📄
Background
Model variations
LSTMs and other RNN-based models can have variations.
Models can also include embedding layers, to preprocess categorical features.
Model
ht
ht
xt
Embedding layer
Numerical features
Categorical features
But first, a bit of context...
Related work
📄
Background
Time-aware models
Vanilla LSTMs still miss a piece of relevant information for time series:
Elapsed time between samples
x0
x1
x2
x3
Δt0
Δt1
Δt2
time
But first, a bit of context...
Related work
📄
Background
Time-aware models
Let's define time-aware models as those that take elapsed time between samples into consideration.
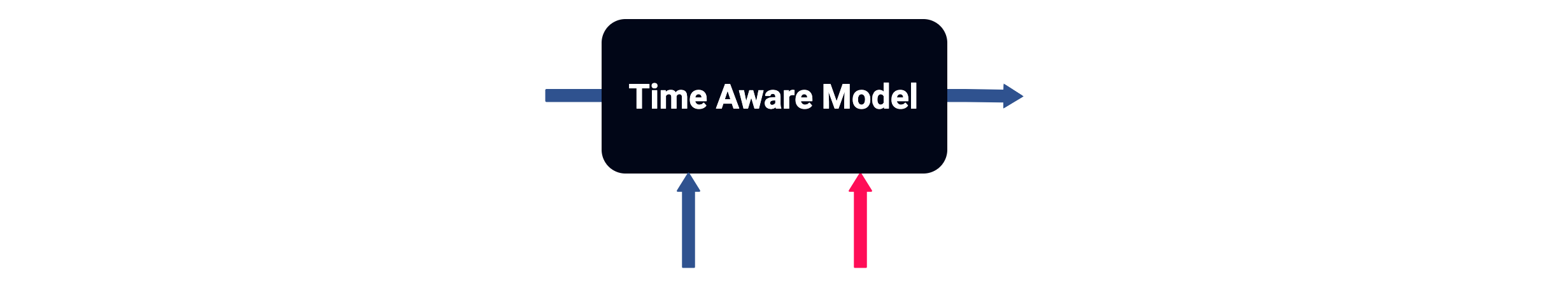
But first, a bit of context...
Related work
📄
Background
Time-aware models
In this thesis, we consider 4 possible solutions:
1˚: Adding elapsed time as a feature;
xt
Usual features
Δt
But first, a bit of context...
Related work
📄
Background
Time-aware models
In this thesis, we consider 4 possible solutions:
1˚: Adding elapsed time as a feature;
2˚: Using a MF1-LSTM cell;

Model from the "Patient Subtyping via Time-Aware LSTM Networks" paper, by I. M. Baytas et. al
But first, a bit of context...
Related work
📄
Background
Time-aware models
In this thesis, we consider 4 possible solutions:
1˚: Adding elapsed time as a feature;
2˚: Using a MF1-LSTM cell;
3˚: Using a MF2-LSTM cell;

Model from the "Patient Subtyping via Time-Aware LSTM Networks" paper, by I. M. Baytas et. al
But first, a bit of context...
Related work
📄
Background
Time-aware models
In this thesis, we consider 4 possible solutions:
1˚: Adding elapsed time as a feature;
2˚: Using a MF1-LSTM cell;
3˚: Using a MF2-LSTM cell;
4˚: Using a T-LSTM cell.

Model from the "Patient Subtyping via Time-Aware LSTM Networks" paper, by I. M. Baytas et. al
But first, a bit of context...
Related work
📄
Background
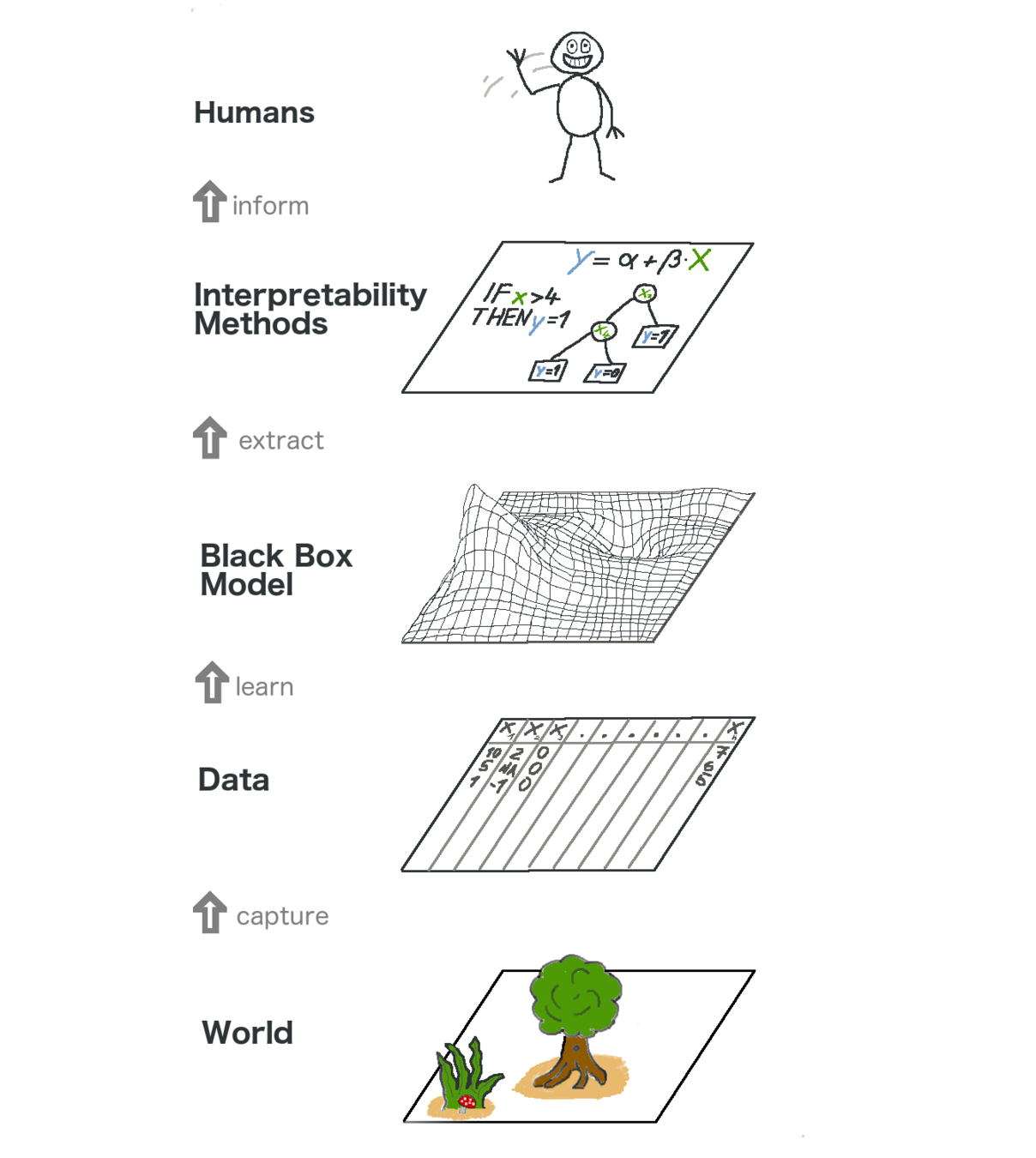
Interpretability
Model interpretability is highly desirable, mainly in critical use cases.
Along this thesis, we define interpretability as:
Interpretability is the degree to which a human can understand the cause of a decision.
Definition extracted from Tim Miller's "Explanation in artificial intelligence: Insights from the social sciences."
But first, a bit of context...
Related work
📄
Background
Interpretability
An easy solution to interpret models is to use ones that are simple to understand.
Such as decision trees...
But first, a bit of context...
Related work
📄
Background
Interpretability
Such as decision trees...
< 0.5
≥ 0.5
True
False
≥ 0.7
< 0.7
feat 1
feat 2
feat 3
No
No
Yes
Yes
But first, a bit of context...
Related work
📄
Background
Interpretability
...or linear regression.
feat 1
feat 2
feat 3
But first, a bit of context...
Related work
📄
Background
Interpretability
More complex models, such as artificial neural networks, can also be used to learn simpler models from their features (mimic learning).
feat 1
feat 2
feat 3
feat 2
feat 3
No
Yes
< 0.5
≥ 0.5
feat 1
True
False
≥ 0.7
< 0.7
No
Yes
feat 1
feat 2
feat 3
But first, a bit of context...
Related work
📄
Background
Interpretability
feat 1
feat 2
feat 3
0.25
0.75
0.5
x
x
x
Attention weights
Artificial neural networks can also be built to be more easily interpretable, such as through attention weights.
But first, a bit of context...
Related work
📄
Background
Interpretability
Despite the intuitiveness of these approaches, they have some issues:
— Constraining the model architecture, to simpler types or with specific components, can limit the performance;
— Attention weights give an incomplete interpretation.
But first, a bit of context...
Related work
📄
Background
Interpretability
An alternative is to use perturbation-based methods.
In this case, the input is modified so as to infer how each part of it affects the output.
No model-type specificities are required.
But first, a bit of context...
Related work
📄
Background
Interpretability — SHAP
SHAP is an interpretability package developed by Scott Lundberg et. al, based on Shapley values, a game theory concept from the 50s.
It unifies several other perturbation-based techniques into the same core logic:
Learn a simpler interpreter model from the original one, through a local linear model.
But first, a bit of context...
Related work
📄
Background
Interpretability — SHAP
Learn a simpler interpreter model from the original one, through a local linear model.
Interpreter model
Nb features
Average output
SHAP value of feature i
(i.e. its value represents the feature's contribution to the output)
But first, a bit of context...
Related work
📄
Background
Interpretability — SHAP
Different feature representation
Learn a simpler interpreter model from the original one, through a local linear model.
But first, a bit of context...
Related work
📄
Background
Interpretability — SHAP
The interpreter model does a local approximation of the original one, around the input but in a representation of (when it corresponds to ) or .
This space constitutes binary values that indicate whether or not a feature is "present".
(i.e. if we are using the original feature value or a background sample that represents the removal of that feature)
0.43
-0.75
0.48
1
1
1
0.56
-0.75
0.16
0
1
0
But first, a bit of context...
Related work
📄
Background
Interpretability — SHAP
Thanks to its Shapley foundations and its core equation, SHAP verifies several desirable properties.
A specially important one is that of local accuracy:
When approximating the original model for a specific input x, local accuracy requires the explanation model to at least match the output of for the original input x:
But first, a bit of context...
Related work
📄
Background
Interpretability — SHAP
Thanks to its Shapley foundations and its core equation, SHAP verifies several desirable properties.
The equation that defines SHAP values according to these properties is the following:
Present features
SHAP value of feature i
Original model
Input
Nb features
Model output on z'
Model output on z' without feature i
Coalition weighting
Calculating the impact of feature i on the output from sample z'
But first, a bit of context...
Related work
📄
Background
Interpretability — SHAP
While SHAP has several possible estimators, the most relevant here is Kernel SHAP, which is model-agnostic.
It trains the local interpreter model with the following loss function:
Square error between the original model and the interpreter
Sample Weighting
Converts z' to z
But first, a bit of context...
Related work
📄
Background
Interpretability — SHAP
This weighting scheme gives higher value to coalitions with a small or large number of features, giving an indication of the feature values’ isolated and global influence.
Now we can order coalition sizes by their weight and train the interpreter model on a subset of samples.
But first, a bit of context...
Related work
📄
Background
Interpretability — SHAP
But first, a bit of context...
Related work
📄
Background
Interpretability — SHAP

Not so fast, we'll see later on some imperfections.
But first, a bit of context...
The core pillars
The core pillars
Performance
Interpretability
🔍
Usability
🏥
Modular
model

Bidirectional

RNN

LSTM

MF1-LSTM

MF2-LSTM

T-LSTM

Embedding layer

Elapsed time as a feature
The core pillars
Performance
Interpretability
🔍
Usability
🏥

Bidirectional

RNN

LSTM

MF1-LSTM

MF2-LSTM

T-LSTM

Embedding layer

Elapsed time as a feature
The core pillars
Performance
Interpretability
🔍
Usability
🏥




The models have been tested in a modular way:
Modular model
The core pillars
Performance
Interpretability
🔍
Usability
🏥




The models have been tested in a modular way:
Modular model
— 5 possible RNN cells: RNN; LSTM; MF1-LSTM; MF2-LSTM; T-LSTM.
The core pillars
Performance
Interpretability
🔍
Usability
🏥



The models have been tested in a modular way:
Modular model
— 5 possible RNN cells: RNN; LSTM; MF1-LSTM; MF2-LSTM; T-LSTM.

— Possibility of being bidirectional.
The core pillars
Performance
Interpretability
🔍
Usability
🏥


The models have been tested in a modular way:
Modular model
— 5 possible RNN cells: RNN; LSTM; MF1-LSTM; MF2-LSTM; T-LSTM.

— Possibility of being bidirectional.

— Possibility of having an embedding layer.
The core pillars
Performance
Interpretability
🔍
Usability
🏥

The models have been tested in a modular way:
Modular model
— 5 possible RNN cells: RNN; LSTM; MF1-LSTM; MF2-LSTM; T-LSTM.

— Possibility of being bidirectional.

— Possibility of having an embedding layer.

— Possibility of using elapsed time as a feature.
(time-awareness)
The core pillars
Performance
Interpretability
🔍
Usability
🏥
The models have been tested in a modular way:
Modular model
— 5 possible RNN cells: RNN; LSTM; MF1-LSTM; MF2-LSTM; T-LSTM.

— Possibility of being bidirectional.

— Possibility of having an embedding layer.

— Possibility of using elapsed time as a feature.
(time-awareness)

The core pillars
Performance
Interpretability
🔍
Usability
🏥
Results
| Model | Avg. Test AUC | Std. Test AUC |
|---|---|---|
| Bidir. LSTM, Δt | 0.937405 | 0.025812 |
| Bidir. LSTM, embed | 0.927344 | 0.025885 |
| Bidir. LSTM | 0.916037 | 0.016473 |
| Bidir. LSTM, embed, Δt | 0.915068 | 0.021216 |
| Bidir. RNN, embed, Δt | 0.896575 | 0.022189 |
| Bidir. RNN | 0.888364 | 0.025372 |
| Bidir. RNN, embed | 0.887089 | 0.028001 |
| Bidir. RNN, Δt | 0.884163 | 0.024578 |
| XGBoost | 0.833373 | 0.035517 |
| LSTM, embed, Δt | 0.822787 | 0.035039 |
| RNN, Δt | 0.798767 | 0.030467 |
| RNN | 0.797243 | 0.014516 |
| LSTM, Δt | 0.795249 | 0.033168 |
| LSTM | 0.793016 | 0.022835 |
| RNN, embed | 0.787666 | 0.025880 |
| LSTM, embedded | 0.785448 | 0.021874 |
| Logistic Regression | 0.781677 | 0.003030 |
| RNN, embed, Δt | 0.777421 | 0.024405 |
| MF1-LSTM | 0.675305 | 0.027665 |
| MF2-LSTM | 0.668578 | 0.023623 |
| MF2-LSTM, embed | 0.653309 | 0.017099 |
| T-LSTM | 0.649320 | 0.023343 |
| T-LSTM, embed | 0.649291 | 0.016514 |
| MF1-LSTM, embed | 0.648051 | 0.008728 |
Models were tested over 3 different random seeds.
The core pillars
Performance
Interpretability
🔍
Usability
🏥
Results
| Model | Avg. Test AUC | Std. Test AUC |
|---|---|---|
| Bidir. LSTM, Δt | 0.937405 | 0.025812 |
| Bidir. LSTM, embed | 0.927344 | 0.025885 |
| Bidir. LSTM | 0.916037 | 0.016473 |
| Bidir. LSTM, embed, Δt | 0.915068 | 0.021216 |
| Bidir. RNN, embed, Δt | 0.896575 | 0.022189 |
| Bidir. RNN | 0.888364 | 0.025372 |
| Bidir. RNN, embed | 0.887089 | 0.028001 |
| Bidir. RNN, Δt | 0.884163 | 0.024578 |
| XGBoost | 0.833373 | 0.035517 |
| LSTM, embed, Δt | 0.822787 | 0.035039 |
| RNN, Δt | 0.798767 | 0.030467 |
| RNN | 0.797243 | 0.014516 |
| LSTM, Δt | 0.795249 | 0.033168 |
| LSTM | 0.793016 | 0.022835 |
| RNN, embed | 0.787666 | 0.025880 |
| LSTM, embedded | 0.785448 | 0.021874 |
| Logistic Regression | 0.781677 | 0.003030 |
| RNN, embed, Δt | 0.777421 | 0.024405 |
| MF1-LSTM | 0.675305 | 0.027665 |
| MF2-LSTM | 0.668578 | 0.023623 |
| MF2-LSTM, embed | 0.653309 | 0.017099 |
| T-LSTM | 0.649320 | 0.023343 |
| T-LSTM, embed | 0.649291 | 0.016514 |
| MF1-LSTM, embed | 0.648051 | 0.008728 |
Models were tested over 3 different random seeds.
Bidirectional LSTM models lead the performance ranking.
The core pillars
Performance
Interpretability
🔍
Usability
🏥
Results
| Model | Avg. Test AUC | Std. Test AUC |
|---|---|---|
| Bidir. LSTM, Δt | 0.937405 | 0.025812 |
| Bidir. LSTM, embed | 0.927344 | 0.025885 |
| Bidir. LSTM | 0.916037 | 0.016473 |
| Bidir. LSTM, embed, Δt | 0.915068 | 0.021216 |
| Bidir. RNN, embed, Δt | 0.896575 | 0.022189 |
| Bidir. RNN | 0.888364 | 0.025372 |
| Bidir. RNN, embed | 0.887089 | 0.028001 |
| Bidir. RNN, Δt | 0.884163 | 0.024578 |
| XGBoost | 0.833373 | 0.035517 |
| LSTM, embed, Δt | 0.822787 | 0.035039 |
| RNN, Δt | 0.798767 | 0.030467 |
| RNN | 0.797243 | 0.014516 |
| LSTM, Δt | 0.795249 | 0.033168 |
| LSTM | 0.793016 | 0.022835 |
| RNN, embed | 0.787666 | 0.025880 |
| LSTM, embedded | 0.785448 | 0.021874 |
| Logistic Regression | 0.781677 | 0.003030 |
| RNN, embed, Δt | 0.777421 | 0.024405 |
| MF1-LSTM | 0.675305 | 0.027665 |
| MF2-LSTM | 0.668578 | 0.023623 |
| MF2-LSTM, embed | 0.653309 | 0.017099 |
| T-LSTM | 0.649320 | 0.023343 |
| T-LSTM, embed | 0.649291 | 0.016514 |
| MF1-LSTM, embed | 0.648051 | 0.008728 |
Models were tested over 3 different random seeds.
Bidirectional LSTM models lead the performance ranking.
XGBoost got comparable results.
The core pillars
Performance
Interpretability
🔍
Usability
🏥
Results
| Model | Avg. Test AUC | Std. Test AUC |
|---|---|---|
| Bidir. LSTM, Δt | 0.937405 | 0.025812 |
| Bidir. LSTM, embed | 0.927344 | 0.025885 |
| Bidir. LSTM | 0.916037 | 0.016473 |
| Bidir. LSTM, embed, Δt | 0.915068 | 0.021216 |
| Bidir. RNN, embed, Δt | 0.896575 | 0.022189 |
| Bidir. RNN | 0.888364 | 0.025372 |
| Bidir. RNN, embed | 0.887089 | 0.028001 |
| Bidir. RNN, Δt | 0.884163 | 0.024578 |
| XGBoost | 0.833373 | 0.035517 |
| LSTM, embed, Δt | 0.822787 | 0.035039 |
| RNN, Δt | 0.798767 | 0.030467 |
| RNN | 0.797243 | 0.014516 |
| LSTM, Δt | 0.795249 | 0.033168 |
| LSTM | 0.793016 | 0.022835 |
| RNN, embed | 0.787666 | 0.025880 |
| LSTM, embedded | 0.785448 | 0.021874 |
| Logistic Regression | 0.781677 | 0.003030 |
| RNN, embed, Δt | 0.777421 | 0.024405 |
| MF1-LSTM | 0.675305 | 0.027665 |
| MF2-LSTM | 0.668578 | 0.023623 |
| MF2-LSTM, embed | 0.653309 | 0.017099 |
| T-LSTM | 0.649320 | 0.023343 |
| T-LSTM, embed | 0.649291 | 0.016514 |
| MF1-LSTM, embed | 0.648051 | 0.008728 |
Models were tested over 3 different random seeds.
Bidirectional LSTM models lead the performance ranking.
XGBoost got comparable results.
Intrinsically time-aware models had the worst performance.
The core pillars
Performance
Interpretability
🔍
Usability
🏥
Component impact
Bidirectionality is the main contributor to performance gains.
LSTMs prove to be better than RNNs, but not by much.
Elapsed time as a feature, in this case, serves as a marginal improvement.
The embedding layer does not benefit the models, on average.
The core pillars
The core pillars
Interpretability
Performance
🚀
Usability
🏥
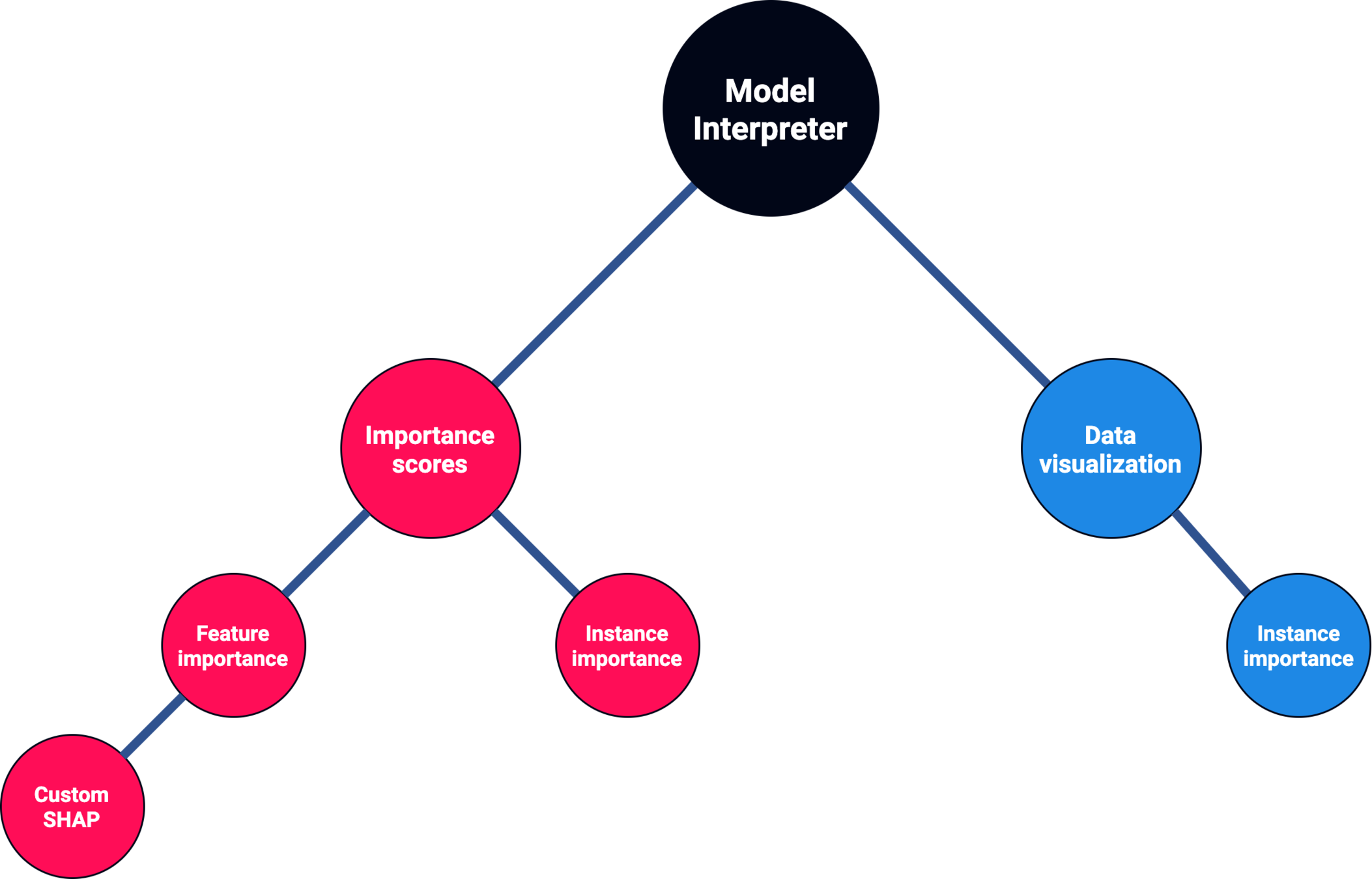
Feature importance
Kernel SHAP's lack of assumptions about the model, that makes it model-agnostic, stops it from working with RNN-based models.
If we apply Kernel SHAP to a RNN model, we see that the sum of SHAP values does not match the model's output:
| ts | real_output | shap_output |
|---|---|---|
| 0 | 0,4068 | 0,4068 |
| 1 | 0,3772 | 0,3848 |
| 2 | 0,3670 | 0,3976 |
| 3 | 0,5840 | 0,5943 |
| 4 | 0,5949 | 0,5851 |
Example from this notebook repository:
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
| ts | real_output | shap_output |
|---|---|---|
| 0 | 0,4068 | 0,4068 |
| 1 | 0,3772 | 0,3848 |
| 2 | 0,3670 | 0,3976 |
| 3 | 0,5840 | 0,5943 |
| 4 | 0,5949 | 0,5851 |
Example from this notebook repository:
It breaks the local accuracy property on RNN-based models.
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
| ts | real_output | shap_output |
|---|---|---|
| 0 | 0,4068 | 0,4068 |
| 1 | 0,3772 | 0,3848 |
| 2 | 0,3670 | 0,3976 |
| 3 | 0,5840 | 0,5943 |
| 4 | 0,5949 | 0,5851 |
Kernel SHAP always tries to explain each sample individually, separate from others.
It considers the samples as being separate sequences of one single instance, eliminating the use of the model’s memory.
Only the first sample of the sequence has matching outputs.
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
To fix this, I adapted Kernel SHAP's code with the following main changes:
— Added parameters that identified the model type, including when it was RNN;
from sklearn.impute import SimpleImputer
from ..common import convert_to_instance, convert_to_model, match_instance_to_data, match_model_to_data, convert_to_instance_with_index, convert_to_link, IdentityLink, convert_to_data, DenseData, SparseData
from scipy.special import binom
from scipy.sparse import issparse
import numpy as np
import pandas as pd
import scipy as sp
import logging
import copy
import itertools
import warnings
from sklearn.linear_model import LassoLarsIC, Lasso, lars_path
from sklearn.cluster import KMeans
from tqdm.auto import tqdm
from .explainer import Explainer
import torch
log = logging.getLogger('shap')
def kmeans(X, k, round_values=True):
""" Summarize a dataset with k mean samples weighted by the number of data points they
each represent.
Parameters
----------
X : numpy.array or pandas.DataFrame or any scipy.sparse matrix
Matrix of data samples to summarize (# samples x # features)
k : int
Number of means to use for approximation.
round_values : bool
For all i, round the ith dimension of each mean sample to match the nearest value
from X[:,i]. This ensures discrete features always get a valid value.
Returns
-------
DenseData object.
"""
group_names = [str(i) for i in range(X.shape[1])]
if str(type(X)).endswith("'pandas.core.frame.DataFrame'>"):
group_names = X.columns
X = X.values
# in case there are any missing values in data impute them
imp = SimpleImputer(missing_values=np.nan, strategy='mean')
X = imp.fit_transform(X)
kmeans = KMeans(n_clusters=k, random_state=0).fit(X)
if round_values:
for i in range(k):
for j in range(X.shape[1]):
xj = X[:,j].toarray().flatten() if issparse(X) else X[:, j]
ind = np.argmin(np.abs(xj - kmeans.cluster_centers_[i,j]))
kmeans.cluster_centers_[i,j] = X[ind,j]
return DenseData(kmeans.cluster_centers_, group_names, None, 1.0*np.bincount(kmeans.labels_))
class KernelExplainer(Explainer):
"""Uses the Kernel SHAP method to explain the output of any function.
Kernel SHAP is a method that uses a special weighted linear regression
to compute the importance of each feature. The computed importance values
are Shapley values from game theory and also coefficents from a local linear
regression.
Parameters
----------
model : function or iml.Model
User supplied function that takes a matrix of samples (# samples x # features) and
computes the output of the model for those samples. The output can be a vector
(# samples) or a matrix (# samples x # model outputs).
data : numpy.array or pandas.DataFrame or shap.common.DenseData or any scipy.sparse matrix
The background dataset to use for integrating out features. To determine the impact
of a feature, that feature is set to "missing" and the change in the model output
is observed. Since most models aren't designed to handle arbitrary missing data at test
time, we simulate "missing" by replacing the feature with the values it takes in the
background dataset. So if the background dataset is a simple sample of all zeros, then
we would approximate a feature being missing by setting it to zero. For small problems
this background dataset can be the whole training set, but for larger problems consider
using a single reference value or using the kmeans function to summarize the dataset.
Note: for sparse case we accept any sparse matrix but convert to lil format for
performance.
link : "identity" or "logit"
A generalized linear model link to connect the feature importance values to the model
output. Since the feature importance values, phi, sum up to the model output, it often makes
sense to connect them to the output with a link function where link(output) = sum(phi).
If the model output is a probability then the LogitLink link function makes the feature
importance values have log-odds units.
isRNN : bool
Boolean that indicates if the model being analyzed is a recurrent neural network (RNN).
If so, it means that sequential data is being used, which requires some modifications
in the way SHAP values are calculated.
if isRNN is True:
id_col_num : int
Number that indicates in which column is the sequence / subject id. Defaults to 0.
ts_col_num : int
Number that indicates in which column is the instance / timestamp. Defaults to 1.
label_col_num : int
Number that indicates in which column is the label, if any. Defaults to None.
recur_layer : torch.nn.LSTM or torch.nn.GRU or torch.nn.RNN, default None
Pointer to the recurrent layer in the model, if it exists. It should
either be a LSTM, GRU or RNN network. If none is specified, the
method will automatically search for a recurrent layer in the model.
isBidir: bool
Boolean that indicates if the recurrent neural network model being analyzed is
bidirectional. If so, it implies a special treatment of the sequences, as the
hidden states can't be moved in the same way of a single direction RNN.
padding_value : numeric
Value to use in the padding, to fill the sequences.
"""
def __init__(self, model, data, link=IdentityLink(), **kwargs):
# convert incoming inputs to standardized iml objects
self.link = convert_to_link(link)
self.model = convert_to_model(model)
self.keep_index = kwargs.get("keep_index", False)
self.keep_index_ordered = kwargs.get("keep_index_ordered", False)
# check if the model is a recurrent neural network
self.isRNN = kwargs.get('isRNN', False)
if self.isRNN and not str(type(data)).endswith("'pandas.core.frame.DataFrame'>"):
# check if the model is a bidirectional recurrent neural network
self.isBidir = kwargs.get('isBidir', False)
# number of the column that corresponds to the sequence / subject id
self.id_col_num = kwargs.get('id_col_num', 0)
# number of the column that corresponds to the instance / timestamp
self.ts_col_num = kwargs.get('ts_col_num', 1)
# number of the column that corresponds to the label
label_col_num = kwargs.get('label_col_num', None)
# padding value
self.padding_value = kwargs.get('padding_value', 999999)
# all columns in the data
self.model_features = list(range(data.shape[1]))
# remove unwanted columns, so that we get only those that actually correspond to model usable features
[self.model_features.remove(col) for col in [self.id_col_num, self.ts_col_num, label_col_num] if col is not None]
# maximum background samples to use
self.max_bkgnd_samples = kwargs.get('max_bkgnd_samples', 20)
if data.shape[0] > self.max_bkgnd_samples:
# use k-means to avoid slow processing of a lot of samples
self.data = kmeans(data[:, self.model_features], k=self.max_bkgnd_samples)
# get the weights corresponding to all the original data
num_samples = data.shape[0]
self.weights = np.ones(num_samples)
self.weights /= np.sum(self.weights)
else:
self.data = convert_to_data(data[:, self.model_features], keep_index=self.keep_index)
self.weights = self.data.weights
# check if the recurrent layer is specified
self.recur_layer = kwargs.get('recur_layer', None)
if self.recur_layer is None:
# get the model object, so as to use its recurrent layer
model_obj = kwargs.get('model_obj', None)
assert model_obj is not None, 'If the model uses a recurrent neural network, either the recurrent layer or the full model object must be specified.'
# search for a recurrent layer
if hasattr(model_obj, 'lstm'):
self.recur_layer = model_obj.lstm
elif hasattr(model_obj, 'gru'):
self.recur_layer = model_obj.gru
elif hasattr(model_obj, 'rnn'):
self.recur_layer = model_obj.rnn
else:
raise Exception('ERROR: No recurrent layer found. Please specify it in the recur_layer argument.')
# get the unique subject ID's in the background data
self.subject_ids = np.unique(data[:, self.id_col_num]).astype(int)
# maximum sequence length in the background data
self.max_seq_len = kwargs.get('max_seq_len', None)
if self.max_seq_len == None:
self.max_seq_len = 1
for id in self.subject_ids:
seq_data = data[np.where((data[:, self.id_col_num] == id))]
cur_seq_length = len(seq_data)
if cur_seq_length > self.max_seq_len:
self.max_seq_len = cur_seq_length
# calculate the output for all the background data
model_null = match_model_to_data(self.model, data, self.isRNN, self.model_features,
self.id_col_num, self.ts_col_num, self.recur_layer,
self.subject_ids, self.max_seq_len, self.model.f,
silent=kwargs.get("silent", False))
else:
self.data = convert_to_data(data, keep_index=self.keep_index)
self.weights = self.data.weights
# calculate the output for all the background data
model_null = match_model_to_data(self.model, self.data)
self.col_names = None
if str(type(data)).endswith("'pandas.core.frame.DataFrame'>"):
# keep the column names so that data can be used in dataframe format
self.col_names = data.columns
# enforce our current input type limitations
assert isinstance(self.data, DenseData) or isinstance(self.data, SparseData), \
"Shap explainer only supports the DenseData and SparseData input currently."
assert not self.data.transposed, "Shap explainer does not support transposed DenseData or SparseData currently."
# warn users about large background data sets
if len(self.data.weights) > 100:
log.warning("Using " + str(len(self.data.weights)) + " background data samples could cause " +
"slower run times. Consider using shap.sample(data, K) or shap.kmeans(data, K) to " +
"summarize the background as K samples.")
# init our parameters
self.N = self.data.data.shape[0]
self.P = self.data.data.shape[1]
self.linkfv = np.vectorize(self.link.f)
self.nsamplesAdded = 0
self.nsamplesRun = 0
# find E_x[f(x)]
if isinstance(model_null, (pd.DataFrame, pd.Series)):
model_null = np.squeeze(model_null.values)
self.fnull = np.sum((model_null.T * self.weights).T, 0)
self.expected_value = self.linkfv(self.fnull)
# see if we have a vector output
self.vector_out = True
if len(self.fnull.shape) == 0:
self.vector_out = False
self.fnull = np.array([self.fnull])
self.D = 1
self.expected_value = float(self.expected_value)
else:
self.D = self.fnull.shape[0]
def shap_values(self, X, **kwargs):
""" Estimate the SHAP values for a set of samples.
Parameters
----------
X : numpy.array or pandas.DataFrame or any scipy.sparse matrix
A matrix of samples (# samples x # features) on which to explain the model's output.
nsamples : "auto" or int
Number of times to re-evaluate the model when explaining each prediction. More samples
lead to lower variance estimates of the SHAP values. The "auto" setting uses
`nsamples = 2 * X.shape[1] + 2048`.
l1_reg : "num_features(int)", "auto" (default for now, but deprecated), "aic", "bic", or float
The l1 regularization to use for feature selection (the estimation procedure is based on
a debiased lasso). The auto option currently uses "aic" when less that 20% of the possible sample
space is enumerated, otherwise it uses no regularization. THE BEHAVIOR OF "auto" WILL CHANGE
in a future version to be based on num_features instead of AIC.
The "aic" and "bic" options use the AIC and BIC rules for regularization.
Using "num_features(int)" selects a fix number of top features. Passing a float directly sets the
"alpha" parameter of the sklearn.linear_model.Lasso model used for feature selection.
Returns
-------
For models with a single output this returns a matrix of SHAP values
(# samples x # features). Each row sums to the difference between the model output for that
sample and the expected value of the model output (which is stored as expected_value
attribute of the explainer). For models with vector outputs this returns a list
of such matrices, one for each output.
"""
# convert dataframes
if str(type(X)).endswith("pandas.core.series.Series'>"):
X = X.values
elif str(type(X)).endswith("'pandas.core.frame.DataFrame'>"):
if self.keep_index:
index_value = X.index.values
index_name = X.index.name
column_name = list(X.columns)
X = X.values
x_type = str(type(X))
arr_type = "'numpy.ndarray'>"
# if sparse, convert to lil for performance
if sp.sparse.issparse(X) and not sp.sparse.isspmatrix_lil(X):
X = X.tolil()
assert x_type.endswith(arr_type) or sp.sparse.isspmatrix_lil(X), "Unknown instance type: " + x_type
assert len(X.shape) == 1 or len(X.shape) == 2 or len(X.shape) == 3, "Instance must have 1, 2 or 3 dimensions!"
if self.isRNN:
# get the unique subject ID's in the test data, in the original order
self.subject_ids, indeces = np.unique(X[:, self.id_col_num], return_index=True)
sorted_idx = np.argsort(indeces)
self.subject_ids = self.subject_ids[sorted_idx].astype(int)
# Remove paddings
self.subject_ids = self.subject_ids[self.subject_ids != self.padding_value]
ts_values = X[:, self.ts_col_num]
ts_values = ts_values[ts_values != self.padding_value]
# maximum sequence length in the test data
max_seq_len = kwargs.get('max_seq_len', None)
if max_seq_len == None:
max_seq_len = 1
for id in self.subject_ids:
seq_data = X[np.where((X[:, self.id_col_num] == id))]
cur_seq_length = len(seq_data)
if cur_seq_length > max_seq_len:
max_seq_len = cur_seq_length
# compare with the maximum sequence length in the background data
if max_seq_len > self.max_seq_len:
# update maximum sequence length
self.max_seq_len = max_seq_len
explanations = np.zeros((len(self.subject_ids), self.max_seq_len, len(self.model_features)))
# count the order of the sequences being iterated
seq_count = 0
# loop through the unique subject ID's
for id in tqdm(self.subject_ids, disable=kwargs.get("silent", False), desc='ID loop'):
# get the data corresponding to the current sequence
seq_data = X[X[:, self.id_col_num] == id]
# get the unique timestamp (or instance index) values of the current sequence
seq_unique_ts = np.unique(seq_data[:, self.ts_col_num]).astype(int)
# count the order of the instances being iterated
ts_count = 0
if self.isBidir is True:
# calculate the full sequence's outputs
torch_seq_data = torch.from_numpy(seq_data[:, self.model_features]).unsqueeze(0).float()
seq_outputs, _ = self.recur_layer(torch_seq_data)
# loop through the possible instances
for ts in tqdm(seq_unique_ts, disable=kwargs.get("silent", False), desc='ts loop', leave=False):
# get the data corresponding to the current instance
inst_data = seq_data[seq_data[:, self.ts_col_num] == ts]
# remove unwanted features (id, ts and label)
inst_data = inst_data[:, self.model_features]
# get the hidden state that the model receives as an input
if ts > 0:
# data from the previous instance(s) in the same sequence
past_data = torch.from_numpy(seq_data[np.where(seq_data[:, self.ts_col_num] < ts)])
if self.isBidir is False:
hidden_state = None
# convert the past data to a 3D tensor
past_data = past_data.unsqueeze(0)
# get the hidden state outputed from the previous recurrent cell
_, hidden_state = self.recur_layer(past_data[:, :, self.model_features].float())
# avoid passing gradients from previous instances
if isinstance(hidden_state, tuple) or isinstance(hidden_state, list):
if isinstance(hidden_state[0], tuple) or isinstance(hidden_state[0], list):
hidden_state = [(hidden_state[i][0].detach(), hidden_state[i][1].detach())
for i in range(len(hidden_state))]
else:
hidden_state = (hidden_state[0].detach(), hidden_state[1].detach())
else:
hidden_state.detach_()
# add the hidden_state to the kwargs
kwargs['hidden_state'] = hidden_state
else:
# add the past_data to the kwargs
kwargs['past_data'] = past_data
if self.isBidir is True:
# add the current instance's output to the kwargs
kwargs['inst_output'] = seq_outputs[ts_count].unsqueeze(0).detach().numpy()
if self.keep_index:
inst_data = convert_to_instance_with_index(inst_data, column_name, seq_count * self.max_seq_len + ts, index_name)
explanations[seq_count, ts_count, :] = self.explain(inst_data, **kwargs).squeeze()
ts_count += 1
seq_count += 1
return explanations
# [TODO] Don't understand the need for this out variable
# # vector-output
# s = explanations[0][0].shape
# if len(s) == 2:
# outs = [np.zeros((X.shape[0], X.shape[1], s[0])) for j in range(s[1])]
# for i in range(X.shape[0]):
# for j in range(X.shape[1]):
# for k in range(s[1]):
# outs[k][i][j] = explanations[i][j][:, k]
# return outs
#
# # single-output
# else:
# out = np.zeros((X.shape[0], X.shape[1], s[0]))
# for i in range(X.shape[0]):
# for j in range(X.shape[1]):
# out[i][j] = explanations[i][j]
# return out
# single instance
elif len(X.shape) == 1:
data = X.reshape((1, X.shape[0]))
if self.keep_index:
data = convert_to_instance_with_index(data, column_name, index_name, index_value)
explanation = self.explain(data, **kwargs)
# vector-output
s = explanation.shape
if len(s) == 2:
outs = [np.zeros(s[0]) for j in range(s[1])]
for j in range(s[1]):
outs[j] = explanation[:, j]
return outs
# single-output
else:
out = np.zeros(s[0])
out[:] = explanation
return out
# explain the whole dataset
elif len(X.shape) == 2:
explanations = []
for i in tqdm(range(X.shape[0]), disable=kwargs.get("silent", False)):
data = X[i:i + 1, :]
if self.keep_index:
data = convert_to_instance_with_index(data, column_name, index_value[i:i + 1], index_name)
explanations.append(self.explain(data, **kwargs))
# vector-output
s = explanations[0].shape
if len(s) == 2:
outs = [np.zeros((X.shape[0], s[0])) for j in range(s[1])]
for i in range(X.shape[0]):
for j in range(s[1]):
outs[j][i] = explanations[i][:, j]
return outs
# single-output
else:
out = np.zeros((X.shape[0], s[0]))
for i in range(X.shape[0]):
out[i] = explanations[i]
return out
def explain(self, incoming_instance, **kwargs):
# convert incoming input to a standardized iml object
instance = convert_to_instance(incoming_instance)
match_instance_to_data(instance, self.data)
# find the feature groups we will test. If a feature does not change from its
# current value then we know it doesn't impact the model
self.varyingInds = self.varying_groups(instance.x)
if self.data.groups is None:
self.varyingFeatureGroups = np.array([i for i in self.varyingInds])
self.M = self.varyingFeatureGroups.shape[0]
else:
self.varyingFeatureGroups = [self.data.groups[i] for i in self.varyingInds]
self.M = len(self.varyingFeatureGroups)
groups = self.data.groups
# convert to numpy array as it is much faster if not jagged array (all groups of same length)
if self.varyingFeatureGroups and all(len(groups[i]) == len(groups[0]) for i in self.varyingInds):
self.varyingFeatureGroups = np.array(self.varyingFeatureGroups)
# further performance optimization in case each group has a single value
if self.varyingFeatureGroups.shape[1] == 1:
self.varyingFeatureGroups = self.varyingFeatureGroups.flatten()
# get the current hidden state, if given
hidden_state = kwargs.get('hidden_state', None)
# get the current past data, if given
past_data = kwargs.get('past_data', None)
# get the current instances' output, if given
inst_output = kwargs.get('inst_output', None)
# find f(x)
if self.keep_index:
if self.isRNN is True:
if self.isBidir is False:
# only provide the hidden state argument if the model is a RNN type model
model_out = self.model.f(instance.convert_to_df(), hidden_state)
else:
model_out = inst_output
else:
model_out = self.model.f(instance.convert_to_df())
else:
if self.isRNN is True:
if self.isBidir is False:
# only provide the hidden state argument if the model is a RNN type model
model_out = self.model.f(instance.x, hidden_state)
else:
model_out = inst_output
else:
model_out = self.model.f(instance.x)
if isinstance(model_out, (pd.DataFrame, pd.Series)):
model_out = model_out.values
self.fx = model_out[0]
if not self.vector_out:
self.fx = np.array([self.fx])
# if no features vary then no feature has an effect
if self.M == 0:
phi = np.zeros((self.data.groups_size, self.D))
phi_var = np.zeros((self.data.groups_size, self.D))
# if only one feature varies then it has all the effect
elif self.M == 1:
phi = np.zeros((self.data.groups_size, self.D))
phi_var = np.zeros((self.data.groups_size, self.D))
diff = self.link.f(self.fx) - self.link.f(self.fnull)
for d in range(self.D):
phi[self.varyingInds[0],d] = diff[d]
# if more than one feature varies then we have to do real work
else:
self.l1_reg = kwargs.get("l1_reg", "auto")
# pick a reasonable number of samples if the user didn't specify how many they wanted
self.nsamples = kwargs.get("nsamples", "auto")
if self.nsamples == "auto":
self.nsamples = 2 * self.M + 2**11
# if we have enough samples to enumerate all subsets then ignore the unneeded samples
self.max_samples = 2 ** 30
if self.M <= 30:
self.max_samples = 2 ** self.M - 2
if self.nsamples > self.max_samples:
self.nsamples = self.max_samples
# reserve space for some of our computations
self.allocate()
# weight the different subset sizes
num_subset_sizes = np.int(np.ceil((self.M - 1) / 2.0))
num_paired_subset_sizes = np.int(np.floor((self.M - 1) / 2.0))
weight_vector = np.array([(self.M - 1.0) / (i * (self.M - i)) for i in range(1, num_subset_sizes + 1)])
weight_vector[:num_paired_subset_sizes] *= 2
weight_vector /= np.sum(weight_vector)
log.debug("weight_vector = {0}".format(weight_vector))
log.debug("num_subset_sizes = {0}".format(num_subset_sizes))
log.debug("num_paired_subset_sizes = {0}".format(num_paired_subset_sizes))
log.debug("M = {0}".format(self.M))
# fill out all the subset sizes we can completely enumerate
# given nsamples*remaining_weight_vector[subset_size]
num_full_subsets = 0
num_samples_left = self.nsamples
group_inds = np.arange(self.M, dtype='int64')
mask = np.zeros(self.M)
remaining_weight_vector = copy.copy(weight_vector)
for subset_size in range(1, num_subset_sizes + 1):
# determine how many subsets (and their complements) are of the current size
nsubsets = binom(self.M, subset_size)
if subset_size <= num_paired_subset_sizes: nsubsets *= 2
log.debug("subset_size = {0}".format(subset_size))
log.debug("nsubsets = {0}".format(nsubsets))
log.debug("self.nsamples*weight_vector[subset_size-1] = {0}".format(
num_samples_left * remaining_weight_vector[subset_size - 1]))
log.debug("self.nsamples*weight_vector[subset_size-1]/nsubsets = {0}".format(
num_samples_left * remaining_weight_vector[subset_size - 1] / nsubsets))
# see if we have enough samples to enumerate all subsets of this size
if num_samples_left * remaining_weight_vector[subset_size - 1] / nsubsets >= 1.0 - 1e-8:
num_full_subsets += 1
num_samples_left -= nsubsets
# rescale what's left of the remaining weight vector to sum to 1
if remaining_weight_vector[subset_size - 1] < 1.0:
remaining_weight_vector /= (1 - remaining_weight_vector[subset_size - 1])
# add all the samples of the current subset size
w = weight_vector[subset_size - 1] / binom(self.M, subset_size)
if subset_size <= num_paired_subset_sizes: w /= 2.0
for inds in itertools.combinations(group_inds, subset_size):
mask[:] = 0.0
mask[np.array(inds, dtype='int64')] = 1.0
self.addsample(instance.x, mask, w)
if subset_size <= num_paired_subset_sizes:
mask[:] = np.abs(mask - 1)
self.addsample(instance.x, mask, w)
else:
break
log.info("num_full_subsets = {0}".format(num_full_subsets))
# add random samples from what is left of the subset space
nfixed_samples = self.nsamplesAdded
samples_left = self.nsamples - self.nsamplesAdded
log.debug("samples_left = {0}".format(samples_left))
if num_full_subsets != num_subset_sizes:
remaining_weight_vector = copy.copy(weight_vector)
remaining_weight_vector[:num_paired_subset_sizes] /= 2 # because we draw two samples each below
remaining_weight_vector = remaining_weight_vector[num_full_subsets:]
remaining_weight_vector /= np.sum(remaining_weight_vector)
log.info("remaining_weight_vector = {0}".format(remaining_weight_vector))
log.info("num_paired_subset_sizes = {0}".format(num_paired_subset_sizes))
ind_set = np.random.choice(len(remaining_weight_vector), 4 * samples_left, p=remaining_weight_vector)
ind_set_pos = 0
used_masks = {}
while samples_left > 0 and ind_set_pos < len(ind_set):
mask.fill(0.0)
ind = ind_set[ind_set_pos] # we call np.random.choice once to save time and then just read it here
ind_set_pos += 1
subset_size = ind + num_full_subsets + 1
mask[np.random.permutation(self.M)[:subset_size]] = 1.0
# only add the sample if we have not seen it before, otherwise just
# increment a previous sample's weight
mask_tuple = tuple(mask)
new_sample = False
if mask_tuple not in used_masks:
new_sample = True
used_masks[mask_tuple] = self.nsamplesAdded
samples_left -= 1
self.addsample(instance.x, mask, 1.0)
else:
self.kernelWeights[used_masks[mask_tuple]] += 1.0
# add the compliment sample
if samples_left > 0 and subset_size <= num_paired_subset_sizes:
mask[:] = np.abs(mask - 1)
# only add the sample if we have not seen it before, otherwise just
# increment a previous sample's weight
if new_sample:
samples_left -= 1
self.addsample(instance.x, mask, 1.0)
else:
# we know the compliment sample is the next one after the original sample, so + 1
self.kernelWeights[used_masks[mask_tuple] + 1] += 1.0
# normalize the kernel weights for the random samples to equal the weight left after
# the fixed enumerated samples have been already counted
weight_left = np.sum(weight_vector[num_full_subsets:])
log.info("weight_left = {0}".format(weight_left))
self.kernelWeights[nfixed_samples:] *= weight_left / self.kernelWeights[nfixed_samples:].sum()
# execute the model on the synthetic samples we have created
self.run(**kwargs)
# solve then expand the feature importance (Shapley value) vector to contain the non-varying features
phi = np.zeros((self.data.groups_size, self.D))
phi_var = np.zeros((self.data.groups_size, self.D))
for d in range(self.D):
vphi, vphi_var = self.solve(self.nsamples / self.max_samples, d)
phi[self.varyingInds, d] = vphi
phi_var[self.varyingInds, d] = vphi_var
if not self.vector_out:
phi = np.squeeze(phi, axis=1)
phi_var = np.squeeze(phi_var, axis=1)
return phi
def varying_groups(self, x):
if not sp.sparse.issparse(x):
varying = np.zeros(self.data.groups_size)
for i in range(0, self.data.groups_size):
inds = self.data.groups[i]
x_group = x[0, inds]
if sp.sparse.issparse(x_group):
if all(j not in x.nonzero()[1] for j in inds):
varying[i] = False
continue
x_group = x_group.todense()
num_mismatches = np.sum(np.invert(np.isclose(x_group, self.data.data[:, inds], equal_nan=True)))
varying[i] = num_mismatches > 0
varying_indices = np.nonzero(varying)[0]
return varying_indices
else:
varying_indices = []
# go over all nonzero columns in background and evaluation data
# if both background and evaluation are zero, the column does not vary
varying_indices = np.unique(np.union1d(self.data.data.nonzero()[1], x.nonzero()[1]))
remove_unvarying_indices = []
for i in range(0, len(varying_indices)):
varying_index = varying_indices[i]
# now verify the nonzero values do vary
data_rows = self.data.data[:, [varying_index]]
nonzero_rows = data_rows.nonzero()[0]
if nonzero_rows.size > 0:
background_data_rows = data_rows[nonzero_rows]
if sp.sparse.issparse(background_data_rows):
background_data_rows = background_data_rows.toarray()
num_mismatches = np.sum(np.abs(background_data_rows - x[0, varying_index]) > 1e-7)
# Note: If feature column non-zero but some background zero, can't remove index
if num_mismatches == 0 and not \
(np.abs(x[0, [varying_index]][0, 0]) > 1e-7 and len(nonzero_rows) < data_rows.shape[0]):
remove_unvarying_indices.append(i)
mask = np.ones(len(varying_indices), dtype=bool)
mask[remove_unvarying_indices] = False
varying_indices = varying_indices[mask]
return varying_indices
def allocate(self):
if sp.sparse.issparse(self.data.data):
# We tile the sparse matrix in csr format but convert it to lil
# for performance when adding samples
shape = self.data.data.shape
nnz = self.data.data.nnz
data_rows, data_cols = shape
rows = data_rows * self.nsamples
shape = rows, data_cols
if nnz == 0:
self.synth_data = sp.sparse.csr_matrix(shape, dtype=self.data.data.dtype).tolil()
else:
data = self.data.data.data
indices = self.data.data.indices
indptr = self.data.data.indptr
last_indptr_idx = indptr[len(indptr) - 1]
indptr_wo_last = indptr[:-1]
new_indptrs = []
for i in range(0, self.nsamples - 1):
new_indptrs.append(indptr_wo_last + (i * last_indptr_idx))
new_indptrs.append(indptr + ((self.nsamples - 1) * last_indptr_idx))
new_indptr = np.concatenate(new_indptrs)
new_data = np.tile(data, self.nsamples)
new_indices = np.tile(indices, self.nsamples)
self.synth_data = sp.sparse.csr_matrix((new_data, new_indices, new_indptr), shape=shape).tolil()
else:
self.synth_data = np.tile(self.data.data, (self.nsamples, 1))
self.maskMatrix = np.zeros((self.nsamples, self.M))
self.kernelWeights = np.zeros(self.nsamples)
self.y = np.zeros((self.nsamples * self.N, self.D))
self.ey = np.zeros((self.nsamples, self.D))
self.lastMask = np.zeros(self.nsamples)
self.nsamplesAdded = 0
self.nsamplesRun = 0
if self.keep_index:
self.synth_data_index = np.tile(self.data.index_value, self.nsamples)
def addsample(self, x, m, w):
offset = self.nsamplesAdded * self.N
if isinstance(self.varyingFeatureGroups, (list,)):
for j in range(self.M):
for k in self.varyingFeatureGroups[j]:
if m[j] == 1.0:
self.synth_data[offset:offset+self.N, k] = x[0, k]
else:
# for non-jagged numpy array we can significantly boost performance
mask = m == 1.0
groups = self.varyingFeatureGroups[mask]
if len(groups.shape) == 2:
for group in groups:
self.synth_data[offset:offset+self.N, group] = x[0, group]
else:
# further performance optimization in case each group has a single feature
evaluation_data = x[0, groups]
# In edge case where background is all dense but evaluation data
# is all sparse, make evaluation data dense
if sp.sparse.issparse(x) and not sp.sparse.issparse(self.synth_data):
evaluation_data = evaluation_data.toarray()
self.synth_data[offset:offset+self.N, groups] = evaluation_data
self.maskMatrix[self.nsamplesAdded, :] = m
self.kernelWeights[self.nsamplesAdded] = w
self.nsamplesAdded += 1
def run(self, **kwargs):
# [TODO] The inefficiency issue is probably derived from here. It seems to want to run the requested number of samples TIMES the TOTAL number of background samples!
num_to_run = self.nsamplesAdded * self.N - self.nsamplesRun * self.N
data = self.synth_data[self.nsamplesRun*self.N:self.nsamplesAdded*self.N,:]
if self.keep_index:
index = self.synth_data_index[self.nsamplesRun*self.N:self.nsamplesAdded*self.N]
index = pd.DataFrame(index, columns=[self.data.index_name])
data = pd.DataFrame(data, columns=self.data.group_names)
data = pd.concat([index, data], axis=1).set_index(self.data.index_name)
if self.keep_index_ordered:
data = data.sort_index()
if self.isRNN is True:
# convert the data to be three-dimensional, considering each
# synthetic sample as a separate sequence
data = torch.from_numpy(data).float().unsqueeze(1)
if self.isBidir is False:
# get the current hidden state, if given
hidden_state = kwargs.get('hidden_state', None)
if hidden_state is not None:
# repeat the hidden state along the batch dimension
if isinstance(hidden_state, torch.Tensor):
hidden_state = hidden_state.repeat(1, data.shape[0], 1)
else:
hidden_state_0 = hidden_state[0].repeat(1, data.shape[0], 1)
hidden_state_1 = hidden_state[1].repeat(1, data.shape[0], 1)
hidden_state = (hidden_state_0, hidden_state_1)
modelOut = self.model.f(data, hidden_state)
else:
# get the current past data, if given
past_data = kwargs.get('past_data', None)
if past_data is not None:
# convert the data to be three-dimensional float tensor and remove ID columns
past_data = past_data[:, 2:].unsqueeze(0).float()
# repeat the past data along the batch dimension
past_data = past_data.repeat(data.shape[0], 1, 1)
# add the previous instances from the same sequence, if there are any
seq_data = torch.cat((past_data, data), dim=1)
else:
seq_data = data
modelOut = self.model.f(seq_data)
if past_data is not None:
# only get the outputs from the last sample, ignoring the previous instances
modelOut = modelOut.reshape([-1, past_data.shape[1]+1, 1])
modelOut = modelOut[:, -1, :]
else:
modelOut = self.model.f(data)
if isinstance(modelOut, (pd.DataFrame, pd.Series)):
modelOut = modelOut.values
self.y[self.nsamplesRun * self.N:self.nsamplesAdded * self.N, :] = np.reshape(modelOut, (num_to_run, self.D))
# find the expected value of each output
for i in range(self.nsamplesRun, self.nsamplesAdded):
eyVal = np.zeros(self.D)
for j in range(0, self.N):
eyVal += self.y[i * self.N + j, :] * self.weights[j]
self.ey[i, :] = eyVal
self.nsamplesRun += 1
def solve(self, fraction_evaluated, dim):
eyAdj = self.linkfv(self.ey[:, dim]) - self.link.f(self.fnull[dim])
s = np.sum(self.maskMatrix, 1)
# do feature selection if we have not well enumerated the space
nonzero_inds = np.arange(self.M)
log.debug("fraction_evaluated = {0}".format(fraction_evaluated))
if self.l1_reg == "auto":
warnings.warn(
"l1_reg=\"auto\" is deprecated and in the next version (v0.29) the behavior will change from a " \
"conditional use of AIC to simply \"num_features(10)\"!"
)
if (self.l1_reg not in ["auto", False, 0]) or (fraction_evaluated < 0.2 and self.l1_reg == "auto"):
w_aug = np.hstack((self.kernelWeights * (self.M - s), self.kernelWeights * s))
log.info("np.sum(w_aug) = {0}".format(np.sum(w_aug)))
log.info("np.sum(self.kernelWeights) = {0}".format(np.sum(self.kernelWeights)))
w_sqrt_aug = np.sqrt(w_aug)
eyAdj_aug = np.hstack((eyAdj, eyAdj - (self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim]))))
eyAdj_aug *= w_sqrt_aug
mask_aug = np.transpose(w_sqrt_aug * np.transpose(np.vstack((self.maskMatrix, self.maskMatrix - 1))))
#var_norms = np.array([np.linalg.norm(mask_aug[:, i]) for i in range(mask_aug.shape[1])])
# select a fixed number of top features
if isinstance(self.l1_reg, str) and self.l1_reg.startswith("num_features("):
r = int(self.l1_reg[len("num_features("):-1])
nonzero_inds = lars_path(mask_aug, eyAdj_aug, max_iter=r)[1]
# use an adaptive regularization method
elif self.l1_reg == "auto" or self.l1_reg == "bic" or self.l1_reg == "aic":
c = "aic" if self.l1_reg == "auto" else self.l1_reg
nonzero_inds = np.nonzero(LassoLarsIC(criterion=c).fit(mask_aug, eyAdj_aug).coef_)[0]
# use a fixed regularization coeffcient
else:
nonzero_inds = np.nonzero(Lasso(alpha=self.l1_reg).fit(mask_aug, eyAdj_aug).coef_)[0]
if len(nonzero_inds) == 0:
return np.zeros(self.M), np.ones(self.M)
# eliminate one variable with the constraint that all features sum to the output
eyAdj2 = eyAdj - self.maskMatrix[:, nonzero_inds[-1]] * (
self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim]))
etmp = np.transpose(np.transpose(self.maskMatrix[:, nonzero_inds[:-1]]) - self.maskMatrix[:, nonzero_inds[-1]])
log.debug("etmp[:4,:] {0}".format(etmp[:4, :]))
# solve a weighted least squares equation to estimate phi
tmp = np.transpose(np.transpose(etmp) * np.transpose(self.kernelWeights))
tmp2 = np.linalg.inv(np.dot(np.transpose(tmp), etmp))
w = np.dot(tmp2, np.dot(np.transpose(tmp), eyAdj2))
log.debug("np.sum(w) = {0}".format(np.sum(w)))
log.debug("self.link(self.fx) - self.link(self.fnull) = {0}".format(
self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim])))
log.debug("self.fx = {0}".format(self.fx[dim]))
log.debug("self.link(self.fx) = {0}".format(self.link.f(self.fx[dim])))
log.debug("self.fnull = {0}".format(self.fnull[dim]))
log.debug("self.link(self.fnull) = {0}".format(self.link.f(self.fnull[dim])))
phi = np.zeros(self.M)
phi[nonzero_inds[:-1]] = w
phi[nonzero_inds[-1]] = (self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim])) - sum(w)
log.info("phi = {0}".format(phi))
# clean up any rounding errors
for i in range(self.M):
if np.abs(phi[i]) < 1e-10:
phi[i] = 0
return phi, np.ones(len(phi))The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
To fix this, I adapted Kernel SHAP's code with the following main changes:
— Added parameters that identified the model type, including when it was RNN;
from sklearn.impute import SimpleImputer
from ..common import convert_to_instance, convert_to_model, match_instance_to_data, match_model_to_data, convert_to_instance_with_index, convert_to_link, IdentityLink, convert_to_data, DenseData, SparseData
from scipy.special import binom
from scipy.sparse import issparse
import numpy as np
import pandas as pd
import scipy as sp
import logging
import copy
import itertools
import warnings
from sklearn.linear_model import LassoLarsIC, Lasso, lars_path
from sklearn.cluster import KMeans
from tqdm.auto import tqdm
from .explainer import Explainer
import torch
log = logging.getLogger('shap')
def kmeans(X, k, round_values=True):
""" Summarize a dataset with k mean samples weighted by the number of data points they
each represent.
Parameters
----------
X : numpy.array or pandas.DataFrame or any scipy.sparse matrix
Matrix of data samples to summarize (# samples x # features)
k : int
Number of means to use for approximation.
round_values : bool
For all i, round the ith dimension of each mean sample to match the nearest value
from X[:,i]. This ensures discrete features always get a valid value.
Returns
-------
DenseData object.
"""
group_names = [str(i) for i in range(X.shape[1])]
if str(type(X)).endswith("'pandas.core.frame.DataFrame'>"):
group_names = X.columns
X = X.values
# in case there are any missing values in data impute them
imp = SimpleImputer(missing_values=np.nan, strategy='mean')
X = imp.fit_transform(X)
kmeans = KMeans(n_clusters=k, random_state=0).fit(X)
if round_values:
for i in range(k):
for j in range(X.shape[1]):
xj = X[:,j].toarray().flatten() if issparse(X) else X[:, j]
ind = np.argmin(np.abs(xj - kmeans.cluster_centers_[i,j]))
kmeans.cluster_centers_[i,j] = X[ind,j]
return DenseData(kmeans.cluster_centers_, group_names, None, 1.0*np.bincount(kmeans.labels_))
class KernelExplainer(Explainer):
"""Uses the Kernel SHAP method to explain the output of any function.
Kernel SHAP is a method that uses a special weighted linear regression
to compute the importance of each feature. The computed importance values
are Shapley values from game theory and also coefficents from a local linear
regression.
Parameters
----------
model : function or iml.Model
User supplied function that takes a matrix of samples (# samples x # features) and
computes the output of the model for those samples. The output can be a vector
(# samples) or a matrix (# samples x # model outputs).
data : numpy.array or pandas.DataFrame or shap.common.DenseData or any scipy.sparse matrix
The background dataset to use for integrating out features. To determine the impact
of a feature, that feature is set to "missing" and the change in the model output
is observed. Since most models aren't designed to handle arbitrary missing data at test
time, we simulate "missing" by replacing the feature with the values it takes in the
background dataset. So if the background dataset is a simple sample of all zeros, then
we would approximate a feature being missing by setting it to zero. For small problems
this background dataset can be the whole training set, but for larger problems consider
using a single reference value or using the kmeans function to summarize the dataset.
Note: for sparse case we accept any sparse matrix but convert to lil format for
performance.
link : "identity" or "logit"
A generalized linear model link to connect the feature importance values to the model
output. Since the feature importance values, phi, sum up to the model output, it often makes
sense to connect them to the output with a link function where link(output) = sum(phi).
If the model output is a probability then the LogitLink link function makes the feature
importance values have log-odds units.
isRNN : bool
Boolean that indicates if the model being analyzed is a recurrent neural network (RNN).
If so, it means that sequential data is being used, which requires some modifications
in the way SHAP values are calculated.
if isRNN is True:
id_col_num : int
Number that indicates in which column is the sequence / subject id. Defaults to 0.
ts_col_num : int
Number that indicates in which column is the instance / timestamp. Defaults to 1.
label_col_num : int
Number that indicates in which column is the label, if any. Defaults to None.
recur_layer : torch.nn.LSTM or torch.nn.GRU or torch.nn.RNN, default None
Pointer to the recurrent layer in the model, if it exists. It should
either be a LSTM, GRU or RNN network. If none is specified, the
method will automatically search for a recurrent layer in the model.
isBidir: bool
Boolean that indicates if the recurrent neural network model being analyzed is
bidirectional. If so, it implies a special treatment of the sequences, as the
hidden states can't be moved in the same way of a single direction RNN.
padding_value : numeric
Value to use in the padding, to fill the sequences.
"""
def __init__(self, model, data, link=IdentityLink(), **kwargs):
# convert incoming inputs to standardized iml objects
self.link = convert_to_link(link)
self.model = convert_to_model(model)
self.keep_index = kwargs.get("keep_index", False)
self.keep_index_ordered = kwargs.get("keep_index_ordered", False)
# check if the model is a recurrent neural network
self.isRNN = kwargs.get('isRNN', False)
if self.isRNN and not str(type(data)).endswith("'pandas.core.frame.DataFrame'>"):
# check if the model is a bidirectional recurrent neural network
self.isBidir = kwargs.get('isBidir', False)
# number of the column that corresponds to the sequence / subject id
self.id_col_num = kwargs.get('id_col_num', 0)
# number of the column that corresponds to the instance / timestamp
self.ts_col_num = kwargs.get('ts_col_num', 1)
# number of the column that corresponds to the label
label_col_num = kwargs.get('label_col_num', None)
# padding value
self.padding_value = kwargs.get('padding_value', 999999)
# all columns in the data
self.model_features = list(range(data.shape[1]))
# remove unwanted columns, so that we get only those that actually correspond to model usable features
[self.model_features.remove(col) for col in [self.id_col_num, self.ts_col_num, label_col_num] if col is not None]
# maximum background samples to use
self.max_bkgnd_samples = kwargs.get('max_bkgnd_samples', 20)
if data.shape[0] > self.max_bkgnd_samples:
# use k-means to avoid slow processing of a lot of samples
self.data = kmeans(data[:, self.model_features], k=self.max_bkgnd_samples)
# get the weights corresponding to all the original data
num_samples = data.shape[0]
self.weights = np.ones(num_samples)
self.weights /= np.sum(self.weights)
else:
self.data = convert_to_data(data[:, self.model_features], keep_index=self.keep_index)
self.weights = self.data.weights
# check if the recurrent layer is specified
self.recur_layer = kwargs.get('recur_layer', None)
if self.recur_layer is None:
# get the model object, so as to use its recurrent layer
model_obj = kwargs.get('model_obj', None)
assert model_obj is not None, 'If the model uses a recurrent neural network, either the recurrent layer or the full model object must be specified.'
# search for a recurrent layer
if hasattr(model_obj, 'lstm'):
self.recur_layer = model_obj.lstm
elif hasattr(model_obj, 'gru'):
self.recur_layer = model_obj.gru
elif hasattr(model_obj, 'rnn'):
self.recur_layer = model_obj.rnn
else:
raise Exception('ERROR: No recurrent layer found. Please specify it in the recur_layer argument.')
# get the unique subject ID's in the background data
self.subject_ids = np.unique(data[:, self.id_col_num]).astype(int)
# maximum sequence length in the background data
self.max_seq_len = kwargs.get('max_seq_len', None)
if self.max_seq_len == None:
self.max_seq_len = 1
for id in self.subject_ids:
seq_data = data[np.where((data[:, self.id_col_num] == id))]
cur_seq_length = len(seq_data)
if cur_seq_length > self.max_seq_len:
self.max_seq_len = cur_seq_length
# calculate the output for all the background data
model_null = match_model_to_data(self.model, data, self.isRNN, self.model_features,
self.id_col_num, self.ts_col_num, self.recur_layer,
self.subject_ids, self.max_seq_len, self.model.f,
silent=kwargs.get("silent", False))
else:
self.data = convert_to_data(data, keep_index=self.keep_index)
self.weights = self.data.weights
# calculate the output for all the background data
model_null = match_model_to_data(self.model, self.data)
self.col_names = None
if str(type(data)).endswith("'pandas.core.frame.DataFrame'>"):
# keep the column names so that data can be used in dataframe format
self.col_names = data.columns
# enforce our current input type limitations
assert isinstance(self.data, DenseData) or isinstance(self.data, SparseData), \
"Shap explainer only supports the DenseData and SparseData input currently."
assert not self.data.transposed, "Shap explainer does not support transposed DenseData or SparseData currently."
# warn users about large background data sets
if len(self.data.weights) > 100:
log.warning("Using " + str(len(self.data.weights)) + " background data samples could cause " +
"slower run times. Consider using shap.sample(data, K) or shap.kmeans(data, K) to " +
"summarize the background as K samples.")
# init our parameters
self.N = self.data.data.shape[0]
self.P = self.data.data.shape[1]
self.linkfv = np.vectorize(self.link.f)
self.nsamplesAdded = 0
self.nsamplesRun = 0
# find E_x[f(x)]
if isinstance(model_null, (pd.DataFrame, pd.Series)):
model_null = np.squeeze(model_null.values)
self.fnull = np.sum((model_null.T * self.weights).T, 0)
self.expected_value = self.linkfv(self.fnull)
# see if we have a vector output
self.vector_out = True
if len(self.fnull.shape) == 0:
self.vector_out = False
self.fnull = np.array([self.fnull])
self.D = 1
self.expected_value = float(self.expected_value)
else:
self.D = self.fnull.shape[0]
def shap_values(self, X, **kwargs):
""" Estimate the SHAP values for a set of samples.
Parameters
----------
X : numpy.array or pandas.DataFrame or any scipy.sparse matrix
A matrix of samples (# samples x # features) on which to explain the model's output.
nsamples : "auto" or int
Number of times to re-evaluate the model when explaining each prediction. More samples
lead to lower variance estimates of the SHAP values. The "auto" setting uses
`nsamples = 2 * X.shape[1] + 2048`.
l1_reg : "num_features(int)", "auto" (default for now, but deprecated), "aic", "bic", or float
The l1 regularization to use for feature selection (the estimation procedure is based on
a debiased lasso). The auto option currently uses "aic" when less that 20% of the possible sample
space is enumerated, otherwise it uses no regularization. THE BEHAVIOR OF "auto" WILL CHANGE
in a future version to be based on num_features instead of AIC.
The "aic" and "bic" options use the AIC and BIC rules for regularization.
Using "num_features(int)" selects a fix number of top features. Passing a float directly sets the
"alpha" parameter of the sklearn.linear_model.Lasso model used for feature selection.
Returns
-------
For models with a single output this returns a matrix of SHAP values
(# samples x # features). Each row sums to the difference between the model output for that
sample and the expected value of the model output (which is stored as expected_value
attribute of the explainer). For models with vector outputs this returns a list
of such matrices, one for each output.
"""
# convert dataframes
if str(type(X)).endswith("pandas.core.series.Series'>"):
X = X.values
elif str(type(X)).endswith("'pandas.core.frame.DataFrame'>"):
if self.keep_index:
index_value = X.index.values
index_name = X.index.name
column_name = list(X.columns)
X = X.values
x_type = str(type(X))
arr_type = "'numpy.ndarray'>"
# if sparse, convert to lil for performance
if sp.sparse.issparse(X) and not sp.sparse.isspmatrix_lil(X):
X = X.tolil()
assert x_type.endswith(arr_type) or sp.sparse.isspmatrix_lil(X), "Unknown instance type: " + x_type
assert len(X.shape) == 1 or len(X.shape) == 2 or len(X.shape) == 3, "Instance must have 1, 2 or 3 dimensions!"
if self.isRNN:
# get the unique subject ID's in the test data, in the original order
self.subject_ids, indeces = np.unique(X[:, self.id_col_num], return_index=True)
sorted_idx = np.argsort(indeces)
self.subject_ids = self.subject_ids[sorted_idx].astype(int)
# Remove paddings
self.subject_ids = self.subject_ids[self.subject_ids != self.padding_value]
ts_values = X[:, self.ts_col_num]
ts_values = ts_values[ts_values != self.padding_value]
# maximum sequence length in the test data
max_seq_len = kwargs.get('max_seq_len', None)
if max_seq_len == None:
max_seq_len = 1
for id in self.subject_ids:
seq_data = X[np.where((X[:, self.id_col_num] == id))]
cur_seq_length = len(seq_data)
if cur_seq_length > max_seq_len:
max_seq_len = cur_seq_length
# compare with the maximum sequence length in the background data
if max_seq_len > self.max_seq_len:
# update maximum sequence length
self.max_seq_len = max_seq_len
explanations = np.zeros((len(self.subject_ids), self.max_seq_len, len(self.model_features)))
# count the order of the sequences being iterated
seq_count = 0
# loop through the unique subject ID's
for id in tqdm(self.subject_ids, disable=kwargs.get("silent", False), desc='ID loop'):
# get the data corresponding to the current sequence
seq_data = X[X[:, self.id_col_num] == id]
# get the unique timestamp (or instance index) values of the current sequence
seq_unique_ts = np.unique(seq_data[:, self.ts_col_num]).astype(int)
# count the order of the instances being iterated
ts_count = 0
if self.isBidir is True:
# calculate the full sequence's outputs
torch_seq_data = torch.from_numpy(seq_data[:, self.model_features]).unsqueeze(0).float()
seq_outputs, _ = self.recur_layer(torch_seq_data)
# loop through the possible instances
for ts in tqdm(seq_unique_ts, disable=kwargs.get("silent", False), desc='ts loop', leave=False):
# get the data corresponding to the current instance
inst_data = seq_data[seq_data[:, self.ts_col_num] == ts]
# remove unwanted features (id, ts and label)
inst_data = inst_data[:, self.model_features]
# get the hidden state that the model receives as an input
if ts > 0:
# data from the previous instance(s) in the same sequence
past_data = torch.from_numpy(seq_data[np.where(seq_data[:, self.ts_col_num] < ts)])
if self.isBidir is False:
hidden_state = None
# convert the past data to a 3D tensor
past_data = past_data.unsqueeze(0)
# get the hidden state outputed from the previous recurrent cell
_, hidden_state = self.recur_layer(past_data[:, :, self.model_features].float())
# avoid passing gradients from previous instances
if isinstance(hidden_state, tuple) or isinstance(hidden_state, list):
if isinstance(hidden_state[0], tuple) or isinstance(hidden_state[0], list):
hidden_state = [(hidden_state[i][0].detach(), hidden_state[i][1].detach())
for i in range(len(hidden_state))]
else:
hidden_state = (hidden_state[0].detach(), hidden_state[1].detach())
else:
hidden_state.detach_()
# add the hidden_state to the kwargs
kwargs['hidden_state'] = hidden_state
else:
# add the past_data to the kwargs
kwargs['past_data'] = past_data
if self.isBidir is True:
# add the current instance's output to the kwargs
kwargs['inst_output'] = seq_outputs[ts_count].unsqueeze(0).detach().numpy()
if self.keep_index:
inst_data = convert_to_instance_with_index(inst_data, column_name, seq_count * self.max_seq_len + ts, index_name)
explanations[seq_count, ts_count, :] = self.explain(inst_data, **kwargs).squeeze()
ts_count += 1
seq_count += 1
return explanations
# [TODO] Don't understand the need for this out variable
# # vector-output
# s = explanations[0][0].shape
# if len(s) == 2:
# outs = [np.zeros((X.shape[0], X.shape[1], s[0])) for j in range(s[1])]
# for i in range(X.shape[0]):
# for j in range(X.shape[1]):
# for k in range(s[1]):
# outs[k][i][j] = explanations[i][j][:, k]
# return outs
#
# # single-output
# else:
# out = np.zeros((X.shape[0], X.shape[1], s[0]))
# for i in range(X.shape[0]):
# for j in range(X.shape[1]):
# out[i][j] = explanations[i][j]
# return out
# single instance
elif len(X.shape) == 1:
data = X.reshape((1, X.shape[0]))
if self.keep_index:
data = convert_to_instance_with_index(data, column_name, index_name, index_value)
explanation = self.explain(data, **kwargs)
# vector-output
s = explanation.shape
if len(s) == 2:
outs = [np.zeros(s[0]) for j in range(s[1])]
for j in range(s[1]):
outs[j] = explanation[:, j]
return outs
# single-output
else:
out = np.zeros(s[0])
out[:] = explanation
return out
# explain the whole dataset
elif len(X.shape) == 2:
explanations = []
for i in tqdm(range(X.shape[0]), disable=kwargs.get("silent", False)):
data = X[i:i + 1, :]
if self.keep_index:
data = convert_to_instance_with_index(data, column_name, index_value[i:i + 1], index_name)
explanations.append(self.explain(data, **kwargs))
# vector-output
s = explanations[0].shape
if len(s) == 2:
outs = [np.zeros((X.shape[0], s[0])) for j in range(s[1])]
for i in range(X.shape[0]):
for j in range(s[1]):
outs[j][i] = explanations[i][:, j]
return outs
# single-output
else:
out = np.zeros((X.shape[0], s[0]))
for i in range(X.shape[0]):
out[i] = explanations[i]
return out
def explain(self, incoming_instance, **kwargs):
# convert incoming input to a standardized iml object
instance = convert_to_instance(incoming_instance)
match_instance_to_data(instance, self.data)
# find the feature groups we will test. If a feature does not change from its
# current value then we know it doesn't impact the model
self.varyingInds = self.varying_groups(instance.x)
if self.data.groups is None:
self.varyingFeatureGroups = np.array([i for i in self.varyingInds])
self.M = self.varyingFeatureGroups.shape[0]
else:
self.varyingFeatureGroups = [self.data.groups[i] for i in self.varyingInds]
self.M = len(self.varyingFeatureGroups)
groups = self.data.groups
# convert to numpy array as it is much faster if not jagged array (all groups of same length)
if self.varyingFeatureGroups and all(len(groups[i]) == len(groups[0]) for i in self.varyingInds):
self.varyingFeatureGroups = np.array(self.varyingFeatureGroups)
# further performance optimization in case each group has a single value
if self.varyingFeatureGroups.shape[1] == 1:
self.varyingFeatureGroups = self.varyingFeatureGroups.flatten()
# get the current hidden state, if given
hidden_state = kwargs.get('hidden_state', None)
# get the current past data, if given
past_data = kwargs.get('past_data', None)
# get the current instances' output, if given
inst_output = kwargs.get('inst_output', None)
# find f(x)
if self.keep_index:
if self.isRNN is True:
if self.isBidir is False:
# only provide the hidden state argument if the model is a RNN type model
model_out = self.model.f(instance.convert_to_df(), hidden_state)
else:
model_out = inst_output
else:
model_out = self.model.f(instance.convert_to_df())
else:
if self.isRNN is True:
if self.isBidir is False:
# only provide the hidden state argument if the model is a RNN type model
model_out = self.model.f(instance.x, hidden_state)
else:
model_out = inst_output
else:
model_out = self.model.f(instance.x)
if isinstance(model_out, (pd.DataFrame, pd.Series)):
model_out = model_out.values
self.fx = model_out[0]
if not self.vector_out:
self.fx = np.array([self.fx])
# if no features vary then no feature has an effect
if self.M == 0:
phi = np.zeros((self.data.groups_size, self.D))
phi_var = np.zeros((self.data.groups_size, self.D))
# if only one feature varies then it has all the effect
elif self.M == 1:
phi = np.zeros((self.data.groups_size, self.D))
phi_var = np.zeros((self.data.groups_size, self.D))
diff = self.link.f(self.fx) - self.link.f(self.fnull)
for d in range(self.D):
phi[self.varyingInds[0],d] = diff[d]
# if more than one feature varies then we have to do real work
else:
self.l1_reg = kwargs.get("l1_reg", "auto")
# pick a reasonable number of samples if the user didn't specify how many they wanted
self.nsamples = kwargs.get("nsamples", "auto")
if self.nsamples == "auto":
self.nsamples = 2 * self.M + 2**11
# if we have enough samples to enumerate all subsets then ignore the unneeded samples
self.max_samples = 2 ** 30
if self.M <= 30:
self.max_samples = 2 ** self.M - 2
if self.nsamples > self.max_samples:
self.nsamples = self.max_samples
# reserve space for some of our computations
self.allocate()
# weight the different subset sizes
num_subset_sizes = np.int(np.ceil((self.M - 1) / 2.0))
num_paired_subset_sizes = np.int(np.floor((self.M - 1) / 2.0))
weight_vector = np.array([(self.M - 1.0) / (i * (self.M - i)) for i in range(1, num_subset_sizes + 1)])
weight_vector[:num_paired_subset_sizes] *= 2
weight_vector /= np.sum(weight_vector)
log.debug("weight_vector = {0}".format(weight_vector))
log.debug("num_subset_sizes = {0}".format(num_subset_sizes))
log.debug("num_paired_subset_sizes = {0}".format(num_paired_subset_sizes))
log.debug("M = {0}".format(self.M))
# fill out all the subset sizes we can completely enumerate
# given nsamples*remaining_weight_vector[subset_size]
num_full_subsets = 0
num_samples_left = self.nsamples
group_inds = np.arange(self.M, dtype='int64')
mask = np.zeros(self.M)
remaining_weight_vector = copy.copy(weight_vector)
for subset_size in range(1, num_subset_sizes + 1):
# determine how many subsets (and their complements) are of the current size
nsubsets = binom(self.M, subset_size)
if subset_size <= num_paired_subset_sizes: nsubsets *= 2
log.debug("subset_size = {0}".format(subset_size))
log.debug("nsubsets = {0}".format(nsubsets))
log.debug("self.nsamples*weight_vector[subset_size-1] = {0}".format(
num_samples_left * remaining_weight_vector[subset_size - 1]))
log.debug("self.nsamples*weight_vector[subset_size-1]/nsubsets = {0}".format(
num_samples_left * remaining_weight_vector[subset_size - 1] / nsubsets))
# see if we have enough samples to enumerate all subsets of this size
if num_samples_left * remaining_weight_vector[subset_size - 1] / nsubsets >= 1.0 - 1e-8:
num_full_subsets += 1
num_samples_left -= nsubsets
# rescale what's left of the remaining weight vector to sum to 1
if remaining_weight_vector[subset_size - 1] < 1.0:
remaining_weight_vector /= (1 - remaining_weight_vector[subset_size - 1])
# add all the samples of the current subset size
w = weight_vector[subset_size - 1] / binom(self.M, subset_size)
if subset_size <= num_paired_subset_sizes: w /= 2.0
for inds in itertools.combinations(group_inds, subset_size):
mask[:] = 0.0
mask[np.array(inds, dtype='int64')] = 1.0
self.addsample(instance.x, mask, w)
if subset_size <= num_paired_subset_sizes:
mask[:] = np.abs(mask - 1)
self.addsample(instance.x, mask, w)
else:
break
log.info("num_full_subsets = {0}".format(num_full_subsets))
# add random samples from what is left of the subset space
nfixed_samples = self.nsamplesAdded
samples_left = self.nsamples - self.nsamplesAdded
log.debug("samples_left = {0}".format(samples_left))
if num_full_subsets != num_subset_sizes:
remaining_weight_vector = copy.copy(weight_vector)
remaining_weight_vector[:num_paired_subset_sizes] /= 2 # because we draw two samples each below
remaining_weight_vector = remaining_weight_vector[num_full_subsets:]
remaining_weight_vector /= np.sum(remaining_weight_vector)
log.info("remaining_weight_vector = {0}".format(remaining_weight_vector))
log.info("num_paired_subset_sizes = {0}".format(num_paired_subset_sizes))
ind_set = np.random.choice(len(remaining_weight_vector), 4 * samples_left, p=remaining_weight_vector)
ind_set_pos = 0
used_masks = {}
while samples_left > 0 and ind_set_pos < len(ind_set):
mask.fill(0.0)
ind = ind_set[ind_set_pos] # we call np.random.choice once to save time and then just read it here
ind_set_pos += 1
subset_size = ind + num_full_subsets + 1
mask[np.random.permutation(self.M)[:subset_size]] = 1.0
# only add the sample if we have not seen it before, otherwise just
# increment a previous sample's weight
mask_tuple = tuple(mask)
new_sample = False
if mask_tuple not in used_masks:
new_sample = True
used_masks[mask_tuple] = self.nsamplesAdded
samples_left -= 1
self.addsample(instance.x, mask, 1.0)
else:
self.kernelWeights[used_masks[mask_tuple]] += 1.0
# add the compliment sample
if samples_left > 0 and subset_size <= num_paired_subset_sizes:
mask[:] = np.abs(mask - 1)
# only add the sample if we have not seen it before, otherwise just
# increment a previous sample's weight
if new_sample:
samples_left -= 1
self.addsample(instance.x, mask, 1.0)
else:
# we know the compliment sample is the next one after the original sample, so + 1
self.kernelWeights[used_masks[mask_tuple] + 1] += 1.0
# normalize the kernel weights for the random samples to equal the weight left after
# the fixed enumerated samples have been already counted
weight_left = np.sum(weight_vector[num_full_subsets:])
log.info("weight_left = {0}".format(weight_left))
self.kernelWeights[nfixed_samples:] *= weight_left / self.kernelWeights[nfixed_samples:].sum()
# execute the model on the synthetic samples we have created
self.run(**kwargs)
# solve then expand the feature importance (Shapley value) vector to contain the non-varying features
phi = np.zeros((self.data.groups_size, self.D))
phi_var = np.zeros((self.data.groups_size, self.D))
for d in range(self.D):
vphi, vphi_var = self.solve(self.nsamples / self.max_samples, d)
phi[self.varyingInds, d] = vphi
phi_var[self.varyingInds, d] = vphi_var
if not self.vector_out:
phi = np.squeeze(phi, axis=1)
phi_var = np.squeeze(phi_var, axis=1)
return phi
def varying_groups(self, x):
if not sp.sparse.issparse(x):
varying = np.zeros(self.data.groups_size)
for i in range(0, self.data.groups_size):
inds = self.data.groups[i]
x_group = x[0, inds]
if sp.sparse.issparse(x_group):
if all(j not in x.nonzero()[1] for j in inds):
varying[i] = False
continue
x_group = x_group.todense()
num_mismatches = np.sum(np.invert(np.isclose(x_group, self.data.data[:, inds], equal_nan=True)))
varying[i] = num_mismatches > 0
varying_indices = np.nonzero(varying)[0]
return varying_indices
else:
varying_indices = []
# go over all nonzero columns in background and evaluation data
# if both background and evaluation are zero, the column does not vary
varying_indices = np.unique(np.union1d(self.data.data.nonzero()[1], x.nonzero()[1]))
remove_unvarying_indices = []
for i in range(0, len(varying_indices)):
varying_index = varying_indices[i]
# now verify the nonzero values do vary
data_rows = self.data.data[:, [varying_index]]
nonzero_rows = data_rows.nonzero()[0]
if nonzero_rows.size > 0:
background_data_rows = data_rows[nonzero_rows]
if sp.sparse.issparse(background_data_rows):
background_data_rows = background_data_rows.toarray()
num_mismatches = np.sum(np.abs(background_data_rows - x[0, varying_index]) > 1e-7)
# Note: If feature column non-zero but some background zero, can't remove index
if num_mismatches == 0 and not \
(np.abs(x[0, [varying_index]][0, 0]) > 1e-7 and len(nonzero_rows) < data_rows.shape[0]):
remove_unvarying_indices.append(i)
mask = np.ones(len(varying_indices), dtype=bool)
mask[remove_unvarying_indices] = False
varying_indices = varying_indices[mask]
return varying_indices
def allocate(self):
if sp.sparse.issparse(self.data.data):
# We tile the sparse matrix in csr format but convert it to lil
# for performance when adding samples
shape = self.data.data.shape
nnz = self.data.data.nnz
data_rows, data_cols = shape
rows = data_rows * self.nsamples
shape = rows, data_cols
if nnz == 0:
self.synth_data = sp.sparse.csr_matrix(shape, dtype=self.data.data.dtype).tolil()
else:
data = self.data.data.data
indices = self.data.data.indices
indptr = self.data.data.indptr
last_indptr_idx = indptr[len(indptr) - 1]
indptr_wo_last = indptr[:-1]
new_indptrs = []
for i in range(0, self.nsamples - 1):
new_indptrs.append(indptr_wo_last + (i * last_indptr_idx))
new_indptrs.append(indptr + ((self.nsamples - 1) * last_indptr_idx))
new_indptr = np.concatenate(new_indptrs)
new_data = np.tile(data, self.nsamples)
new_indices = np.tile(indices, self.nsamples)
self.synth_data = sp.sparse.csr_matrix((new_data, new_indices, new_indptr), shape=shape).tolil()
else:
self.synth_data = np.tile(self.data.data, (self.nsamples, 1))
self.maskMatrix = np.zeros((self.nsamples, self.M))
self.kernelWeights = np.zeros(self.nsamples)
self.y = np.zeros((self.nsamples * self.N, self.D))
self.ey = np.zeros((self.nsamples, self.D))
self.lastMask = np.zeros(self.nsamples)
self.nsamplesAdded = 0
self.nsamplesRun = 0
if self.keep_index:
self.synth_data_index = np.tile(self.data.index_value, self.nsamples)
def addsample(self, x, m, w):
offset = self.nsamplesAdded * self.N
if isinstance(self.varyingFeatureGroups, (list,)):
for j in range(self.M):
for k in self.varyingFeatureGroups[j]:
if m[j] == 1.0:
self.synth_data[offset:offset+self.N, k] = x[0, k]
else:
# for non-jagged numpy array we can significantly boost performance
mask = m == 1.0
groups = self.varyingFeatureGroups[mask]
if len(groups.shape) == 2:
for group in groups:
self.synth_data[offset:offset+self.N, group] = x[0, group]
else:
# further performance optimization in case each group has a single feature
evaluation_data = x[0, groups]
# In edge case where background is all dense but evaluation data
# is all sparse, make evaluation data dense
if sp.sparse.issparse(x) and not sp.sparse.issparse(self.synth_data):
evaluation_data = evaluation_data.toarray()
self.synth_data[offset:offset+self.N, groups] = evaluation_data
self.maskMatrix[self.nsamplesAdded, :] = m
self.kernelWeights[self.nsamplesAdded] = w
self.nsamplesAdded += 1
def run(self, **kwargs):
# [TODO] The inefficiency issue is probably derived from here. It seems to want to run the requested number of samples TIMES the TOTAL number of background samples!
num_to_run = self.nsamplesAdded * self.N - self.nsamplesRun * self.N
data = self.synth_data[self.nsamplesRun*self.N:self.nsamplesAdded*self.N,:]
if self.keep_index:
index = self.synth_data_index[self.nsamplesRun*self.N:self.nsamplesAdded*self.N]
index = pd.DataFrame(index, columns=[self.data.index_name])
data = pd.DataFrame(data, columns=self.data.group_names)
data = pd.concat([index, data], axis=1).set_index(self.data.index_name)
if self.keep_index_ordered:
data = data.sort_index()
if self.isRNN is True:
# convert the data to be three-dimensional, considering each
# synthetic sample as a separate sequence
data = torch.from_numpy(data).float().unsqueeze(1)
if self.isBidir is False:
# get the current hidden state, if given
hidden_state = kwargs.get('hidden_state', None)
if hidden_state is not None:
# repeat the hidden state along the batch dimension
if isinstance(hidden_state, torch.Tensor):
hidden_state = hidden_state.repeat(1, data.shape[0], 1)
else:
hidden_state_0 = hidden_state[0].repeat(1, data.shape[0], 1)
hidden_state_1 = hidden_state[1].repeat(1, data.shape[0], 1)
hidden_state = (hidden_state_0, hidden_state_1)
modelOut = self.model.f(data, hidden_state)
else:
# get the current past data, if given
past_data = kwargs.get('past_data', None)
if past_data is not None:
# convert the data to be three-dimensional float tensor and remove ID columns
past_data = past_data[:, 2:].unsqueeze(0).float()
# repeat the past data along the batch dimension
past_data = past_data.repeat(data.shape[0], 1, 1)
# add the previous instances from the same sequence, if there are any
seq_data = torch.cat((past_data, data), dim=1)
else:
seq_data = data
modelOut = self.model.f(seq_data)
if past_data is not None:
# only get the outputs from the last sample, ignoring the previous instances
modelOut = modelOut.reshape([-1, past_data.shape[1]+1, 1])
modelOut = modelOut[:, -1, :]
else:
modelOut = self.model.f(data)
if isinstance(modelOut, (pd.DataFrame, pd.Series)):
modelOut = modelOut.values
self.y[self.nsamplesRun * self.N:self.nsamplesAdded * self.N, :] = np.reshape(modelOut, (num_to_run, self.D))
# find the expected value of each output
for i in range(self.nsamplesRun, self.nsamplesAdded):
eyVal = np.zeros(self.D)
for j in range(0, self.N):
eyVal += self.y[i * self.N + j, :] * self.weights[j]
self.ey[i, :] = eyVal
self.nsamplesRun += 1
def solve(self, fraction_evaluated, dim):
eyAdj = self.linkfv(self.ey[:, dim]) - self.link.f(self.fnull[dim])
s = np.sum(self.maskMatrix, 1)
# do feature selection if we have not well enumerated the space
nonzero_inds = np.arange(self.M)
log.debug("fraction_evaluated = {0}".format(fraction_evaluated))
if self.l1_reg == "auto":
warnings.warn(
"l1_reg=\"auto\" is deprecated and in the next version (v0.29) the behavior will change from a " \
"conditional use of AIC to simply \"num_features(10)\"!"
)
if (self.l1_reg not in ["auto", False, 0]) or (fraction_evaluated < 0.2 and self.l1_reg == "auto"):
w_aug = np.hstack((self.kernelWeights * (self.M - s), self.kernelWeights * s))
log.info("np.sum(w_aug) = {0}".format(np.sum(w_aug)))
log.info("np.sum(self.kernelWeights) = {0}".format(np.sum(self.kernelWeights)))
w_sqrt_aug = np.sqrt(w_aug)
eyAdj_aug = np.hstack((eyAdj, eyAdj - (self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim]))))
eyAdj_aug *= w_sqrt_aug
mask_aug = np.transpose(w_sqrt_aug * np.transpose(np.vstack((self.maskMatrix, self.maskMatrix - 1))))
#var_norms = np.array([np.linalg.norm(mask_aug[:, i]) for i in range(mask_aug.shape[1])])
# select a fixed number of top features
if isinstance(self.l1_reg, str) and self.l1_reg.startswith("num_features("):
r = int(self.l1_reg[len("num_features("):-1])
nonzero_inds = lars_path(mask_aug, eyAdj_aug, max_iter=r)[1]
# use an adaptive regularization method
elif self.l1_reg == "auto" or self.l1_reg == "bic" or self.l1_reg == "aic":
c = "aic" if self.l1_reg == "auto" else self.l1_reg
nonzero_inds = np.nonzero(LassoLarsIC(criterion=c).fit(mask_aug, eyAdj_aug).coef_)[0]
# use a fixed regularization coeffcient
else:
nonzero_inds = np.nonzero(Lasso(alpha=self.l1_reg).fit(mask_aug, eyAdj_aug).coef_)[0]
if len(nonzero_inds) == 0:
return np.zeros(self.M), np.ones(self.M)
# eliminate one variable with the constraint that all features sum to the output
eyAdj2 = eyAdj - self.maskMatrix[:, nonzero_inds[-1]] * (
self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim]))
etmp = np.transpose(np.transpose(self.maskMatrix[:, nonzero_inds[:-1]]) - self.maskMatrix[:, nonzero_inds[-1]])
log.debug("etmp[:4,:] {0}".format(etmp[:4, :]))
# solve a weighted least squares equation to estimate phi
tmp = np.transpose(np.transpose(etmp) * np.transpose(self.kernelWeights))
tmp2 = np.linalg.inv(np.dot(np.transpose(tmp), etmp))
w = np.dot(tmp2, np.dot(np.transpose(tmp), eyAdj2))
log.debug("np.sum(w) = {0}".format(np.sum(w)))
log.debug("self.link(self.fx) - self.link(self.fnull) = {0}".format(
self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim])))
log.debug("self.fx = {0}".format(self.fx[dim]))
log.debug("self.link(self.fx) = {0}".format(self.link.f(self.fx[dim])))
log.debug("self.fnull = {0}".format(self.fnull[dim]))
log.debug("self.link(self.fnull) = {0}".format(self.link.f(self.fnull[dim])))
phi = np.zeros(self.M)
phi[nonzero_inds[:-1]] = w
phi[nonzero_inds[-1]] = (self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim])) - sum(w)
log.info("phi = {0}".format(phi))
# clean up any rounding errors
for i in range(self.M):
if np.abs(phi[i]) < 1e-10:
phi[i] = 0
return phi, np.ones(len(phi))— Started going sequence-by-sequence, instead of sample-by-sample;
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
To fix this, I adapted Kernel SHAP's code with the following main changes:
— Added parameters that identified the model type, including when it was RNN;
from sklearn.impute import SimpleImputer
from ..common import convert_to_instance, convert_to_model, match_instance_to_data, match_model_to_data, convert_to_instance_with_index, convert_to_link, IdentityLink, convert_to_data, DenseData, SparseData
from scipy.special import binom
from scipy.sparse import issparse
import numpy as np
import pandas as pd
import scipy as sp
import logging
import copy
import itertools
import warnings
from sklearn.linear_model import LassoLarsIC, Lasso, lars_path
from sklearn.cluster import KMeans
from tqdm.auto import tqdm
from .explainer import Explainer
import torch
log = logging.getLogger('shap')
def kmeans(X, k, round_values=True):
""" Summarize a dataset with k mean samples weighted by the number of data points they
each represent.
Parameters
----------
X : numpy.array or pandas.DataFrame or any scipy.sparse matrix
Matrix of data samples to summarize (# samples x # features)
k : int
Number of means to use for approximation.
round_values : bool
For all i, round the ith dimension of each mean sample to match the nearest value
from X[:,i]. This ensures discrete features always get a valid value.
Returns
-------
DenseData object.
"""
group_names = [str(i) for i in range(X.shape[1])]
if str(type(X)).endswith("'pandas.core.frame.DataFrame'>"):
group_names = X.columns
X = X.values
# in case there are any missing values in data impute them
imp = SimpleImputer(missing_values=np.nan, strategy='mean')
X = imp.fit_transform(X)
kmeans = KMeans(n_clusters=k, random_state=0).fit(X)
if round_values:
for i in range(k):
for j in range(X.shape[1]):
xj = X[:,j].toarray().flatten() if issparse(X) else X[:, j]
ind = np.argmin(np.abs(xj - kmeans.cluster_centers_[i,j]))
kmeans.cluster_centers_[i,j] = X[ind,j]
return DenseData(kmeans.cluster_centers_, group_names, None, 1.0*np.bincount(kmeans.labels_))
class KernelExplainer(Explainer):
"""Uses the Kernel SHAP method to explain the output of any function.
Kernel SHAP is a method that uses a special weighted linear regression
to compute the importance of each feature. The computed importance values
are Shapley values from game theory and also coefficents from a local linear
regression.
Parameters
----------
model : function or iml.Model
User supplied function that takes a matrix of samples (# samples x # features) and
computes the output of the model for those samples. The output can be a vector
(# samples) or a matrix (# samples x # model outputs).
data : numpy.array or pandas.DataFrame or shap.common.DenseData or any scipy.sparse matrix
The background dataset to use for integrating out features. To determine the impact
of a feature, that feature is set to "missing" and the change in the model output
is observed. Since most models aren't designed to handle arbitrary missing data at test
time, we simulate "missing" by replacing the feature with the values it takes in the
background dataset. So if the background dataset is a simple sample of all zeros, then
we would approximate a feature being missing by setting it to zero. For small problems
this background dataset can be the whole training set, but for larger problems consider
using a single reference value or using the kmeans function to summarize the dataset.
Note: for sparse case we accept any sparse matrix but convert to lil format for
performance.
link : "identity" or "logit"
A generalized linear model link to connect the feature importance values to the model
output. Since the feature importance values, phi, sum up to the model output, it often makes
sense to connect them to the output with a link function where link(output) = sum(phi).
If the model output is a probability then the LogitLink link function makes the feature
importance values have log-odds units.
isRNN : bool
Boolean that indicates if the model being analyzed is a recurrent neural network (RNN).
If so, it means that sequential data is being used, which requires some modifications
in the way SHAP values are calculated.
if isRNN is True:
id_col_num : int
Number that indicates in which column is the sequence / subject id. Defaults to 0.
ts_col_num : int
Number that indicates in which column is the instance / timestamp. Defaults to 1.
label_col_num : int
Number that indicates in which column is the label, if any. Defaults to None.
recur_layer : torch.nn.LSTM or torch.nn.GRU or torch.nn.RNN, default None
Pointer to the recurrent layer in the model, if it exists. It should
either be a LSTM, GRU or RNN network. If none is specified, the
method will automatically search for a recurrent layer in the model.
isBidir: bool
Boolean that indicates if the recurrent neural network model being analyzed is
bidirectional. If so, it implies a special treatment of the sequences, as the
hidden states can't be moved in the same way of a single direction RNN.
padding_value : numeric
Value to use in the padding, to fill the sequences.
"""
def __init__(self, model, data, link=IdentityLink(), **kwargs):
# convert incoming inputs to standardized iml objects
self.link = convert_to_link(link)
self.model = convert_to_model(model)
self.keep_index = kwargs.get("keep_index", False)
self.keep_index_ordered = kwargs.get("keep_index_ordered", False)
# check if the model is a recurrent neural network
self.isRNN = kwargs.get('isRNN', False)
if self.isRNN and not str(type(data)).endswith("'pandas.core.frame.DataFrame'>"):
# check if the model is a bidirectional recurrent neural network
self.isBidir = kwargs.get('isBidir', False)
# number of the column that corresponds to the sequence / subject id
self.id_col_num = kwargs.get('id_col_num', 0)
# number of the column that corresponds to the instance / timestamp
self.ts_col_num = kwargs.get('ts_col_num', 1)
# number of the column that corresponds to the label
label_col_num = kwargs.get('label_col_num', None)
# padding value
self.padding_value = kwargs.get('padding_value', 999999)
# all columns in the data
self.model_features = list(range(data.shape[1]))
# remove unwanted columns, so that we get only those that actually correspond to model usable features
[self.model_features.remove(col) for col in [self.id_col_num, self.ts_col_num, label_col_num] if col is not None]
# maximum background samples to use
self.max_bkgnd_samples = kwargs.get('max_bkgnd_samples', 20)
if data.shape[0] > self.max_bkgnd_samples:
# use k-means to avoid slow processing of a lot of samples
self.data = kmeans(data[:, self.model_features], k=self.max_bkgnd_samples)
# get the weights corresponding to all the original data
num_samples = data.shape[0]
self.weights = np.ones(num_samples)
self.weights /= np.sum(self.weights)
else:
self.data = convert_to_data(data[:, self.model_features], keep_index=self.keep_index)
self.weights = self.data.weights
# check if the recurrent layer is specified
self.recur_layer = kwargs.get('recur_layer', None)
if self.recur_layer is None:
# get the model object, so as to use its recurrent layer
model_obj = kwargs.get('model_obj', None)
assert model_obj is not None, 'If the model uses a recurrent neural network, either the recurrent layer or the full model object must be specified.'
# search for a recurrent layer
if hasattr(model_obj, 'lstm'):
self.recur_layer = model_obj.lstm
elif hasattr(model_obj, 'gru'):
self.recur_layer = model_obj.gru
elif hasattr(model_obj, 'rnn'):
self.recur_layer = model_obj.rnn
else:
raise Exception('ERROR: No recurrent layer found. Please specify it in the recur_layer argument.')
# get the unique subject ID's in the background data
self.subject_ids = np.unique(data[:, self.id_col_num]).astype(int)
# maximum sequence length in the background data
self.max_seq_len = kwargs.get('max_seq_len', None)
if self.max_seq_len == None:
self.max_seq_len = 1
for id in self.subject_ids:
seq_data = data[np.where((data[:, self.id_col_num] == id))]
cur_seq_length = len(seq_data)
if cur_seq_length > self.max_seq_len:
self.max_seq_len = cur_seq_length
# calculate the output for all the background data
model_null = match_model_to_data(self.model, data, self.isRNN, self.model_features,
self.id_col_num, self.ts_col_num, self.recur_layer,
self.subject_ids, self.max_seq_len, self.model.f,
silent=kwargs.get("silent", False))
else:
self.data = convert_to_data(data, keep_index=self.keep_index)
self.weights = self.data.weights
# calculate the output for all the background data
model_null = match_model_to_data(self.model, self.data)
self.col_names = None
if str(type(data)).endswith("'pandas.core.frame.DataFrame'>"):
# keep the column names so that data can be used in dataframe format
self.col_names = data.columns
# enforce our current input type limitations
assert isinstance(self.data, DenseData) or isinstance(self.data, SparseData), \
"Shap explainer only supports the DenseData and SparseData input currently."
assert not self.data.transposed, "Shap explainer does not support transposed DenseData or SparseData currently."
# warn users about large background data sets
if len(self.data.weights) > 100:
log.warning("Using " + str(len(self.data.weights)) + " background data samples could cause " +
"slower run times. Consider using shap.sample(data, K) or shap.kmeans(data, K) to " +
"summarize the background as K samples.")
# init our parameters
self.N = self.data.data.shape[0]
self.P = self.data.data.shape[1]
self.linkfv = np.vectorize(self.link.f)
self.nsamplesAdded = 0
self.nsamplesRun = 0
# find E_x[f(x)]
if isinstance(model_null, (pd.DataFrame, pd.Series)):
model_null = np.squeeze(model_null.values)
self.fnull = np.sum((model_null.T * self.weights).T, 0)
self.expected_value = self.linkfv(self.fnull)
# see if we have a vector output
self.vector_out = True
if len(self.fnull.shape) == 0:
self.vector_out = False
self.fnull = np.array([self.fnull])
self.D = 1
self.expected_value = float(self.expected_value)
else:
self.D = self.fnull.shape[0]
def shap_values(self, X, **kwargs):
""" Estimate the SHAP values for a set of samples.
Parameters
----------
X : numpy.array or pandas.DataFrame or any scipy.sparse matrix
A matrix of samples (# samples x # features) on which to explain the model's output.
nsamples : "auto" or int
Number of times to re-evaluate the model when explaining each prediction. More samples
lead to lower variance estimates of the SHAP values. The "auto" setting uses
`nsamples = 2 * X.shape[1] + 2048`.
l1_reg : "num_features(int)", "auto" (default for now, but deprecated), "aic", "bic", or float
The l1 regularization to use for feature selection (the estimation procedure is based on
a debiased lasso). The auto option currently uses "aic" when less that 20% of the possible sample
space is enumerated, otherwise it uses no regularization. THE BEHAVIOR OF "auto" WILL CHANGE
in a future version to be based on num_features instead of AIC.
The "aic" and "bic" options use the AIC and BIC rules for regularization.
Using "num_features(int)" selects a fix number of top features. Passing a float directly sets the
"alpha" parameter of the sklearn.linear_model.Lasso model used for feature selection.
Returns
-------
For models with a single output this returns a matrix of SHAP values
(# samples x # features). Each row sums to the difference between the model output for that
sample and the expected value of the model output (which is stored as expected_value
attribute of the explainer). For models with vector outputs this returns a list
of such matrices, one for each output.
"""
# convert dataframes
if str(type(X)).endswith("pandas.core.series.Series'>"):
X = X.values
elif str(type(X)).endswith("'pandas.core.frame.DataFrame'>"):
if self.keep_index:
index_value = X.index.values
index_name = X.index.name
column_name = list(X.columns)
X = X.values
x_type = str(type(X))
arr_type = "'numpy.ndarray'>"
# if sparse, convert to lil for performance
if sp.sparse.issparse(X) and not sp.sparse.isspmatrix_lil(X):
X = X.tolil()
assert x_type.endswith(arr_type) or sp.sparse.isspmatrix_lil(X), "Unknown instance type: " + x_type
assert len(X.shape) == 1 or len(X.shape) == 2 or len(X.shape) == 3, "Instance must have 1, 2 or 3 dimensions!"
if self.isRNN:
# get the unique subject ID's in the test data, in the original order
self.subject_ids, indeces = np.unique(X[:, self.id_col_num], return_index=True)
sorted_idx = np.argsort(indeces)
self.subject_ids = self.subject_ids[sorted_idx].astype(int)
# Remove paddings
self.subject_ids = self.subject_ids[self.subject_ids != self.padding_value]
ts_values = X[:, self.ts_col_num]
ts_values = ts_values[ts_values != self.padding_value]
# maximum sequence length in the test data
max_seq_len = kwargs.get('max_seq_len', None)
if max_seq_len == None:
max_seq_len = 1
for id in self.subject_ids:
seq_data = X[np.where((X[:, self.id_col_num] == id))]
cur_seq_length = len(seq_data)
if cur_seq_length > max_seq_len:
max_seq_len = cur_seq_length
# compare with the maximum sequence length in the background data
if max_seq_len > self.max_seq_len:
# update maximum sequence length
self.max_seq_len = max_seq_len
explanations = np.zeros((len(self.subject_ids), self.max_seq_len, len(self.model_features)))
# count the order of the sequences being iterated
seq_count = 0
# loop through the unique subject ID's
for id in tqdm(self.subject_ids, disable=kwargs.get("silent", False), desc='ID loop'):
# get the data corresponding to the current sequence
seq_data = X[X[:, self.id_col_num] == id]
# get the unique timestamp (or instance index) values of the current sequence
seq_unique_ts = np.unique(seq_data[:, self.ts_col_num]).astype(int)
# count the order of the instances being iterated
ts_count = 0
if self.isBidir is True:
# calculate the full sequence's outputs
torch_seq_data = torch.from_numpy(seq_data[:, self.model_features]).unsqueeze(0).float()
seq_outputs, _ = self.recur_layer(torch_seq_data)
# loop through the possible instances
for ts in tqdm(seq_unique_ts, disable=kwargs.get("silent", False), desc='ts loop', leave=False):
# get the data corresponding to the current instance
inst_data = seq_data[seq_data[:, self.ts_col_num] == ts]
# remove unwanted features (id, ts and label)
inst_data = inst_data[:, self.model_features]
# get the hidden state that the model receives as an input
if ts > 0:
# data from the previous instance(s) in the same sequence
past_data = torch.from_numpy(seq_data[np.where(seq_data[:, self.ts_col_num] < ts)])
if self.isBidir is False:
hidden_state = None
# convert the past data to a 3D tensor
past_data = past_data.unsqueeze(0)
# get the hidden state outputed from the previous recurrent cell
_, hidden_state = self.recur_layer(past_data[:, :, self.model_features].float())
# avoid passing gradients from previous instances
if isinstance(hidden_state, tuple) or isinstance(hidden_state, list):
if isinstance(hidden_state[0], tuple) or isinstance(hidden_state[0], list):
hidden_state = [(hidden_state[i][0].detach(), hidden_state[i][1].detach())
for i in range(len(hidden_state))]
else:
hidden_state = (hidden_state[0].detach(), hidden_state[1].detach())
else:
hidden_state.detach_()
# add the hidden_state to the kwargs
kwargs['hidden_state'] = hidden_state
else:
# add the past_data to the kwargs
kwargs['past_data'] = past_data
if self.isBidir is True:
# add the current instance's output to the kwargs
kwargs['inst_output'] = seq_outputs[ts_count].unsqueeze(0).detach().numpy()
if self.keep_index:
inst_data = convert_to_instance_with_index(inst_data, column_name, seq_count * self.max_seq_len + ts, index_name)
explanations[seq_count, ts_count, :] = self.explain(inst_data, **kwargs).squeeze()
ts_count += 1
seq_count += 1
return explanations
# [TODO] Don't understand the need for this out variable
# # vector-output
# s = explanations[0][0].shape
# if len(s) == 2:
# outs = [np.zeros((X.shape[0], X.shape[1], s[0])) for j in range(s[1])]
# for i in range(X.shape[0]):
# for j in range(X.shape[1]):
# for k in range(s[1]):
# outs[k][i][j] = explanations[i][j][:, k]
# return outs
#
# # single-output
# else:
# out = np.zeros((X.shape[0], X.shape[1], s[0]))
# for i in range(X.shape[0]):
# for j in range(X.shape[1]):
# out[i][j] = explanations[i][j]
# return out
# single instance
elif len(X.shape) == 1:
data = X.reshape((1, X.shape[0]))
if self.keep_index:
data = convert_to_instance_with_index(data, column_name, index_name, index_value)
explanation = self.explain(data, **kwargs)
# vector-output
s = explanation.shape
if len(s) == 2:
outs = [np.zeros(s[0]) for j in range(s[1])]
for j in range(s[1]):
outs[j] = explanation[:, j]
return outs
# single-output
else:
out = np.zeros(s[0])
out[:] = explanation
return out
# explain the whole dataset
elif len(X.shape) == 2:
explanations = []
for i in tqdm(range(X.shape[0]), disable=kwargs.get("silent", False)):
data = X[i:i + 1, :]
if self.keep_index:
data = convert_to_instance_with_index(data, column_name, index_value[i:i + 1], index_name)
explanations.append(self.explain(data, **kwargs))
# vector-output
s = explanations[0].shape
if len(s) == 2:
outs = [np.zeros((X.shape[0], s[0])) for j in range(s[1])]
for i in range(X.shape[0]):
for j in range(s[1]):
outs[j][i] = explanations[i][:, j]
return outs
# single-output
else:
out = np.zeros((X.shape[0], s[0]))
for i in range(X.shape[0]):
out[i] = explanations[i]
return out
def explain(self, incoming_instance, **kwargs):
# convert incoming input to a standardized iml object
instance = convert_to_instance(incoming_instance)
match_instance_to_data(instance, self.data)
# find the feature groups we will test. If a feature does not change from its
# current value then we know it doesn't impact the model
self.varyingInds = self.varying_groups(instance.x)
if self.data.groups is None:
self.varyingFeatureGroups = np.array([i for i in self.varyingInds])
self.M = self.varyingFeatureGroups.shape[0]
else:
self.varyingFeatureGroups = [self.data.groups[i] for i in self.varyingInds]
self.M = len(self.varyingFeatureGroups)
groups = self.data.groups
# convert to numpy array as it is much faster if not jagged array (all groups of same length)
if self.varyingFeatureGroups and all(len(groups[i]) == len(groups[0]) for i in self.varyingInds):
self.varyingFeatureGroups = np.array(self.varyingFeatureGroups)
# further performance optimization in case each group has a single value
if self.varyingFeatureGroups.shape[1] == 1:
self.varyingFeatureGroups = self.varyingFeatureGroups.flatten()
# get the current hidden state, if given
hidden_state = kwargs.get('hidden_state', None)
# get the current past data, if given
past_data = kwargs.get('past_data', None)
# get the current instances' output, if given
inst_output = kwargs.get('inst_output', None)
# find f(x)
if self.keep_index:
if self.isRNN is True:
if self.isBidir is False:
# only provide the hidden state argument if the model is a RNN type model
model_out = self.model.f(instance.convert_to_df(), hidden_state)
else:
model_out = inst_output
else:
model_out = self.model.f(instance.convert_to_df())
else:
if self.isRNN is True:
if self.isBidir is False:
# only provide the hidden state argument if the model is a RNN type model
model_out = self.model.f(instance.x, hidden_state)
else:
model_out = inst_output
else:
model_out = self.model.f(instance.x)
if isinstance(model_out, (pd.DataFrame, pd.Series)):
model_out = model_out.values
self.fx = model_out[0]
if not self.vector_out:
self.fx = np.array([self.fx])
# if no features vary then no feature has an effect
if self.M == 0:
phi = np.zeros((self.data.groups_size, self.D))
phi_var = np.zeros((self.data.groups_size, self.D))
# if only one feature varies then it has all the effect
elif self.M == 1:
phi = np.zeros((self.data.groups_size, self.D))
phi_var = np.zeros((self.data.groups_size, self.D))
diff = self.link.f(self.fx) - self.link.f(self.fnull)
for d in range(self.D):
phi[self.varyingInds[0],d] = diff[d]
# if more than one feature varies then we have to do real work
else:
self.l1_reg = kwargs.get("l1_reg", "auto")
# pick a reasonable number of samples if the user didn't specify how many they wanted
self.nsamples = kwargs.get("nsamples", "auto")
if self.nsamples == "auto":
self.nsamples = 2 * self.M + 2**11
# if we have enough samples to enumerate all subsets then ignore the unneeded samples
self.max_samples = 2 ** 30
if self.M <= 30:
self.max_samples = 2 ** self.M - 2
if self.nsamples > self.max_samples:
self.nsamples = self.max_samples
# reserve space for some of our computations
self.allocate()
# weight the different subset sizes
num_subset_sizes = np.int(np.ceil((self.M - 1) / 2.0))
num_paired_subset_sizes = np.int(np.floor((self.M - 1) / 2.0))
weight_vector = np.array([(self.M - 1.0) / (i * (self.M - i)) for i in range(1, num_subset_sizes + 1)])
weight_vector[:num_paired_subset_sizes] *= 2
weight_vector /= np.sum(weight_vector)
log.debug("weight_vector = {0}".format(weight_vector))
log.debug("num_subset_sizes = {0}".format(num_subset_sizes))
log.debug("num_paired_subset_sizes = {0}".format(num_paired_subset_sizes))
log.debug("M = {0}".format(self.M))
# fill out all the subset sizes we can completely enumerate
# given nsamples*remaining_weight_vector[subset_size]
num_full_subsets = 0
num_samples_left = self.nsamples
group_inds = np.arange(self.M, dtype='int64')
mask = np.zeros(self.M)
remaining_weight_vector = copy.copy(weight_vector)
for subset_size in range(1, num_subset_sizes + 1):
# determine how many subsets (and their complements) are of the current size
nsubsets = binom(self.M, subset_size)
if subset_size <= num_paired_subset_sizes: nsubsets *= 2
log.debug("subset_size = {0}".format(subset_size))
log.debug("nsubsets = {0}".format(nsubsets))
log.debug("self.nsamples*weight_vector[subset_size-1] = {0}".format(
num_samples_left * remaining_weight_vector[subset_size - 1]))
log.debug("self.nsamples*weight_vector[subset_size-1]/nsubsets = {0}".format(
num_samples_left * remaining_weight_vector[subset_size - 1] / nsubsets))
# see if we have enough samples to enumerate all subsets of this size
if num_samples_left * remaining_weight_vector[subset_size - 1] / nsubsets >= 1.0 - 1e-8:
num_full_subsets += 1
num_samples_left -= nsubsets
# rescale what's left of the remaining weight vector to sum to 1
if remaining_weight_vector[subset_size - 1] < 1.0:
remaining_weight_vector /= (1 - remaining_weight_vector[subset_size - 1])
# add all the samples of the current subset size
w = weight_vector[subset_size - 1] / binom(self.M, subset_size)
if subset_size <= num_paired_subset_sizes: w /= 2.0
for inds in itertools.combinations(group_inds, subset_size):
mask[:] = 0.0
mask[np.array(inds, dtype='int64')] = 1.0
self.addsample(instance.x, mask, w)
if subset_size <= num_paired_subset_sizes:
mask[:] = np.abs(mask - 1)
self.addsample(instance.x, mask, w)
else:
break
log.info("num_full_subsets = {0}".format(num_full_subsets))
# add random samples from what is left of the subset space
nfixed_samples = self.nsamplesAdded
samples_left = self.nsamples - self.nsamplesAdded
log.debug("samples_left = {0}".format(samples_left))
if num_full_subsets != num_subset_sizes:
remaining_weight_vector = copy.copy(weight_vector)
remaining_weight_vector[:num_paired_subset_sizes] /= 2 # because we draw two samples each below
remaining_weight_vector = remaining_weight_vector[num_full_subsets:]
remaining_weight_vector /= np.sum(remaining_weight_vector)
log.info("remaining_weight_vector = {0}".format(remaining_weight_vector))
log.info("num_paired_subset_sizes = {0}".format(num_paired_subset_sizes))
ind_set = np.random.choice(len(remaining_weight_vector), 4 * samples_left, p=remaining_weight_vector)
ind_set_pos = 0
used_masks = {}
while samples_left > 0 and ind_set_pos < len(ind_set):
mask.fill(0.0)
ind = ind_set[ind_set_pos] # we call np.random.choice once to save time and then just read it here
ind_set_pos += 1
subset_size = ind + num_full_subsets + 1
mask[np.random.permutation(self.M)[:subset_size]] = 1.0
# only add the sample if we have not seen it before, otherwise just
# increment a previous sample's weight
mask_tuple = tuple(mask)
new_sample = False
if mask_tuple not in used_masks:
new_sample = True
used_masks[mask_tuple] = self.nsamplesAdded
samples_left -= 1
self.addsample(instance.x, mask, 1.0)
else:
self.kernelWeights[used_masks[mask_tuple]] += 1.0
# add the compliment sample
if samples_left > 0 and subset_size <= num_paired_subset_sizes:
mask[:] = np.abs(mask - 1)
# only add the sample if we have not seen it before, otherwise just
# increment a previous sample's weight
if new_sample:
samples_left -= 1
self.addsample(instance.x, mask, 1.0)
else:
# we know the compliment sample is the next one after the original sample, so + 1
self.kernelWeights[used_masks[mask_tuple] + 1] += 1.0
# normalize the kernel weights for the random samples to equal the weight left after
# the fixed enumerated samples have been already counted
weight_left = np.sum(weight_vector[num_full_subsets:])
log.info("weight_left = {0}".format(weight_left))
self.kernelWeights[nfixed_samples:] *= weight_left / self.kernelWeights[nfixed_samples:].sum()
# execute the model on the synthetic samples we have created
self.run(**kwargs)
# solve then expand the feature importance (Shapley value) vector to contain the non-varying features
phi = np.zeros((self.data.groups_size, self.D))
phi_var = np.zeros((self.data.groups_size, self.D))
for d in range(self.D):
vphi, vphi_var = self.solve(self.nsamples / self.max_samples, d)
phi[self.varyingInds, d] = vphi
phi_var[self.varyingInds, d] = vphi_var
if not self.vector_out:
phi = np.squeeze(phi, axis=1)
phi_var = np.squeeze(phi_var, axis=1)
return phi
def varying_groups(self, x):
if not sp.sparse.issparse(x):
varying = np.zeros(self.data.groups_size)
for i in range(0, self.data.groups_size):
inds = self.data.groups[i]
x_group = x[0, inds]
if sp.sparse.issparse(x_group):
if all(j not in x.nonzero()[1] for j in inds):
varying[i] = False
continue
x_group = x_group.todense()
num_mismatches = np.sum(np.invert(np.isclose(x_group, self.data.data[:, inds], equal_nan=True)))
varying[i] = num_mismatches > 0
varying_indices = np.nonzero(varying)[0]
return varying_indices
else:
varying_indices = []
# go over all nonzero columns in background and evaluation data
# if both background and evaluation are zero, the column does not vary
varying_indices = np.unique(np.union1d(self.data.data.nonzero()[1], x.nonzero()[1]))
remove_unvarying_indices = []
for i in range(0, len(varying_indices)):
varying_index = varying_indices[i]
# now verify the nonzero values do vary
data_rows = self.data.data[:, [varying_index]]
nonzero_rows = data_rows.nonzero()[0]
if nonzero_rows.size > 0:
background_data_rows = data_rows[nonzero_rows]
if sp.sparse.issparse(background_data_rows):
background_data_rows = background_data_rows.toarray()
num_mismatches = np.sum(np.abs(background_data_rows - x[0, varying_index]) > 1e-7)
# Note: If feature column non-zero but some background zero, can't remove index
if num_mismatches == 0 and not \
(np.abs(x[0, [varying_index]][0, 0]) > 1e-7 and len(nonzero_rows) < data_rows.shape[0]):
remove_unvarying_indices.append(i)
mask = np.ones(len(varying_indices), dtype=bool)
mask[remove_unvarying_indices] = False
varying_indices = varying_indices[mask]
return varying_indices
def allocate(self):
if sp.sparse.issparse(self.data.data):
# We tile the sparse matrix in csr format but convert it to lil
# for performance when adding samples
shape = self.data.data.shape
nnz = self.data.data.nnz
data_rows, data_cols = shape
rows = data_rows * self.nsamples
shape = rows, data_cols
if nnz == 0:
self.synth_data = sp.sparse.csr_matrix(shape, dtype=self.data.data.dtype).tolil()
else:
data = self.data.data.data
indices = self.data.data.indices
indptr = self.data.data.indptr
last_indptr_idx = indptr[len(indptr) - 1]
indptr_wo_last = indptr[:-1]
new_indptrs = []
for i in range(0, self.nsamples - 1):
new_indptrs.append(indptr_wo_last + (i * last_indptr_idx))
new_indptrs.append(indptr + ((self.nsamples - 1) * last_indptr_idx))
new_indptr = np.concatenate(new_indptrs)
new_data = np.tile(data, self.nsamples)
new_indices = np.tile(indices, self.nsamples)
self.synth_data = sp.sparse.csr_matrix((new_data, new_indices, new_indptr), shape=shape).tolil()
else:
self.synth_data = np.tile(self.data.data, (self.nsamples, 1))
self.maskMatrix = np.zeros((self.nsamples, self.M))
self.kernelWeights = np.zeros(self.nsamples)
self.y = np.zeros((self.nsamples * self.N, self.D))
self.ey = np.zeros((self.nsamples, self.D))
self.lastMask = np.zeros(self.nsamples)
self.nsamplesAdded = 0
self.nsamplesRun = 0
if self.keep_index:
self.synth_data_index = np.tile(self.data.index_value, self.nsamples)
def addsample(self, x, m, w):
offset = self.nsamplesAdded * self.N
if isinstance(self.varyingFeatureGroups, (list,)):
for j in range(self.M):
for k in self.varyingFeatureGroups[j]:
if m[j] == 1.0:
self.synth_data[offset:offset+self.N, k] = x[0, k]
else:
# for non-jagged numpy array we can significantly boost performance
mask = m == 1.0
groups = self.varyingFeatureGroups[mask]
if len(groups.shape) == 2:
for group in groups:
self.synth_data[offset:offset+self.N, group] = x[0, group]
else:
# further performance optimization in case each group has a single feature
evaluation_data = x[0, groups]
# In edge case where background is all dense but evaluation data
# is all sparse, make evaluation data dense
if sp.sparse.issparse(x) and not sp.sparse.issparse(self.synth_data):
evaluation_data = evaluation_data.toarray()
self.synth_data[offset:offset+self.N, groups] = evaluation_data
self.maskMatrix[self.nsamplesAdded, :] = m
self.kernelWeights[self.nsamplesAdded] = w
self.nsamplesAdded += 1
def run(self, **kwargs):
# [TODO] The inefficiency issue is probably derived from here. It seems to want to run the requested number of samples TIMES the TOTAL number of background samples!
num_to_run = self.nsamplesAdded * self.N - self.nsamplesRun * self.N
data = self.synth_data[self.nsamplesRun*self.N:self.nsamplesAdded*self.N,:]
if self.keep_index:
index = self.synth_data_index[self.nsamplesRun*self.N:self.nsamplesAdded*self.N]
index = pd.DataFrame(index, columns=[self.data.index_name])
data = pd.DataFrame(data, columns=self.data.group_names)
data = pd.concat([index, data], axis=1).set_index(self.data.index_name)
if self.keep_index_ordered:
data = data.sort_index()
if self.isRNN is True:
# convert the data to be three-dimensional, considering each
# synthetic sample as a separate sequence
data = torch.from_numpy(data).float().unsqueeze(1)
if self.isBidir is False:
# get the current hidden state, if given
hidden_state = kwargs.get('hidden_state', None)
if hidden_state is not None:
# repeat the hidden state along the batch dimension
if isinstance(hidden_state, torch.Tensor):
hidden_state = hidden_state.repeat(1, data.shape[0], 1)
else:
hidden_state_0 = hidden_state[0].repeat(1, data.shape[0], 1)
hidden_state_1 = hidden_state[1].repeat(1, data.shape[0], 1)
hidden_state = (hidden_state_0, hidden_state_1)
modelOut = self.model.f(data, hidden_state)
else:
# get the current past data, if given
past_data = kwargs.get('past_data', None)
if past_data is not None:
# convert the data to be three-dimensional float tensor and remove ID columns
past_data = past_data[:, 2:].unsqueeze(0).float()
# repeat the past data along the batch dimension
past_data = past_data.repeat(data.shape[0], 1, 1)
# add the previous instances from the same sequence, if there are any
seq_data = torch.cat((past_data, data), dim=1)
else:
seq_data = data
modelOut = self.model.f(seq_data)
if past_data is not None:
# only get the outputs from the last sample, ignoring the previous instances
modelOut = modelOut.reshape([-1, past_data.shape[1]+1, 1])
modelOut = modelOut[:, -1, :]
else:
modelOut = self.model.f(data)
if isinstance(modelOut, (pd.DataFrame, pd.Series)):
modelOut = modelOut.values
self.y[self.nsamplesRun * self.N:self.nsamplesAdded * self.N, :] = np.reshape(modelOut, (num_to_run, self.D))
# find the expected value of each output
for i in range(self.nsamplesRun, self.nsamplesAdded):
eyVal = np.zeros(self.D)
for j in range(0, self.N):
eyVal += self.y[i * self.N + j, :] * self.weights[j]
self.ey[i, :] = eyVal
self.nsamplesRun += 1
def solve(self, fraction_evaluated, dim):
eyAdj = self.linkfv(self.ey[:, dim]) - self.link.f(self.fnull[dim])
s = np.sum(self.maskMatrix, 1)
# do feature selection if we have not well enumerated the space
nonzero_inds = np.arange(self.M)
log.debug("fraction_evaluated = {0}".format(fraction_evaluated))
if self.l1_reg == "auto":
warnings.warn(
"l1_reg=\"auto\" is deprecated and in the next version (v0.29) the behavior will change from a " \
"conditional use of AIC to simply \"num_features(10)\"!"
)
if (self.l1_reg not in ["auto", False, 0]) or (fraction_evaluated < 0.2 and self.l1_reg == "auto"):
w_aug = np.hstack((self.kernelWeights * (self.M - s), self.kernelWeights * s))
log.info("np.sum(w_aug) = {0}".format(np.sum(w_aug)))
log.info("np.sum(self.kernelWeights) = {0}".format(np.sum(self.kernelWeights)))
w_sqrt_aug = np.sqrt(w_aug)
eyAdj_aug = np.hstack((eyAdj, eyAdj - (self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim]))))
eyAdj_aug *= w_sqrt_aug
mask_aug = np.transpose(w_sqrt_aug * np.transpose(np.vstack((self.maskMatrix, self.maskMatrix - 1))))
#var_norms = np.array([np.linalg.norm(mask_aug[:, i]) for i in range(mask_aug.shape[1])])
# select a fixed number of top features
if isinstance(self.l1_reg, str) and self.l1_reg.startswith("num_features("):
r = int(self.l1_reg[len("num_features("):-1])
nonzero_inds = lars_path(mask_aug, eyAdj_aug, max_iter=r)[1]
# use an adaptive regularization method
elif self.l1_reg == "auto" or self.l1_reg == "bic" or self.l1_reg == "aic":
c = "aic" if self.l1_reg == "auto" else self.l1_reg
nonzero_inds = np.nonzero(LassoLarsIC(criterion=c).fit(mask_aug, eyAdj_aug).coef_)[0]
# use a fixed regularization coeffcient
else:
nonzero_inds = np.nonzero(Lasso(alpha=self.l1_reg).fit(mask_aug, eyAdj_aug).coef_)[0]
if len(nonzero_inds) == 0:
return np.zeros(self.M), np.ones(self.M)
# eliminate one variable with the constraint that all features sum to the output
eyAdj2 = eyAdj - self.maskMatrix[:, nonzero_inds[-1]] * (
self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim]))
etmp = np.transpose(np.transpose(self.maskMatrix[:, nonzero_inds[:-1]]) - self.maskMatrix[:, nonzero_inds[-1]])
log.debug("etmp[:4,:] {0}".format(etmp[:4, :]))
# solve a weighted least squares equation to estimate phi
tmp = np.transpose(np.transpose(etmp) * np.transpose(self.kernelWeights))
tmp2 = np.linalg.inv(np.dot(np.transpose(tmp), etmp))
w = np.dot(tmp2, np.dot(np.transpose(tmp), eyAdj2))
log.debug("np.sum(w) = {0}".format(np.sum(w)))
log.debug("self.link(self.fx) - self.link(self.fnull) = {0}".format(
self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim])))
log.debug("self.fx = {0}".format(self.fx[dim]))
log.debug("self.link(self.fx) = {0}".format(self.link.f(self.fx[dim])))
log.debug("self.fnull = {0}".format(self.fnull[dim]))
log.debug("self.link(self.fnull) = {0}".format(self.link.f(self.fnull[dim])))
phi = np.zeros(self.M)
phi[nonzero_inds[:-1]] = w
phi[nonzero_inds[-1]] = (self.link.f(self.fx[dim]) - self.link.f(self.fnull[dim])) - sum(w)
log.info("phi = {0}".format(phi))
# clean up any rounding errors
for i in range(self.M):
if np.abs(phi[i]) < 1e-10:
phi[i] = 0
return phi, np.ones(len(phi))— Started going sequence-by-sequence, instead of sample-by-sample;
— Preserved the model's memory in each sequence.
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
| ts | real_output | shap_output |
|---|---|---|
| 0 | 0,4068 | 0,4068 |
| 1 | 0,3772 | 0,3848 |
| 2 | 0,3670 | 0,3976 |
| 3 | 0,5840 | 0,5943 |
| 4 | 0,5949 | 0,5851 |
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
Now we can see that the local accuracy property is valid again:
| ts | real_output | shap_output |
|---|---|---|
| 0 | 0,4068 | 0,4068 |
| 1 | 0,3772 | 0,3772 |
| 2 | 0,3670 | 0,3670 |
| 3 | 0,5840 | 0,5840 |
| 4 | 0,5949 | 0,5949 |
Custom SHAP
| ts | real_output | shap_output |
|---|---|---|
| 0 | 0,4068 | 0,4068 |
| 1 | 0,3772 | 0,3848 |
| 2 | 0,3670 | 0,3976 |
| 3 | 0,5840 | 0,5943 |
| 4 | 0,5949 | 0,5851 |
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
The original Kernel SHAP approach still had one problem: it was unpracticaly slow.
A computationally heavy part of the process is the iteration through multiple combinations of samples from the background data.
If we were to use only 50 background samples, interpreting a RNN model on all the ALS dataset would take around 27 hours.
A solution for this lies in SHAP's marginal expectation formula:
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
A solution for this lies in SHAP's marginal expectation formula:
Model
Present features
Absent features
If SHAP gets the expected value of the absent features, we can just use each feature's average value as the single reference value.
This reference value simply becomes a vector of all zeros, if we normalize the data through z-scores:
Normalized data
Input
Mean
Standard deviation
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
Applying this custom SHAP package, we get the following overall feature importance:
Feature importance for birdir. LSTM Δt
Feature importance for XGBoost
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
Feature importance for birdir. LSTM Δt
Feature importance for XGBoost
Similar distribution of most important features, with dominance of 3r.
As expected, respiratory (e.g. 3r, p10, r, 2r) and mobility (e.g. p7, p5) symptoms have the highest influence in the prediction of non-invasive ventilation.
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Feature importance
The custom SHAP package is publicly available on GitHub:
The core pillars
Interpretability
Performance
🚀
Usability
🏥
In a multivariate time series context, feature importance does not give a complete interpretation.
feat 1
feat 2
feat 3
Features in timestamp 2
The core pillars
Interpretability
Performance
🚀
Usability
🏥
feat 1
feat 2
feat 3
Features in timestamp 2
In a multivariate time series context, feature importance does not give a complete interpretation.
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Time series
t0
t1
t2
t3
t4
time
In a multivariate time series context, feature importance does not give a complete interpretation.
We also need to interpret the influence of each instance (i.e. each event in time).
Instance importance
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Instance importance
An initial approach comes rather naturally: just remove the instance of which we want to get an importance score and see how it affects the final output.
We can call this the occlusion score.
Index of the last instance
Set of instances in the sequence
Instance that we are interpreting
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Instance importance
Occlusion is likely not enough. There tend to be certain moments when something new happens that can have repercussions or be repeated in the following events.
An interesting alternative might be using the variation that the instance brings to the output: the output variation score.
The core pillars
Interpretability
Performance
🚀
Usability
🏥
Instance importance
As both scores can be relevant in many scenarios, the ideal solution is to combine both in a weighted sum.
Considering the more straightforward approach of occlusion, and some empirical analysis, we get to the instance importance score.
The core pillars
The core pillars
Usability
Performance
🚀
Interpretability
🔍
Regarding AI in healthcare, there are two parts where usability can be addressed:
— Coding packages
— No-code interfaces / dashboards
The core pillars
Usability
Performance
🚀
Interpretability
🔍
Packages
For most data processing and machine learning tasks:
The core pillars
Usability
Performance
🚀
Interpretability
🔍
Packages
For model interpretation:
The core pillars
Usability
Performance
🚀
Interpretability
🔍
If you can't see the dashboard above, to go to the screenshots
Dashboard
The core pillars
Usability
Performance
🚀
Interpretability
🔍
If you can't see the dashboard above, to go to the screenshots
Dashboard
The core pillars
Usability
Performance
🚀
Interpretability
🔍
The previous two slides require that you have the HAI dashboard running in Python.
In case you want to experiment with the dashboard, clone the following repository:
Otherwise, you can just follow along the next slides, which contain screenshots.
Dashboard
The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
The dashboard has a homepage that gives an overview of the main characteristics and links to the subpages.
The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

It has a navigation bar for quick access to each page.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

There are dataset and model selectors.


The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
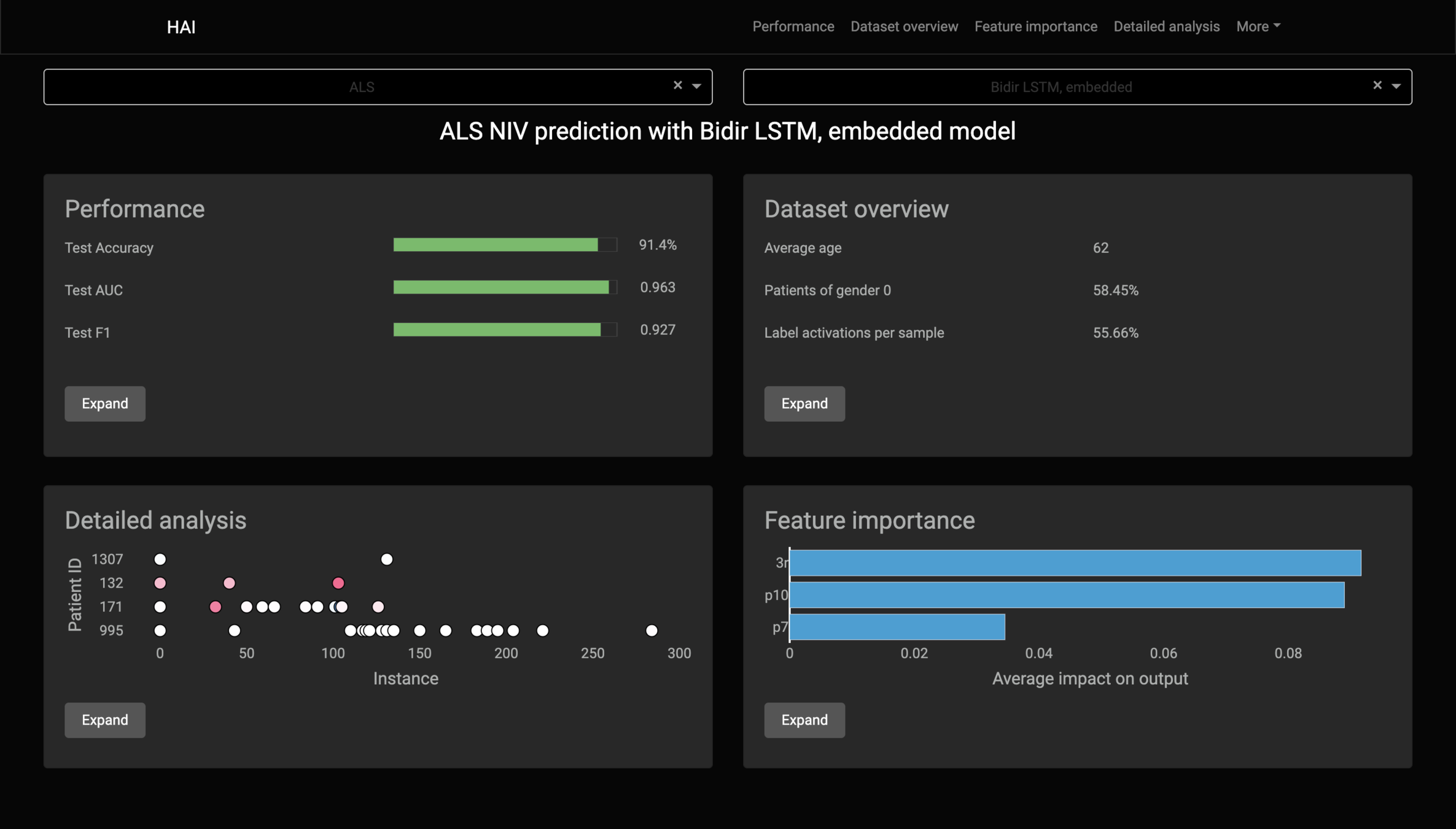
A dynamic title gives us the context of what dataset and model are being analysed.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

Each subpage has a preview card, that also links to it.




The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
The performance page gives an overview of the chosen model.
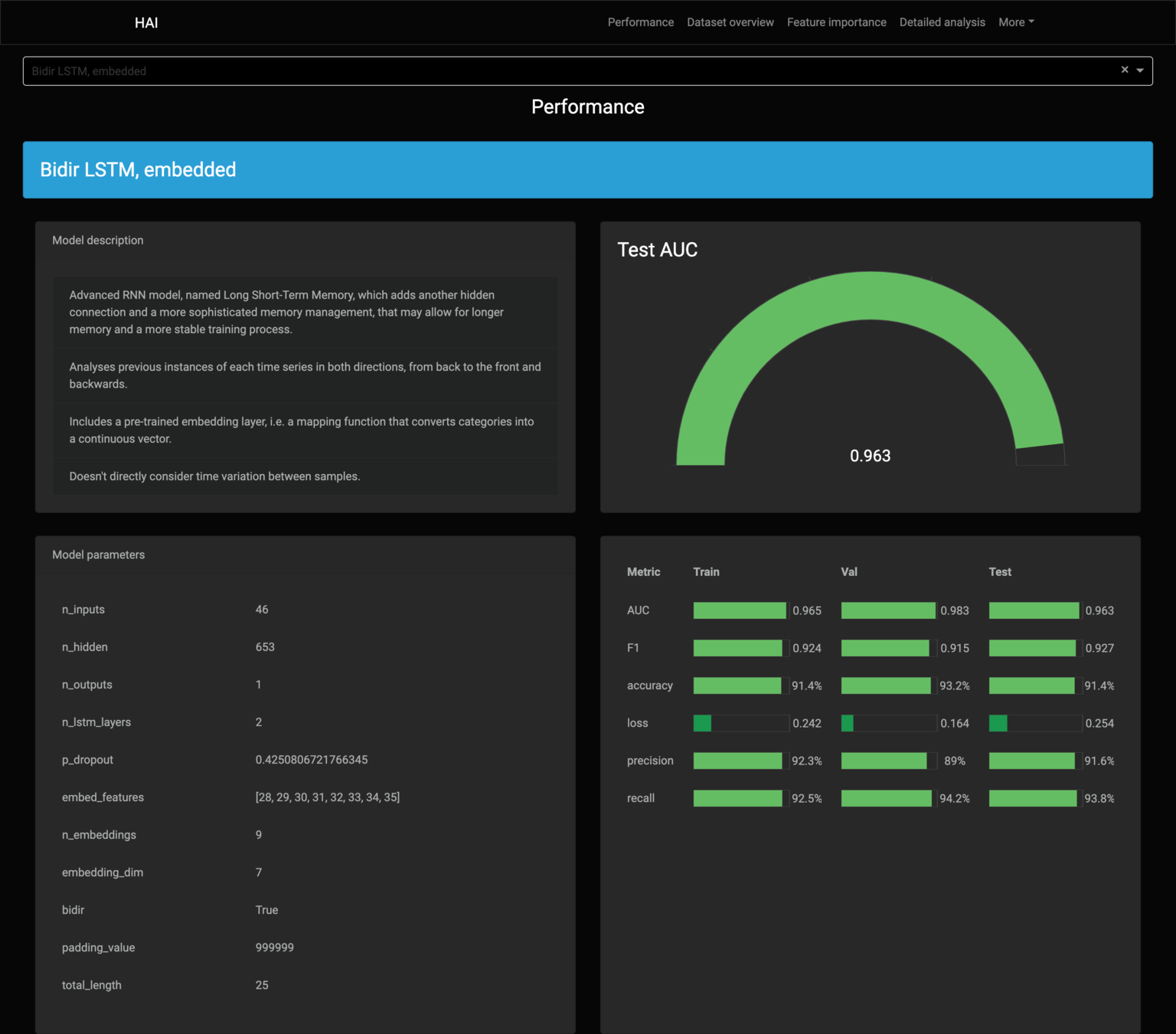
The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

The selected model's name is highlighted.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

An high-level description of the model, based on its components, is presented first.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

All the hyperparameters are presented below.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

Test AUC, the main metric in this thesis, is highlighted.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

All the remaining calculated metrics are shown below.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
"Dataset overview" gives brief insights on the chosen dataset.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

The selected dataset's name is highlighted.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

3 distinct tabs are available.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

The first one, "Size", shows basic stats of the dataset, including overall size and sequence length.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

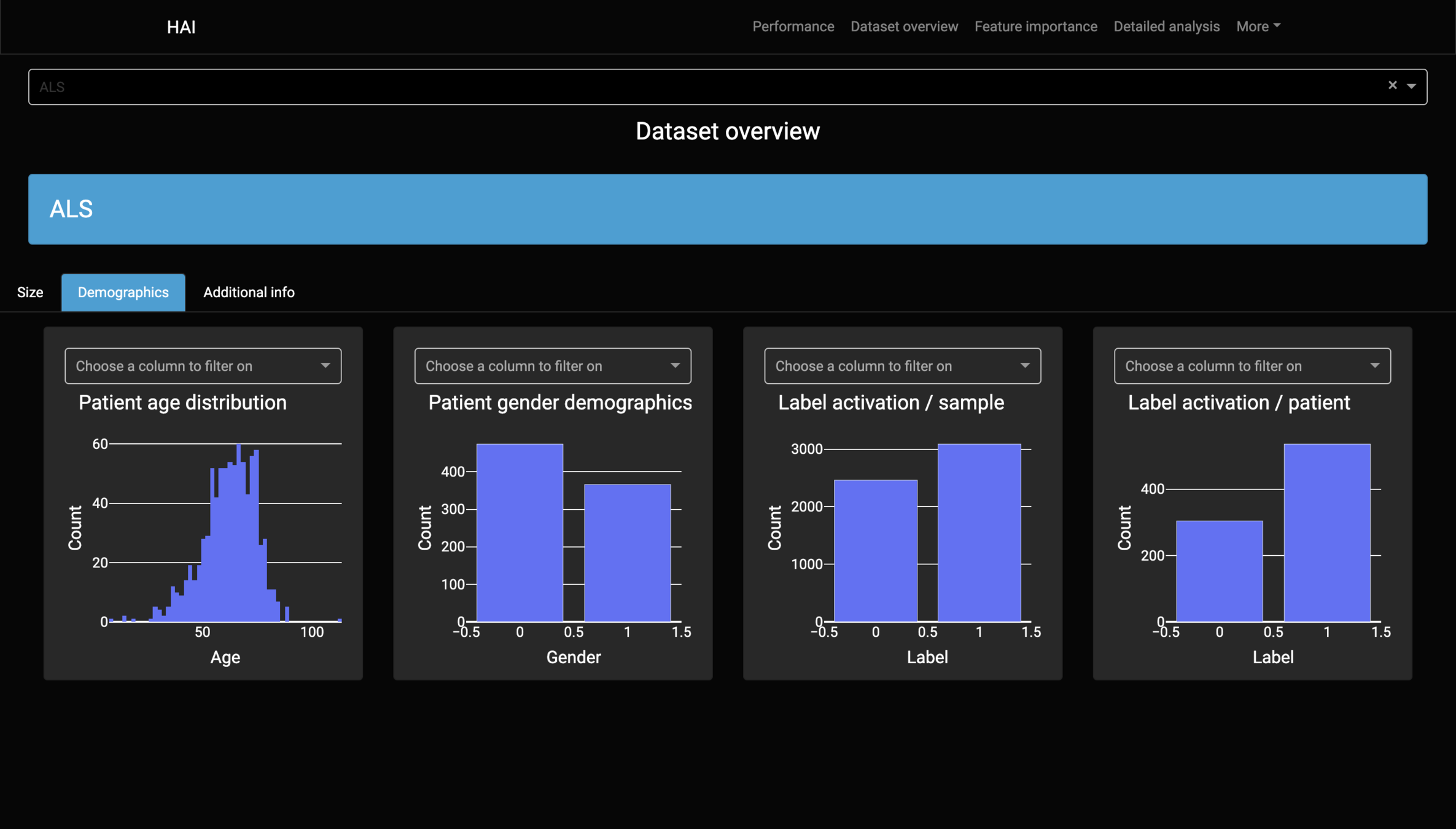
The second tab focuses on demographics.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard

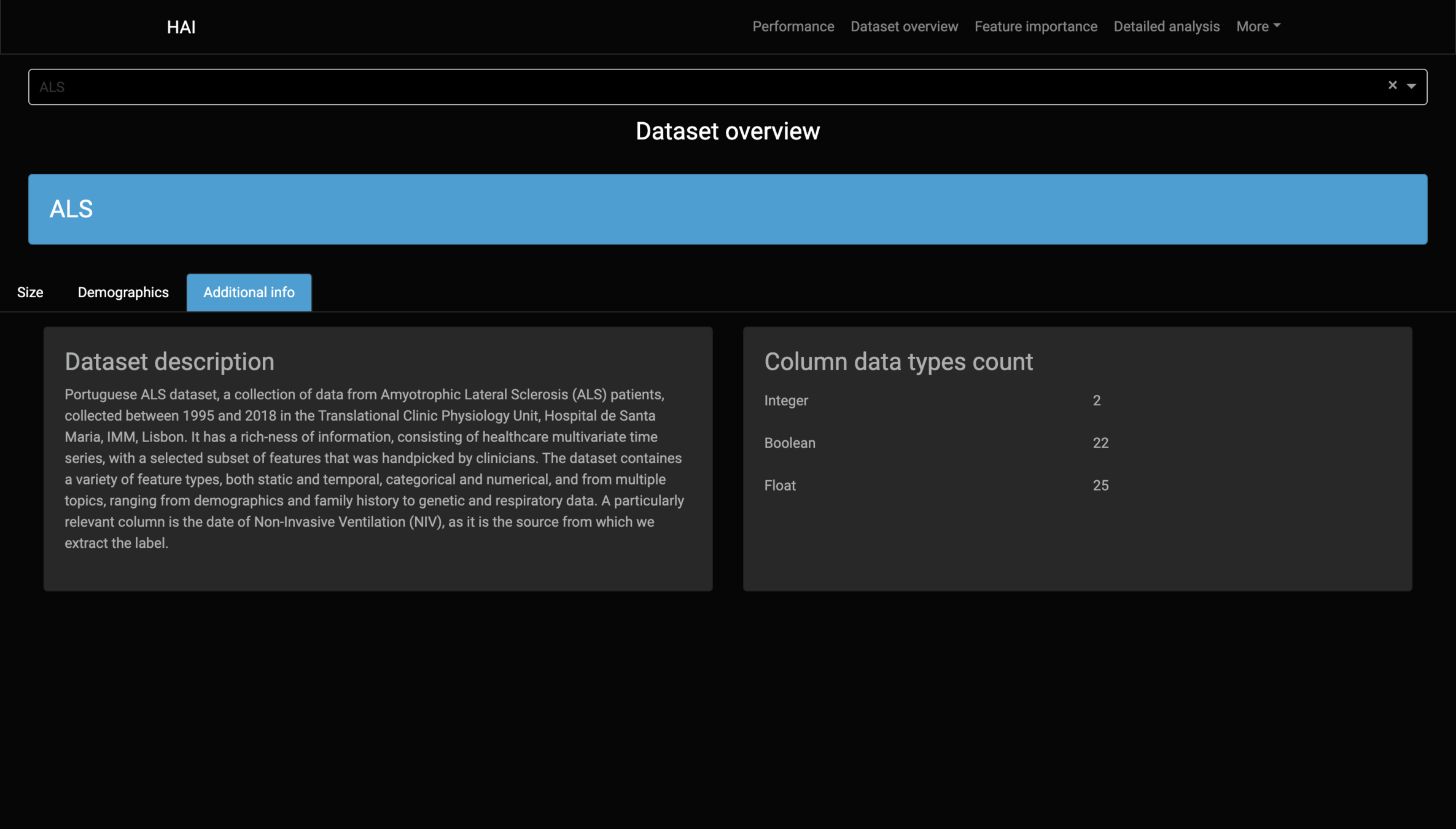

The third and final tab displays additional information, as a written description and column types.
The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
In this page, we can see aggregate feature importance, with the possibility of filtering by feature values.
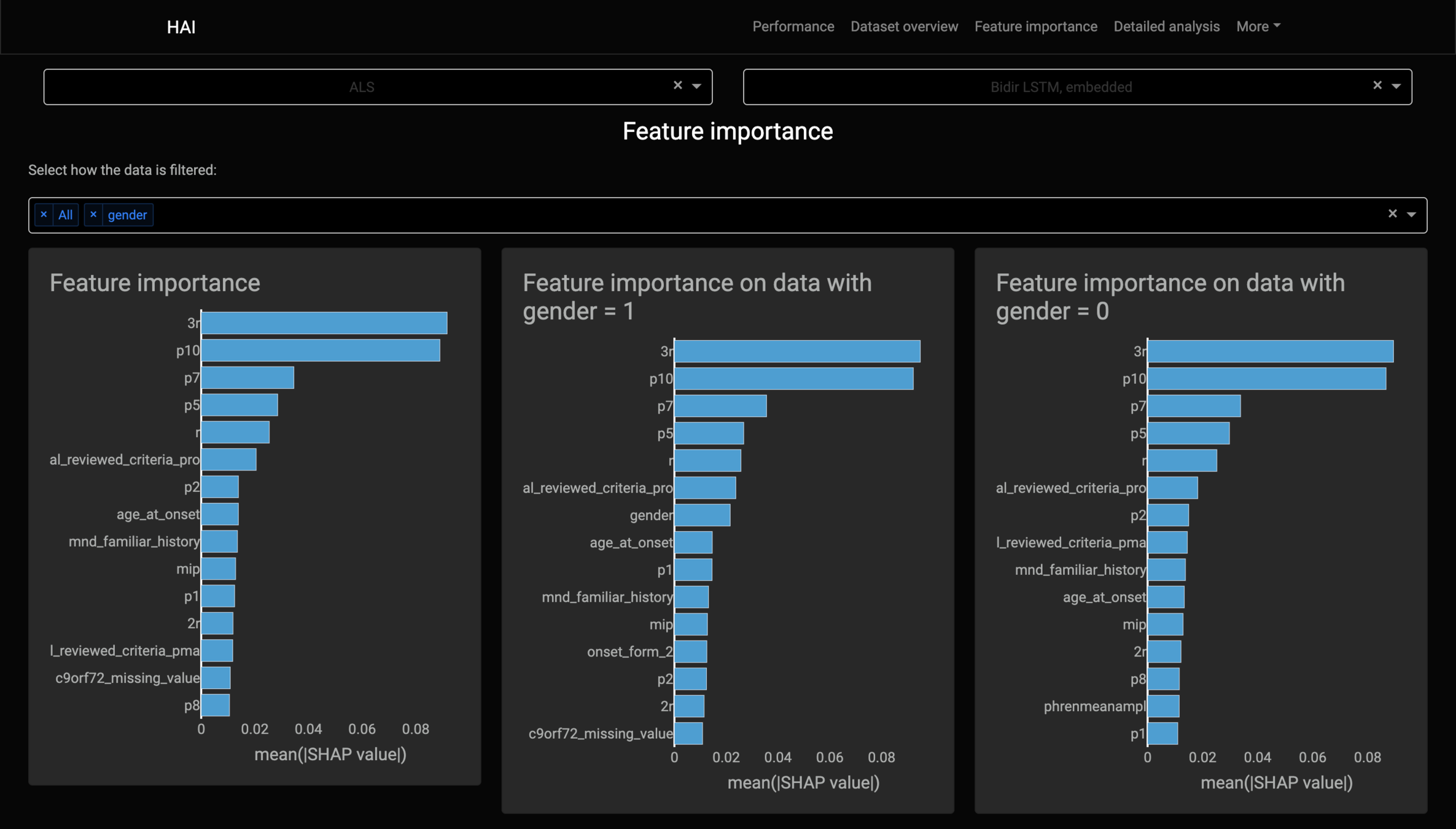
The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
"Detailed analysis" combines most concepts in a more in-depth look at patients' data.
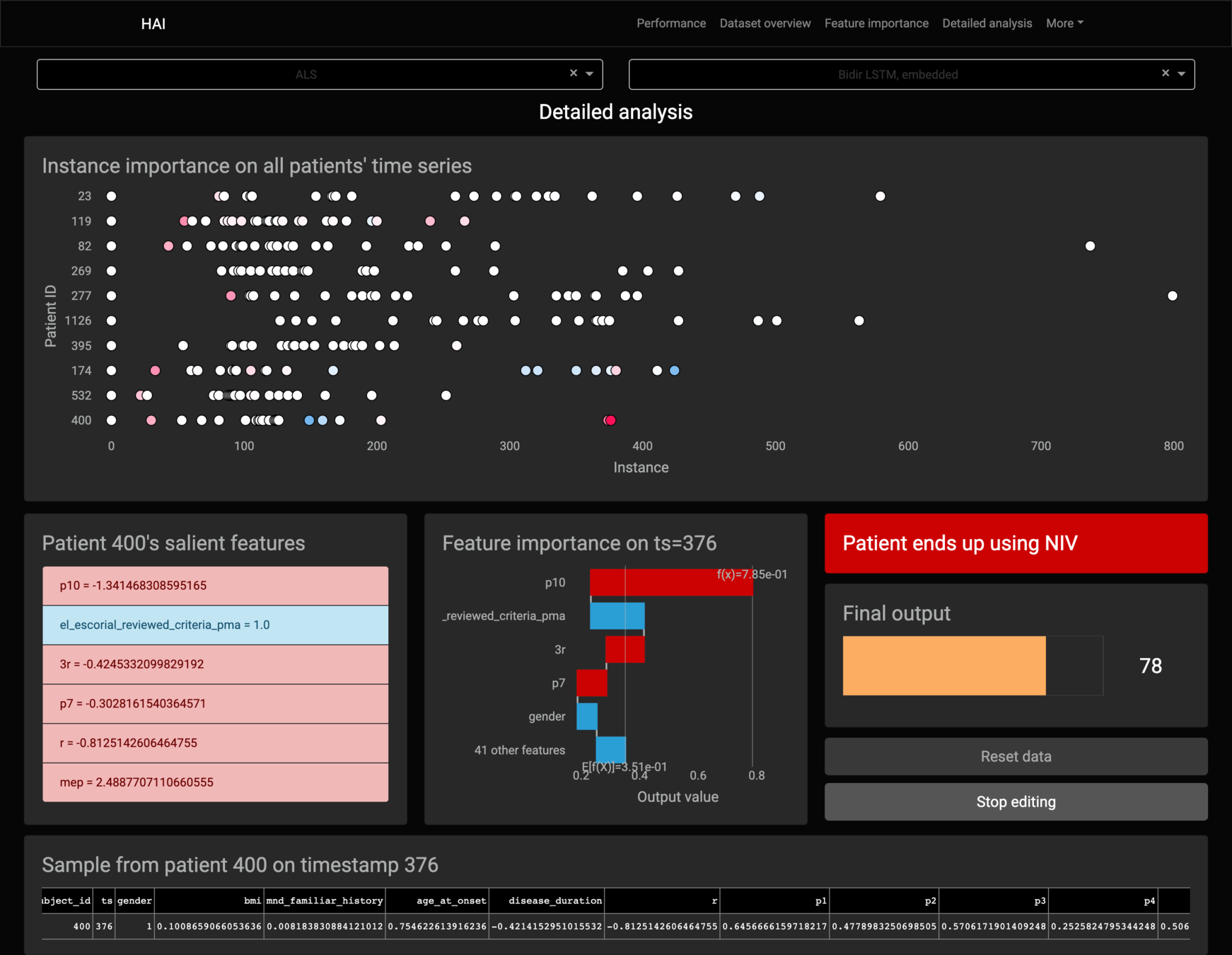
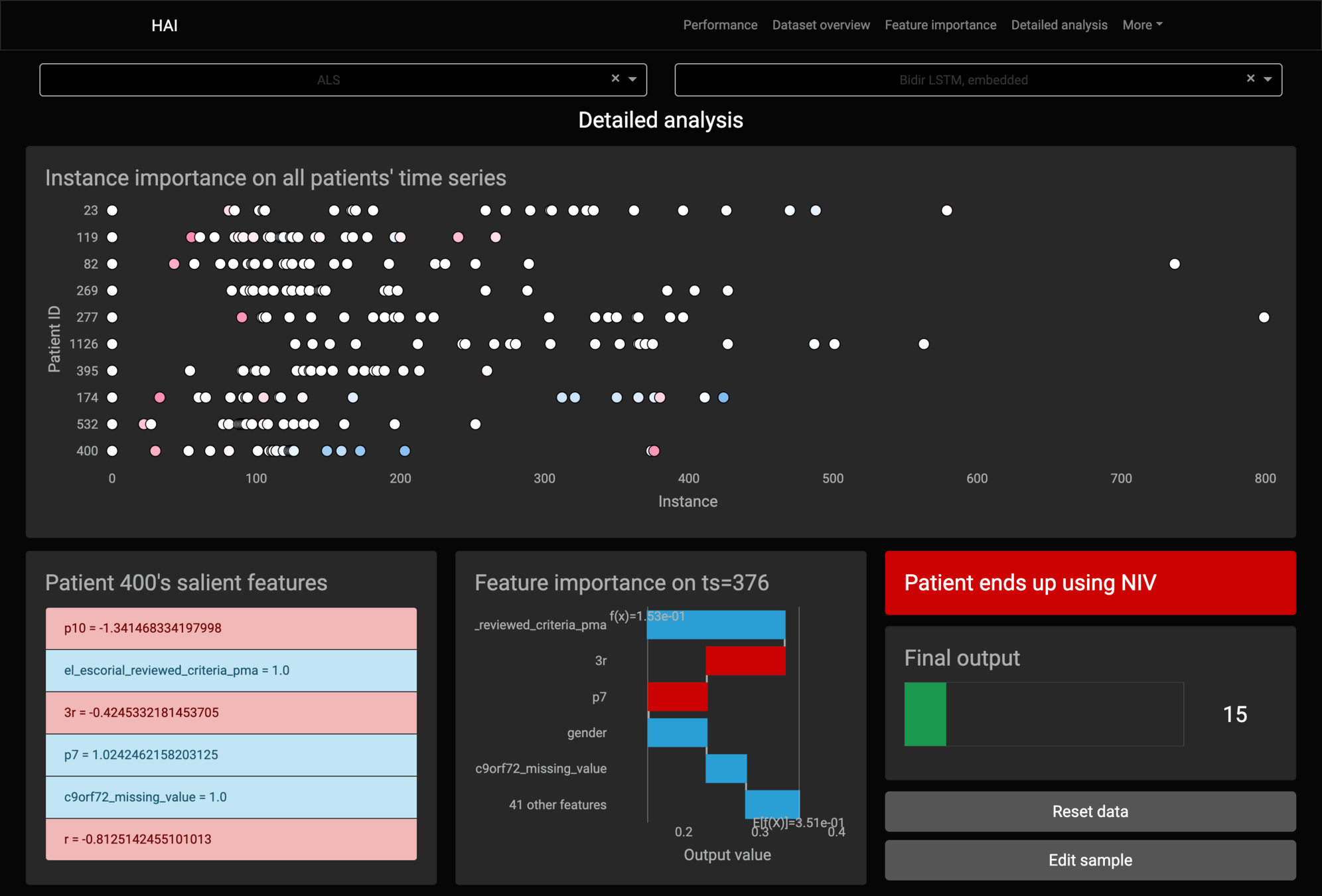
The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
The larger card shows all patients' data and the instance importance scores.
Each row corresponds to a patient's time series; each circle is a sample.
Colors indicate the importance score: Blue → Low; Red → High.
Red
Blue



The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
The most salient feature values of the selected patient's time series (i.e. the ones with higher absolute feature importance) are displayed on the bottom left.



The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
The current sample's feature importance shows in the middle card.



The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
To the right, we have the patient's final outcome.



The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
To the right, we have the patient's final outcome.

As well as the model's final prediction probability.



The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
Then we can also edit each sample and see how it affects the output and the importance scores.



The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
Then we can also edit each sample and see how it affects the output and the importance scores.

The core pillars
Usability
Performance
🚀
Interpretability
🔍
Dashboard
Then we can also edit each sample and see how it affects the output and the importance scores.

The core pillars
LSTM-based models surpassed baseline by over 0.1 test AUC.
Kernel SHAP was adapted to RNN-type models.
A new instance importance score was introduced.
Two packages were released to facilitate coding.
A dashboard was presented as a proof of concept.
Bidirectionality gave a significant performance boost.
☑️
☑️
☑️
☑️
☑️
☑️
the core pillars
With the efforts made on
we now have a guideline on how to facilitate
trustworthy, high-performance AI in
in healthcare.
In this particular example, we can visualize a
predictive medicine tool, to assist physicians.
Future work
Experiment with larger and more complex datasets.
Test other, potentially better performing model types, such as transformers and Neural ODEs.
Explore faster and/or more reliable interpretability techniques.
(this thesis' code already contemplates large data processing and distributed training pipelines)
Present work on scientific conferences.


☑️

The End
But first, a bit of context...
But first, a bit of context...
Related work
Background
📖
Google AI's behemoth
+
+
-
-
-
+
Uses heterogeneous data, including text notes;
Achieved state-of-the-art results in 4 distinct tasks;
Identifies events with the biggest impact on the model's decision, using attention and embeddings weighting;
Does not calculate feature contribution scores and only interprets part of the model;
The ensemble model is computationally heavy;
Some confidentiality in the model architecture, applied methods and the data stats.
But first, a bit of context...
Related work
Background
📖
RETAIN & RetainVis
+
Analytically calculable contribution scores;
-
Interpretability depends on a highly specific architecture;
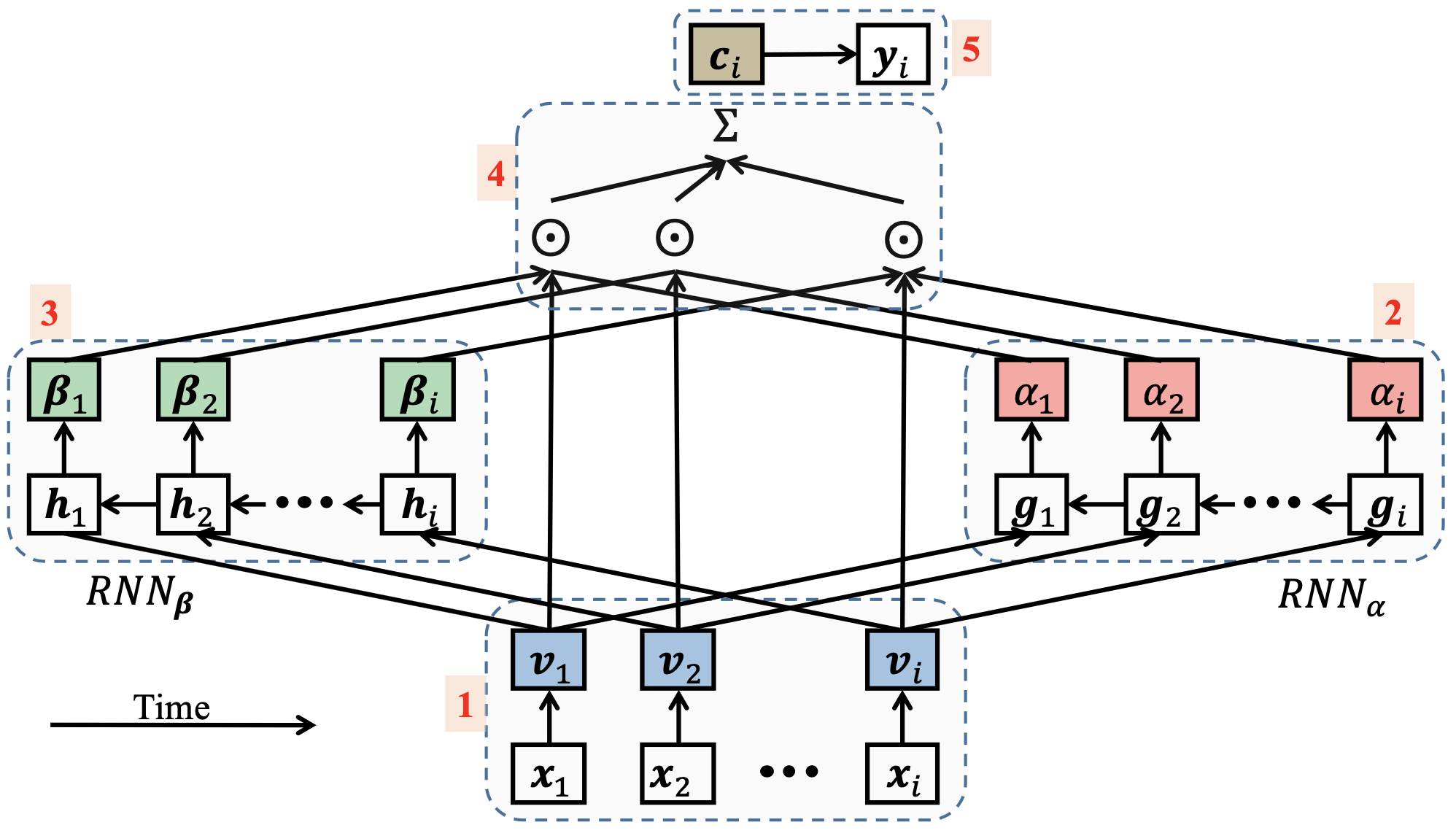
+
Incorporates time interval information;
+
Developed an intuitive dashboard;
-
Comparable performance to simpler RNN models.
But first, a bit of context...
Related work
Background
📖
But first, a bit of context...
Related work
📄
Background
Model variations
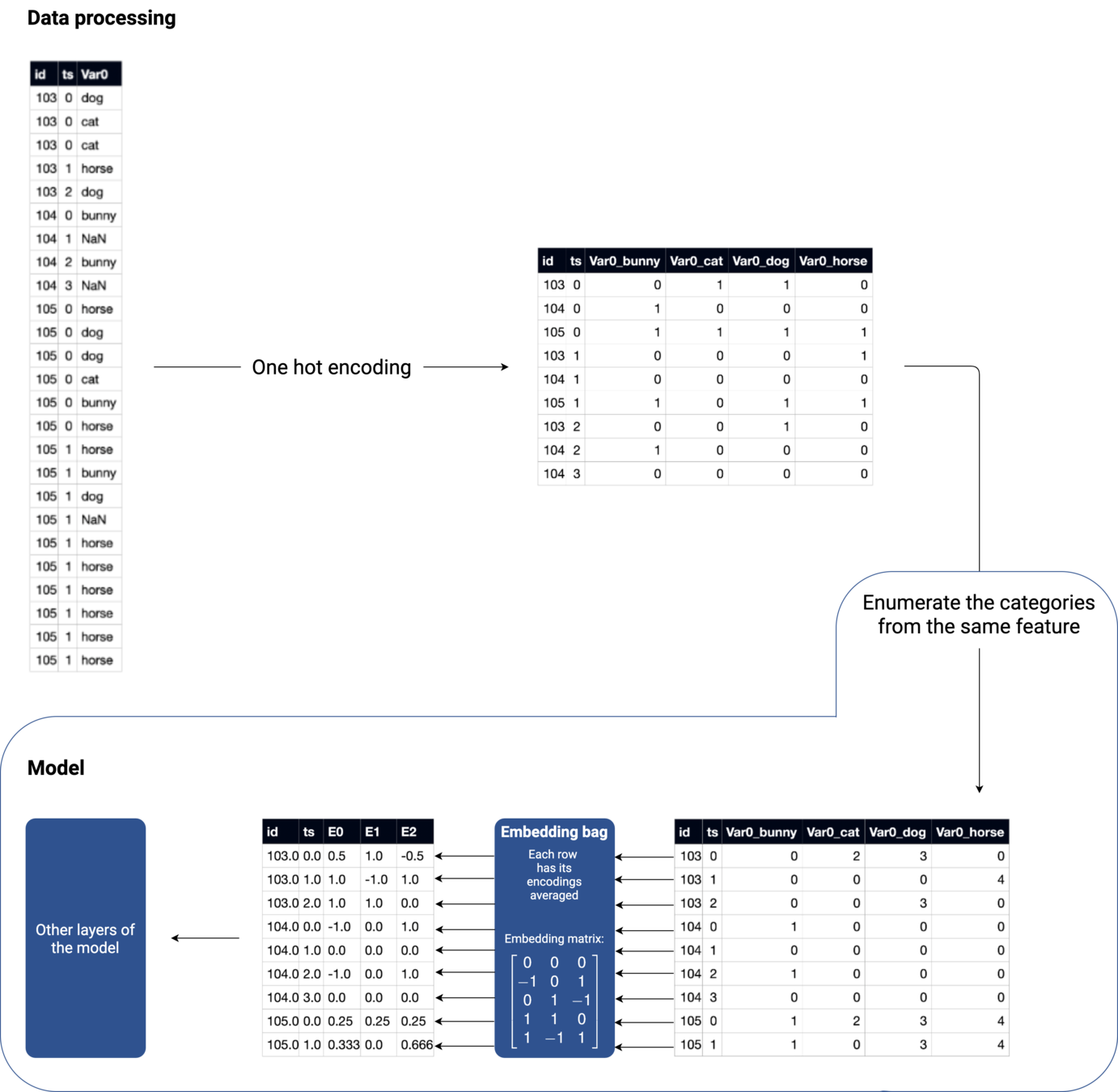
The embedding process, adapted to multivariate time series scenarios, can be seen here.
Master's Thesis - Predictive Medicine Using Interpretable Recurrent Neural Networks
By André Cristóvão Neves Ferreira
Master's Thesis - Predictive Medicine Using Interpretable Recurrent Neural Networks
Presentation for the master's thesis defense on the topic "Predictive Medicine Using Interpretable Recurrent Neural Networks".
- 533