Soss: Lightweight Probabilistic Programming in Julia
Chad Scherrer
June 16, 2023
Making Rational Decisions
Managed Uncertainty
Rational Decisions
Bayesian Analysis
Probabilistic Programming
- Physical systems
- Hypothesis testing
- Modeling as simulation
- Medicine
- Finance
- Insurance
Risk
Custom models
The Two-Language Problem
A disconnect between the "user language" and "developer language"
X
3
Python
C
Deep Learning Framework
- Harder for beginner users
- Barrier to entry for developers
- Limits extensibility
?
Introducing Soss
- Simple high-level syntax
- Uses GeneralizedGenerated.jl for flexible staged compilation
- Model type parameter includes type-level representation of itself
- Allows specialized code generation for each primitive(model, data)
A (Very) Simple Example
P(\mu,\sigma|x)\propto P(\mu,\sigma)P(x|\mu,\sigma)
\begin{aligned}
\mu &\sim \text{Normal}(0,5)\\
\sigma &\sim \text{Cauchy}_+(0,3) \\
x_j &\sim \text{Normal}(\mu,\sigma)
\end{aligned}
Theory
Soss
julia> Soss.sourceLogpdf()(m)
quote
_ℓ = 0.0
_ℓ += logpdf(HalfCauchy(3), σ)
_ℓ += logpdf(Normal(0, 5), μ)
_ℓ += logpdf(Normal(μ, σ) |> iid(N), x)
return _ℓ
end@model N begin
μ ~ Normal(0, 5)
σ ~ HalfCauchy(3)
x ~ Normal(μ, σ) |> iid(N)
endBuilding a Linear Model
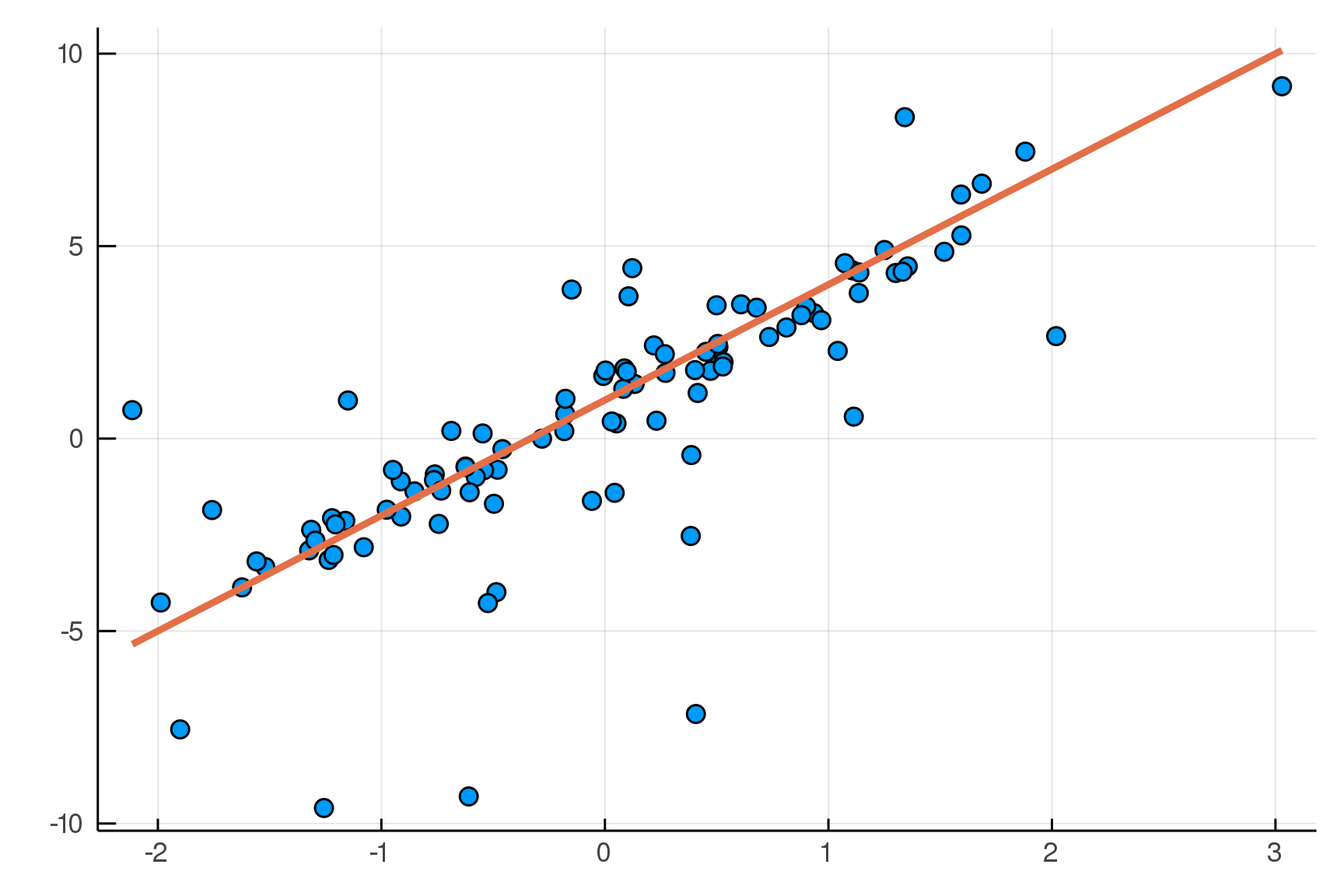
Building a Linear Model
m = @model x begin
α ~ Normal()
β ~ Normal()
σ ~ HalfNormal()
yhat = α .+ β .* x
n = length(x)
y ~ For(n) do j
Normal(yhat[j], σ)
end
endjulia> m(x=truth.x)
Joint Distribution
Bound arguments: [x]
Variables: [σ, β, α, yhat, n, y]
@model x begin
σ ~ HalfNormal()
β ~ Normal()
α ~ Normal()
yhat = α .+ β .* x
n = length(x)
y ~ For(n) do j
Normal(yhat[j], σ)
end
endObserved data is not specified yet!
Sampling from the Posterior Distribution
julia> post = dynamicHMC(m(x=truth.x), (y=truth.y,)) |> particles
(σ = 2.02 ± 0.15, β = 2.99 ± 0.19, α = 0.788 ± 0.2)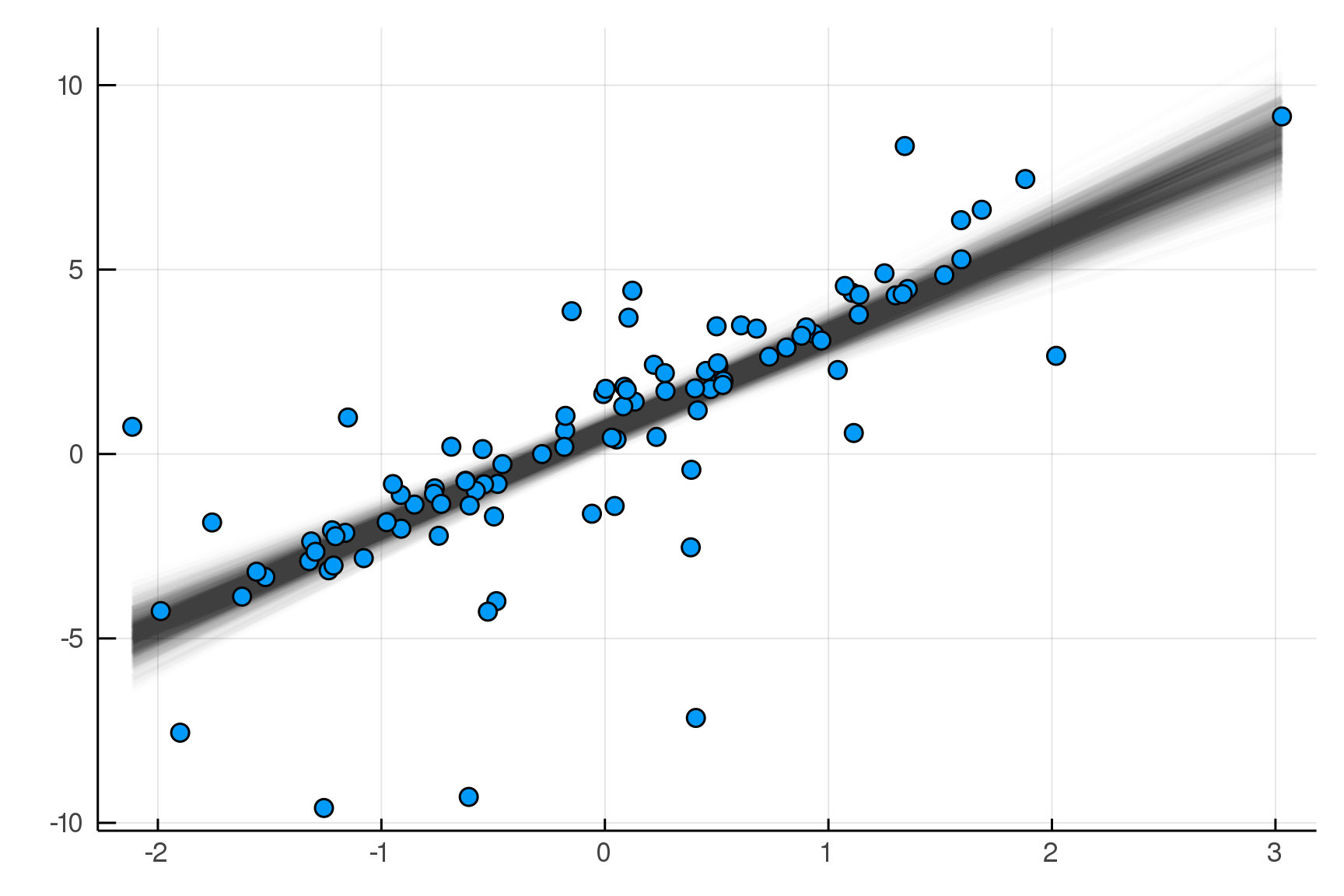
Posterior distribution
Possible best-fit lines
The Posterior Predictive Distribution
Start with Data
Sample Parameters|Data
Sample Data|Parameters
Real Data
Replicated Fake Data
Compare
Posterior
Distribution
Predictive
Distribution
The Posterior Predictive Distribution
pred = predictive(m, :α, :β, :σ)
@model (x, α, β, σ) begin
yhat = α .+ β .* x
n = length(x)
y ~ For(n) do j
Normal(yhat[j], σ)
end
endm = @model x begin
α ~ Normal()
β ~ Normal()
σ ~ HalfNormal()
yhat = α .+ β .* x
n = length(x)
y ~ For(n) do j
Normal(yhat[j], σ)
end
endpostpred = map(post) do θ
delete(rand(pred(θ)((x=x,))), :n, :x)
end |> particlespredictive makes a new model!
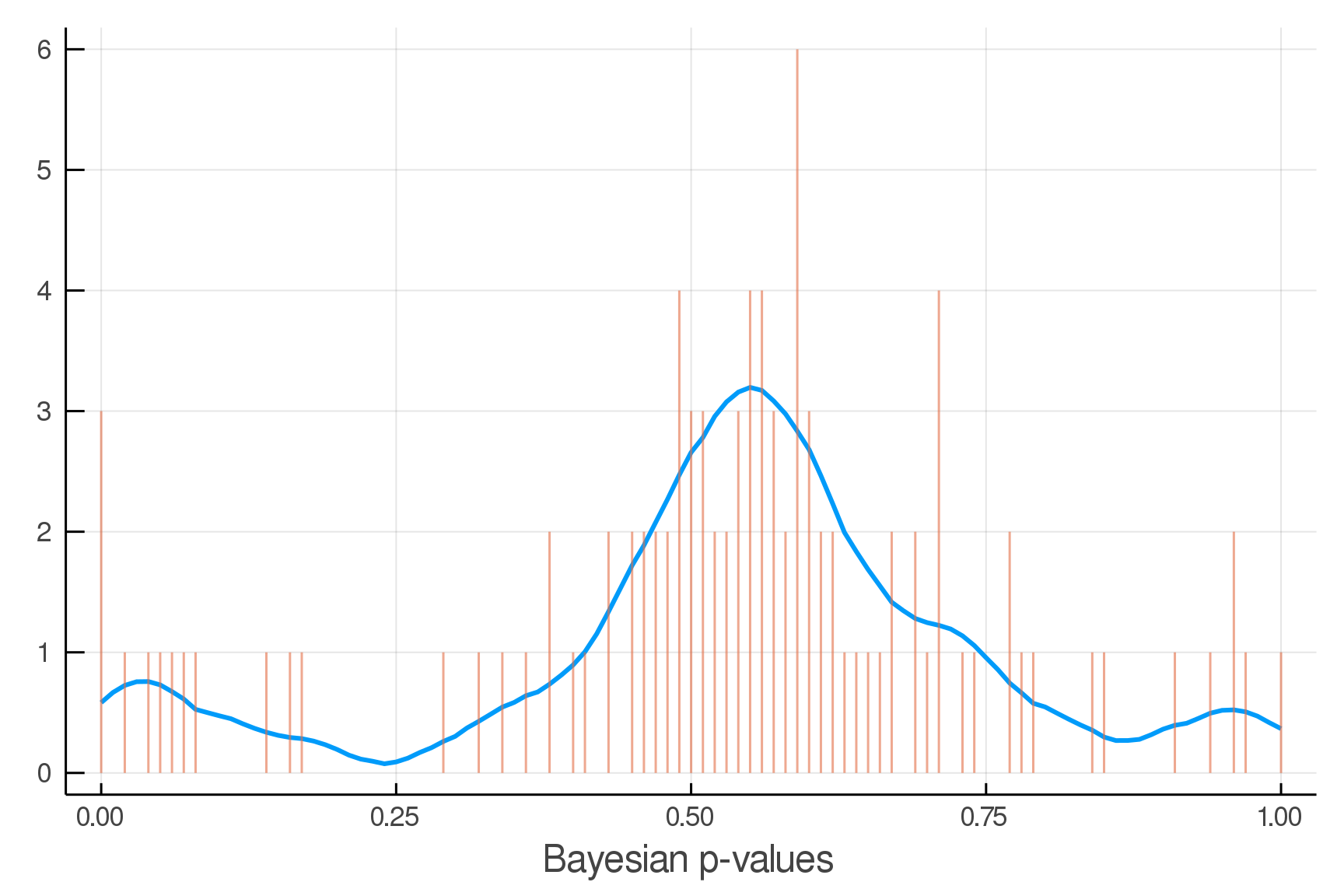
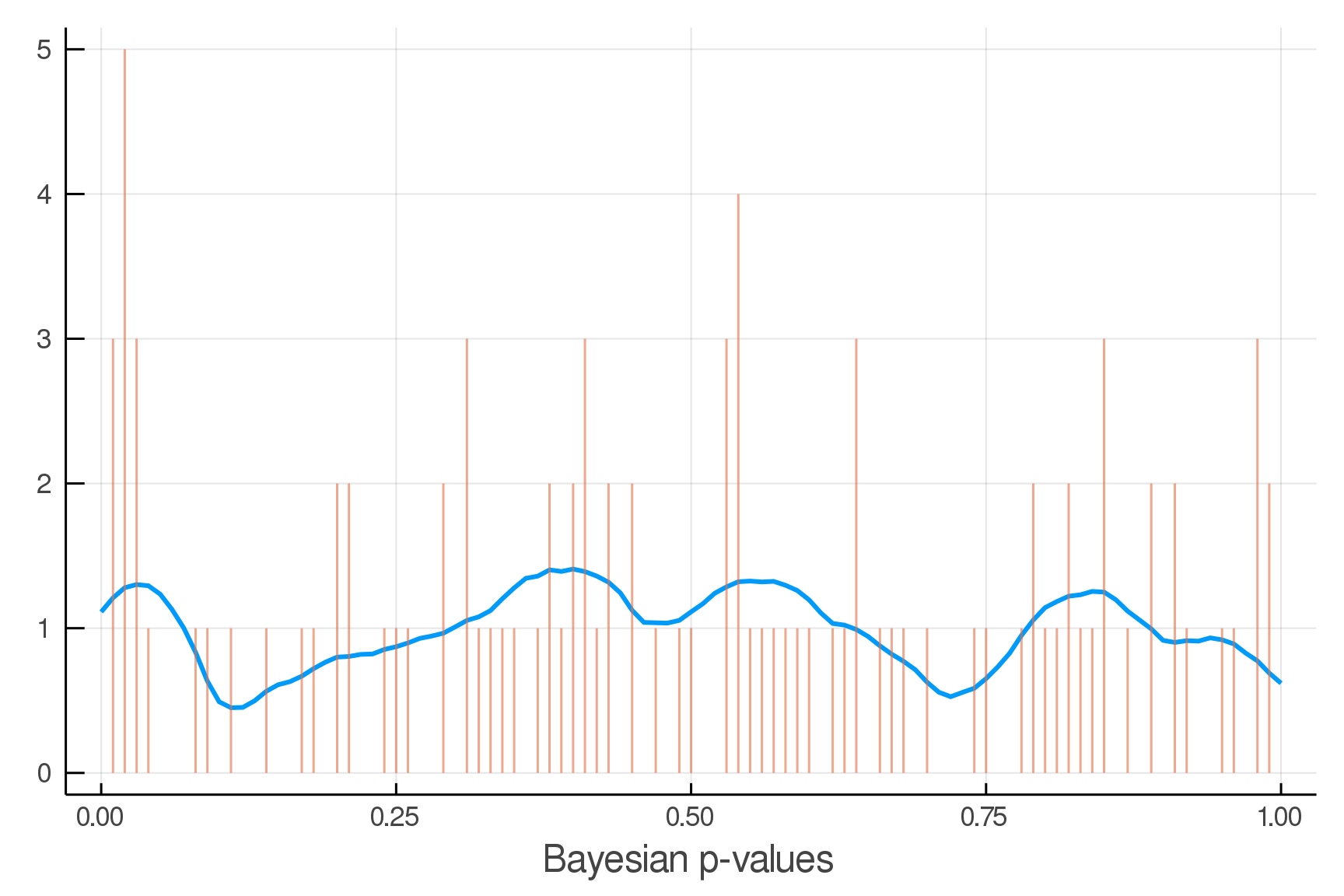
Posterior Predictive Checks
pvals = mean.(truth.y .> postpred.y)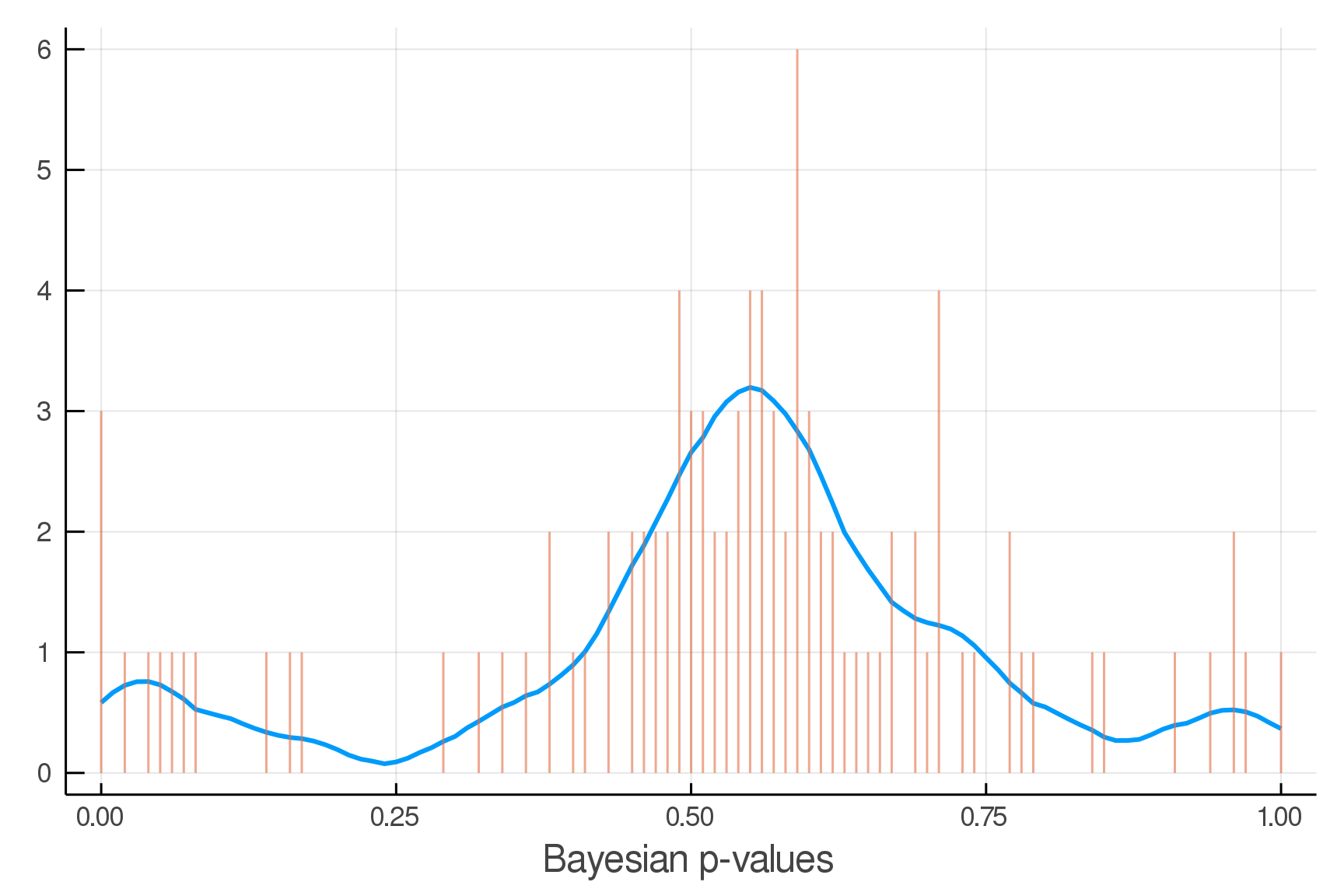
Where we expect the data
Where we see the data
Making it Robust
m2 = @model x begin
α ~ Normal()
β ~ Normal()
σ ~ HalfNormal()
yhat = α .+ β .* x
νinv ~ HalfNormal()
ν = 1/νinv
n = length(x)
y ~ For(n) do j
StudentT(ν,yhat[j],σ)
end
endjulia> post2 = dynamicHMC(m2(x=truth.x), (y=truth.y,)) |> particles
( σ = 0.57 ± 0.09, νinv = 0.609 ± 0.14
, β = 2.73 ± 0.073, α = 0.893 ± 0.077)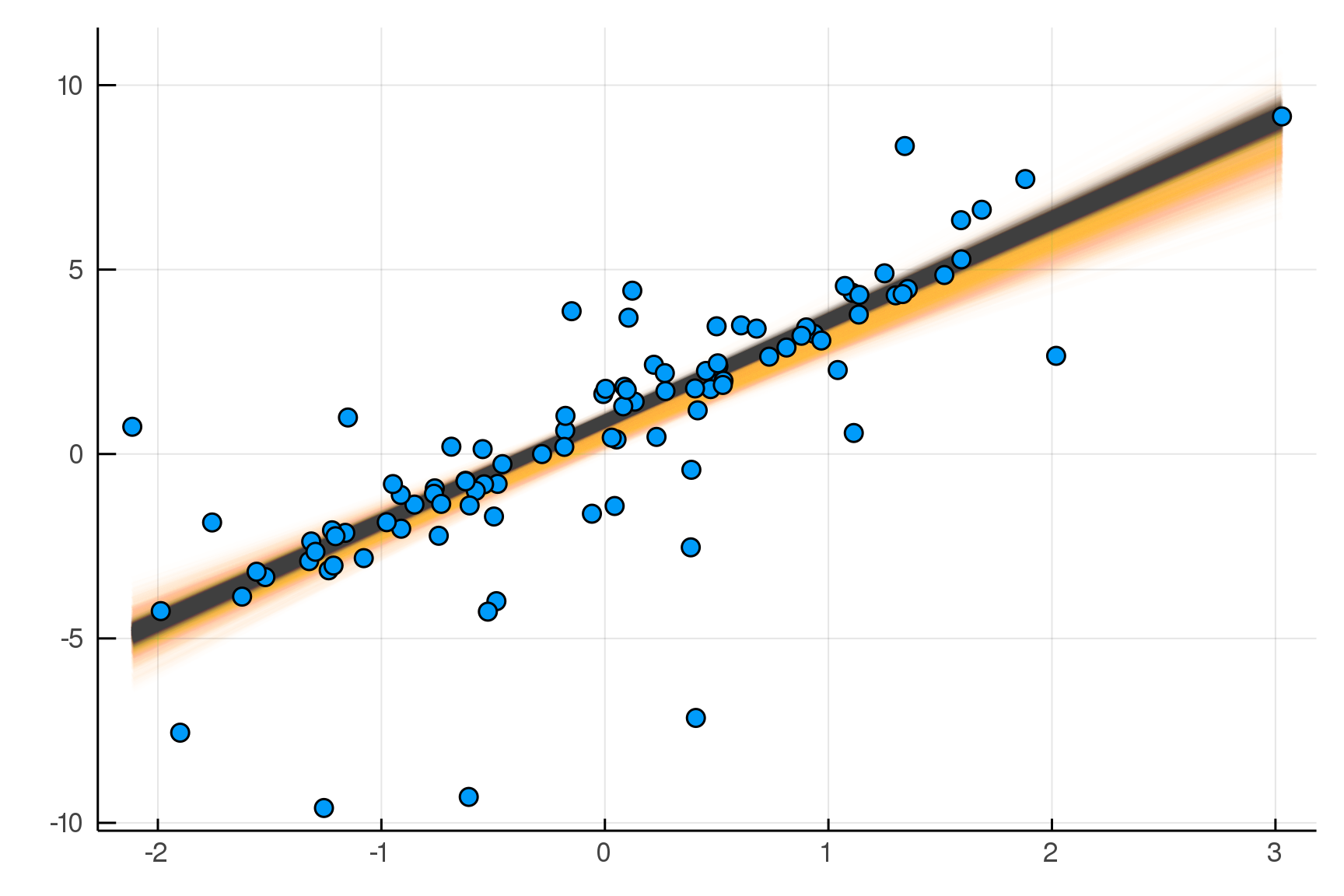
Updated Posterior Predictive Checks
Inference Primitive: rand
julia> Soss.sourceRand()(m)
quote
σ = rand(HalfNormal())
β = rand(Normal())
α = rand(Normal())
yhat = α .+ β .* x
n = length(x)
y = rand(For(((j,)->begin
Normal(yhat[j], σ)
end), n))
(x = x, yhat = yhat, n = n
, α = α, β = β, σ = σ, y = y)
end@model x begin
σ ~ HalfNormal()
β ~ Normal()
α ~ Normal()
yhat = α .+ β .* x
n = length(x)
y ~ For(n) do j
Normal(yhat[j], σ)
end
endInference Primitive: logpdf
julia> Soss.sourceLogpdf()(m)
quote
_ℓ = 0.0
_ℓ += logpdf(HalfNormal(), σ)
_ℓ += logpdf(Normal(), β)
_ℓ += logpdf(Normal(), α)
yhat = α .+ β .* x
n = length(x)
_ℓ += logpdf(For(n) do j
Normal(yhat[j], σ)
end, y)
return _ℓ
end@model x begin
σ ~ HalfNormal()
β ~ Normal()
α ~ Normal()
yhat = α .+ β .* x
n = length(x)
y ~ For(n) do j
Normal(yhat[j], σ)
end
endInference Primitive: symlogpdf
julia> Soss.sourceSymlogpdf()(m)
quote
_ℓ = 0.0
x = sympy.IndexedBase(:x)
yhat = sympy.IndexedBase(:yhat)
n = sympy.IndexedBase(:n)
α = sympy.IndexedBase(:α)
β = sympy.IndexedBase(:β)
σ = sympy.IndexedBase(:σ)
y = sympy.IndexedBase(:y)
_ℓ += symlogpdf(HalfNormal(), σ)
_ℓ += symlogpdf(Normal(), β)
_ℓ += symlogpdf(Normal(), α)
yhat = sympy.IndexedBase(:yhat)
n = sympy.IndexedBase(:n)
_ℓ += symlogpdf(For(n) do j
Normal(yhat[j], σ)
end, y)
return _ℓ
end@model x begin
σ ~ HalfNormal()
β ~ Normal()
α ~ Normal()
yhat = α .+ β .* x
n = length(x)
y ~ For(n) do j
Normal(yhat[j], σ)
end
endSymbolic Simplification
julia> symlogpdf(m)julia> symlogpdf(m) |> expandSums-3.7-0.5α^{2}-0.5β^{2}-σ^{2}+\sum_{j_{1}=1}^{n}\left(-0.92-\logσ-\frac{0.5\left(y_{j_{1}}-\hat{y}_{j_{1}}\right)^{2}}{σ^{2}}\right)
-3.7-0.5α^{2}-0.5β^{2}-σ^{2}-0.92n-n\logσ-\frac{0.5}{\sigma^{2}}\sum_{j_{1}=1}^{n}\left(y_{j_{1}}-\hat{y}_{j_{1}}\right)^{2}
Code Generation
julia> symlogpdf(m()) |> expandSums |> foldConstants |> codegen
quote
var"##add#643" = 0.0
var"##add#643" += -3.6757541328186907
var"##add#643" += begin
var"##mul#644" = 1.0
var"##mul#644" *= -0.5
var"##mul#644" *= begin
var"##arg1#646" = α
var"##arg2#647" = 2
var"##symfunc#645" = (Soss._pow)(var"##arg1#646", var"##arg2#647")
var"##symfunc#645"
end
var"##mul#644"
end
var"##add#643" += begin
var"##mul#648" = 1.0
var"##mul#648" *= -0.5
var"##mul#648" *= begin
var"##arg1#650" = β
var"##arg2#651" = 2
var"##symfunc#649" = (Soss._pow)(var"##arg1#650", var"##arg2#651")
var"##symfunc#649"
end
var"##mul#648"
end
var"##add#643" += begin
var"##mul#652" = 1.0
var"##mul#652" *= -1.0
var"##mul#652" *= begin
var"##arg1#654" = σ
var"##arg2#655" = 2
var"##symfunc#653" = (Soss._pow)(var"##arg1#654", var"##arg2#655")
var"##symfunc#653"
end
var"##mul#652"
end
var"##add#643" += begin
var"##mul#656" = 1.0
var"##mul#656" *= -0.9189385332046727
var"##mul#656" *= n
var"##mul#656"
end
var"##add#643" += begin
var"##mul#657" = 1.0
var"##mul#657" *= -0.5
var"##mul#657" *= begin
var"##arg1#659" = σ
var"##arg2#660" = -2
var"##symfunc#658" = (Soss._pow)(var"##arg1#659", var"##arg2#660")
var"##symfunc#658"
end
var"##mul#657" *= begin
let
var"##sum#661" = 0.0
begin
var"##lo#663" = 1
var"##hi#664" = n
@inbounds for _j1 = var"##lo#663":var"##hi#664"
begin
var"##Δsum#662" = begin
var"##arg1#666" = begin
var"##add#668" = 0.0
var"##add#668" += begin
var"##mul#669" = 1.0
var"##mul#669" *= -1.0
var"##mul#669" *= begin
var"##arg1#671" = yhat
var"##arg2#672" = _j1
var"##symfunc#670" = (getindex)(var"##arg1#671", var"##arg2#672")
var"##symfunc#670"
end
var"##mul#669"
end
var"##add#668" += begin
var"##arg1#674" = y
var"##arg2#675" = _j1
var"##symfunc#673" = (getindex)(var"##arg1#674", var"##arg2#675")
var"##symfunc#673"
end
var"##add#668"
end
var"##arg2#667" = 2
var"##symfunc#665" = (Soss._pow)(var"##arg1#666", var"##arg2#667")
var"##symfunc#665"
end
var"##sum#661" += var"##Δsum#662"
end
end
end
var"##sum#661"
end
end
var"##mul#657"
end
var"##add#643" += begin
var"##mul#676" = 1.0
var"##mul#676" *= -1.0
var"##mul#676" *= n
var"##mul#676" *= begin
var"##arg1#678" = σ
var"##symfunc#677" = (log)(var"##arg1#678")
var"##symfunc#677"
end
var"##mul#676"
end
var"##add#643"
endjulia> @btime logpdf($m(x=x), $truth)
1.911 μs (25 allocations: 1.42 KiB)
-901.7607073245318julia> @btime logpdf($m(x=x), $truth, codegen)
144.671 ns (1 allocation: 896 bytes)
-903.4977930382969Default
Code Generation
- New feature, still in development
- Speedup depends on lots of things
First-Class Models
julia> m = @model begin
a ~ @model begin
x ~ Normal()
end
end;
julia> rand(m())
(a = (x = -0.20051706307697828,),)julia> m2 = @model anotherModel begin
y ~ anotherModel
z ~ anotherModel
w ~ Normal(y.a.x / z.a.x, 1)
end;
julia> rand(m2(anotherModel=m)).w
-1.822683102320004Higher-order Model Example
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endArguments
Body
- Models are declarative and function-like
- Input the arguments
- Output everything (as a named tuple)
Suspiciously missing:
Which variables are observed?
Getting the DAG
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endjulia> digraph(m).N # forward
Dict{Symbol,Set{Symbol}} with 6 entries:
:σ => Set(Symbol[:y])
:y => Set(Symbol[])
:yhat => Set(Symbol[:y])
:β => Set(Symbol[:yhat])
:Prior => Set(Symbol[:β])
:x => Set(Symbol[:y, :yhat])
julia> digraph(m).NN # reverse
Dict{Symbol,Set{Symbol}} with 6 entries:
:σ => Set(Symbol[])
:y => Set(Symbol[:σ, :yhat, :x])
:yhat => Set(Symbol[:β, :x])
:β => Set(Symbol[:Prior])
:Prior => Set(Symbol[])
:x => Set(Symbol[])x
\beta
\hat{y}
y
\text{Prior}
\sigma
using Ed Scheinerman's SimpleWorld.jl
The Predictive Distribution
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endjulia> pred = predictive(m, :β)
@model (x, σ, β) begin
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endx
\beta
\hat{y}
y
\text{Prior}
\sigma
julia> predictive(m, :yhat)
@model (x, σ, yhat) begin
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endusing Taine Zhao's MLStyle.jl
Model Types
struct Model{Args, Body}
args :: Vector{Symbol}
vals :: NamedTuple
dists :: NamedTuple
retn :: Union{Nothing, Symbol, Expr}
endusing Taine Zhao's GeneralizedGenerated.jl
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endjulia> typeof(m)
Model{
NamedTuple{(:Prior, :x, :σ),T}
where T<:Tuple,
begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
end
}Thanks to Taine Zhao for this improvement
Importance Sampling
p = @model begin
α ~ Normal(1,1)
β ~ Normal(α^2,1)
end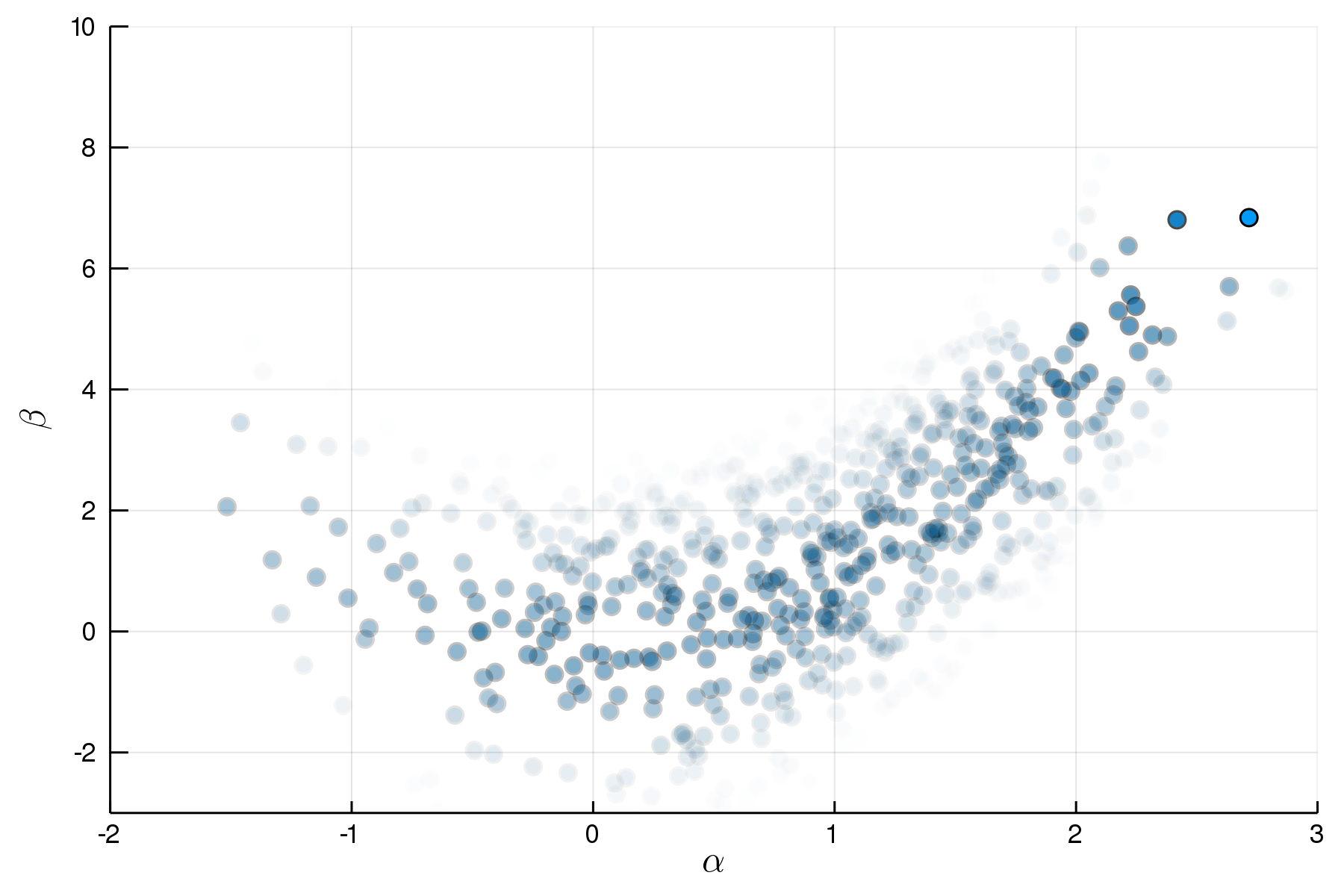
q = @model μα,σα,μβ,σβ begin
α ~ Normal(μα,σα)
β ~ Normal(μβ,σβ)
end julia> sourceParticleImportance(p,q)
:(function ##particlemportance#737(##N#736, pars)
@unpack (μα, σα, μβ, σβ) = pars
ℓ = 0.0 * Particles(##N#736, Uniform())
α = Particles(##N#736, Normal(μα, σα))
ℓ -= logpdf(Normal(μα, σα), α)
β = Particles(##N#736, Normal(μβ, σβ))
ℓ -= logpdf(Normal(μβ, σβ), β)
ℓ += logpdf(Normal(1, 1), α)
ℓ += logpdf(Normal(α ^ 2, 1), β)
return (ℓ, (α = α, β = β))
end)=> Variational Inference
\ell(x) = \log p(x) - \log q(x)
\text{Sample } x \sim q\text{, then evaluate}
Hidden Markov Models, Step by Step
hmmStep = @model s0,step,noise begin
s1 ~ EqualMix(step.(s0))
y ~ noise(s1.x)
end;julia> s0 = rand(Normal(0,10), 100);
julia> particles(s0)
Part100(0.4518 ± 9.93)
julia> rand(hmmStep(args)) |> pairs
pairs(::NamedTuple) with 5 entries:
:s0 => [7.9962, -4.30039, 8.0346, -0.628184, … 20.3864…]
:step => step
:noise => noise
:s1 => (s = 18.0924, ν = 2.0472, x = 16.8622)
:y => (s = 16.8622, y = 15.0147)
julia> dynamicHMC(hmmStep(args), (y=(y=1.0,),)) |> particles
(s1 = (ν = 5.48 ± 26.0, x = 0.792 ± 0.95),)function step(s)
m = @model s begin
ν ~ HalfCauchy()
x ~ StudentT(ν, s,1)
end
m(s=s)
end;function noise(s)
m = @model s begin
y ~ Normal(s,1)
end
m(s=s)
end;Thank You!
A Running Example
\begin{aligned}
\text{Prior} &\in \mathbb{P}(\mathbb{R}) \\
x &\in \mathbb{R}^n \\
\sigma &\in \mathbb{R}_+ \\
\\
\beta &\sim \text{Prior} \\
\hat{y} &= \beta x \\
y &\sim \text{Normal}(\hat{y},\sigma)
\end{aligned}
- Build the model
- Sample from generative model
- Evaluate log density
- Sample from posterior
- Determine predictive distribution
- Sample from posterior predictive distribution
Goals
Sampling from the Model
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endjulia> Prior = Normal()
Normal{Float64}(μ=0.0, σ=1.0)
julia> args = (Prior=Prior, x=x, σ=2.0);
julia> truth = rand(m(args));
julia> pairs(truth)
pairs(::NamedTuple) with 6 entries:
:Prior => Normal{Float64}(μ=0.0, σ=1.0)
:x => [-0.556027, -0.444383, 0.0271553]
:σ => 2.0
:yhat => [0.166521, 0.133086, -0.00813259]
:β => -0.299484
:y => [3.72224, -2.15672, -0.945344]Joint Distributions
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endjulia> pairs(args)
pairs(::NamedTuple) with 3 entries:
:Prior => Normal{Float64}(μ=0.0, σ=1.0)
:x => [-0.556027, -0.444383, 0.0271553]
:σ => 2.0
julia> m(args)
Joint Distribution
Bound arguments: [Prior, x, σ]
Variables: [β, yhat, y]
@model (Prior, x, σ) begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
end
Sampling from the Posterior
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endjulia> pairs(args)
pairs(::NamedTuple) with 3 entries:
:Prior => Normal{Float64}(μ=0.0, σ=1.0)
:x => [-0.556027, -0.444383, 0.0271553]
:σ => 2.0
julia> post = dynamicHMC(m(args), (y=truth.y,));
julia> particles(post)
(β = -0.181 ± 0.92,)using Tamas Papp's DynamicHMC.jl
and Fredrik Bagge Carlson's MonteCarloMeasurements.jl
The Predictive Distribution
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endjulia> argspost = merge(args, particles(post));
julia> pairs(argspost)
pairs(::NamedTuple) with 4 entries:
:Prior => Normal{Float64}(μ=0.0, σ=1.0)
:x => [-0.556027, -0.444383, 0.0271553]
:σ => 2.0
:β => -0.18 ± 0.9julia> pred = predictive(m, :β)
@model (x, σ, β) begin
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endThe Predictive Distribution
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endjulia> argspost = merge(args, particles(post));
julia> pairs(argspost)
pairs(::NamedTuple) with 4 entries:
:Prior => Normal{Float64}(μ=0.0, σ=1.0)
:x => [-0.556027, -0.444383, 0.0271553]
:σ => 2.0
:β => -0.18 ± 0.9
julia> postpred = pred(argspost) |> rand;
julia> pairs(postpred)
pairs(::NamedTuple) with 5 entries:
:x => [-0.556027, -0.444383, 0.0271553]
:σ => 2.0
:β => -0.18 ± 0.9
:yhat => [0.1 ± 0.5, 0.0801 ± 0.4, -0.0049 ± 0.024]
:y => [-5.18 ± 0.5, 2.09 ± 0.4, 2.16 ± 0.024]
julia> pred = predictive(m, :β)
@model (x, σ, β) begin
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endUnder the Hood
Canonical Representation
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endjulia> canonical(m)
@model (Prior, x, σ) begin
β ~ Prior
yhat = β .* x
y ~ For(((j,)->begin
Normal(yhat[j], σ)
end), (eachindex(x),))
endInference Primitive: rand
julia> Soss.sourceRand()(m)
quote
β = rand(Prior)
yhat = β .* x
y = rand(For(((j,)->begin
Normal(yhat[j], σ)
end),
(eachindex(x),)))
(Prior = Prior, x = x, σ = σ,
yhat = yhat, β = β, y = y)
endm = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endInference Primitive: logpdf
julia> Soss.sourceLogpdf()(m)
quote
_ℓ = 0.0
_ℓ += logpdf(Prior, β)
yhat = β .* x
_ℓ += logpdf(For(eachindex(x)) do j
Normal(yhat[j], σ)
end, y)
return _ℓ
endm = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endInference Primitive: weightedSample
julia> Soss.sourceWeightedSample((y=truth.y,))(m)
quote
_ℓ = 0.0
β = rand(Prior)
yhat = β .* x
_ℓ += logpdf(For(eachindex(x)) do j
Normal(yhat[j], σ)
end, y)
return (_ℓ,
(Prior = Prior, x = x, σ = σ,
yhat = yhat, β = β, y = y)
)
end
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endInference Primitive: xform
m = @model Prior,x,σ begin
β ~ Prior
yhat = β .* x
y ~ For(eachindex(x)) do j
Normal(yhat[j], σ)
end
endjulia> xform(jd1, (y=truth.y,))
TransformVariables.TransformTuple{
NamedTuple{(:β,),Tuple{TransformVariables.Identity}}
}((β = asℝ,), 1)
julia> xform(jd2, (y=truth.y,))
TransformVariables.TransformTuple{
NamedTuple{(:β,),Tuple{TransformVariables.ShiftedExp{true,Float64}}}
}((β = asℝ₊,), 1)julia> jd1 = m(Prior=Normal(), x=x, σ=2.0);
julia> jd2 = m(Prior=Exponential(), x=x, σ=2.0);using Tamas Papp's TransformVariables.jl
Copy of 2023-03-Basis-LabMeeting
By Chad Scherrer
Copy of 2023-03-Basis-LabMeeting
- 527



