Motivation
Explosion of -omics data
Can we combine transcriptomics data and metabolic network modelization in order to infer "metabolic goals" ?
Difficulty to get metabolic flux measurements
- Transcriptomic to flux
- Flux to metabolic goal
SPOT
InvFBA
Can we observe differences between conditions ?
Workflow



| GENE EXPRESSION TO "REACTION EXPRESSION" (GPR association) |
|---|
Gene expression
Metabolic network
External database
| Sample 1 | Sample 2 | |
|---|---|---|
| reaction 1 | 8 | 7 |
| reaction 2 | 4 | 2 |
| SPOT |
|---|
| Flux 1 | Flux 2 | |
|---|---|---|
| reaction 1 | 0,0017 | 0,001 |
| reaction 2 | 0,02 | 0,01 |
| InvFBA |
|---|


METABOLIC GOALS
"Reaction expression"
Fluxes
SPOT
Gene-Protein-Reaction association
rxn01964_c0= ((fig|32051.5.peg.751 and fig|32051.5.peg.2372) or fig|32051.5.peg.2168)
Reaction rule
| fig|32051.5.peg.751 | fig|32051.5.peg.2372 | fig|32051.5.peg.2168 | |
|---|---|---|---|
| Gene expression | 7 | 15 | 23 |
Gene expression
Reaction value
- "Unknown" genes in reactions rules
- Reactions with no genes associated
- "NA" values in gene expression
246/1095 reactions
in Synechococcus
SPOT
Compute metabolic flux from transcriptomic data
Validation on 20 samples (E.coli and S.cerevisiae) where flux measurements (via C-MFA) and transcriptomic data were available
No a priori assumption on objective function
Simplified Pearson cOrrelation with Transcriptomic data
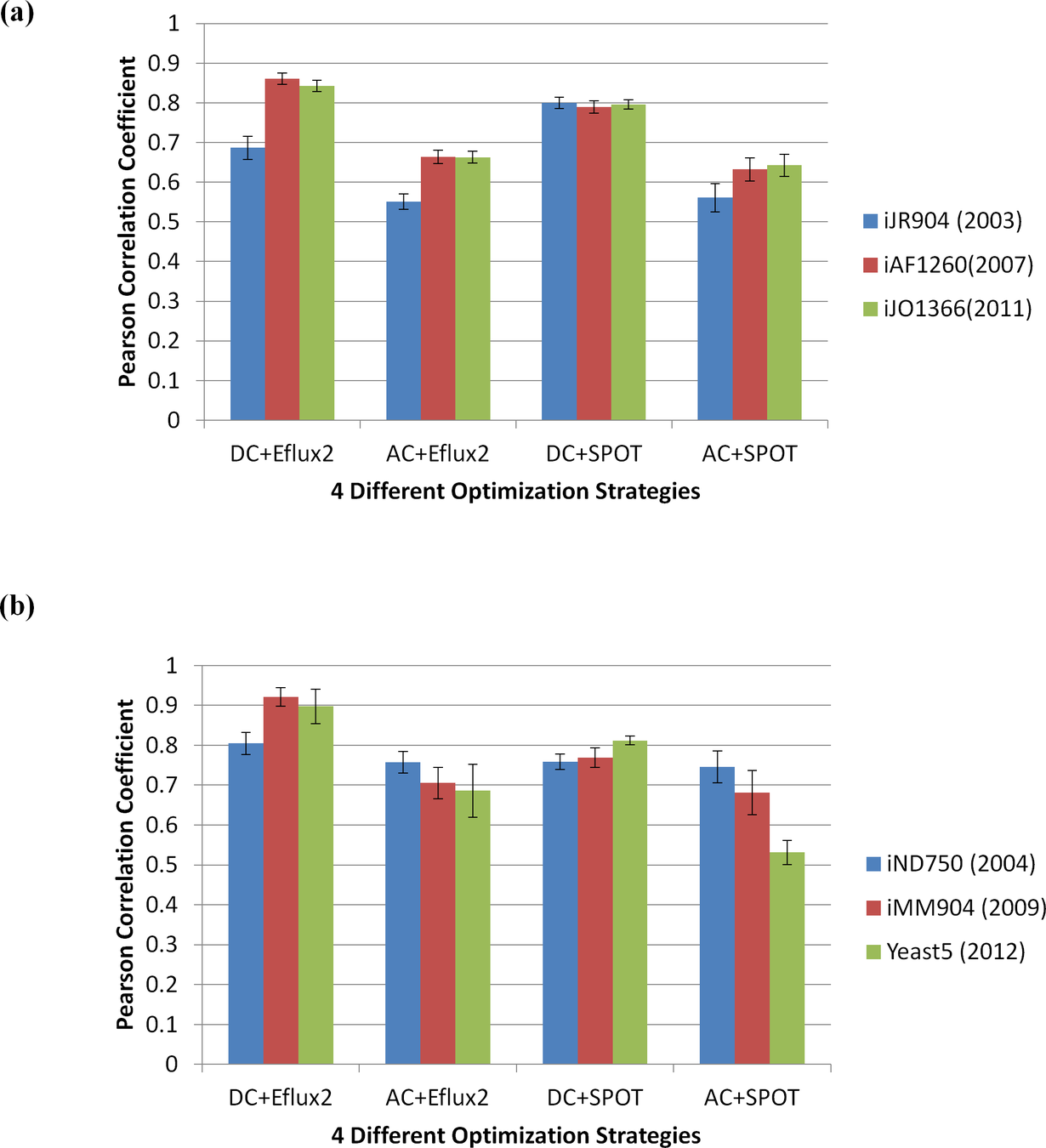
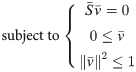
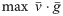
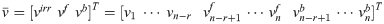
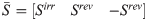
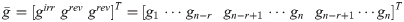
Decompose reversible reactions
InvFBA
Models


Find an objective function (c) that minimizes the gap
between optimal flux and observed flux
Get sparser objective function
optimal solution of FBA
i.e., find a vector c that lead to near optimality of
Infer organism goal from observed fluxes
Linear Programming, duality theory
GUROBI
OVA
Objective Variability Analysis

If OVA ran on all reactions, we observe that there are plenty of reactions coefficients that can take the maximum value
OVA determines the range of each coefficient in the objective function
There exists an infinity of invFBA solutions
InvFBA
Experimentations
1) Simulated e-coli fluxes from FBA with 3 different carbon source limitations
4) Different E-coli strains measured fluxes
Biomass function retrieved
(under reasonable noise)
3) Fluxes inferred with TEAM
2) Noisy simulated e-coli fluxes from FBA
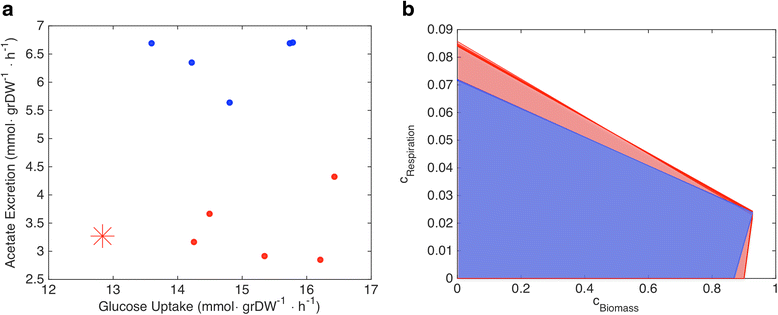
Analyzed with OVA
Tradeoff
Focusing on specific reactions (all non exchange reactions set to zero)
Outline
Rewrite SPOT and invFBA methods in Python
Validate each implementation by comparing it to its MATLAB version
Associate the two methods together and check results
Validation methods ?
Weird preliminary results
Adapt invFBA in order to fit the SPOT decomposition of reversible reactions
Package methods under conda environment and deploy it
InvFBA
Validation
Biomass reaction successfully retrieved on ec_iJO1366 with MATLAB and with Python => same coefficients in both cases
Test on COBRA samples where we apply FBA (with biomass reversible reactions set as an objective)
Forward biomass reaction retrieved in : salmonella, ecoli
not in ecoli_core
EX_cpd11416_c0 : 0.427558420744
rxn01116_c0 : 0.131712221776
rxn00459_c0 : 0.131712221776
rxn05319_c0 : 0.111448803041
EX_cpd00067_e0 : 0.0658561108879
EX_cpd00007_e0 : 0.0607902562042
rxn00260_c0 : 0.0506585468369
EX_cpd00041_e0 : 0.0202634187348
Test on WH7803 (Synechococcus)
and simulated flux (via FBA)
-1.0*biomass0_reverse_9c18d + 1.0*biomass0
Objective function
Sample included with InvFBA
... but there exists a solution where the
biomass reaction coefficient equals 1
Results change at
each run
OVA
Objective Variability Analysis
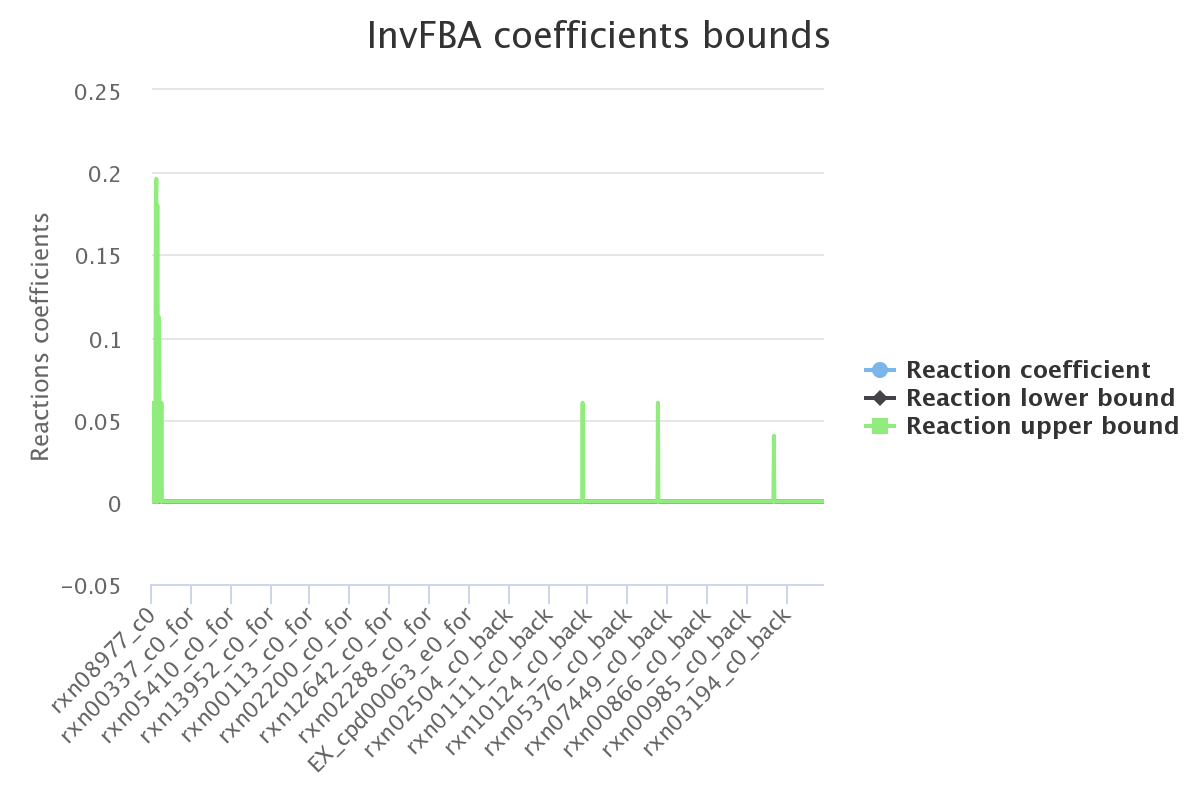
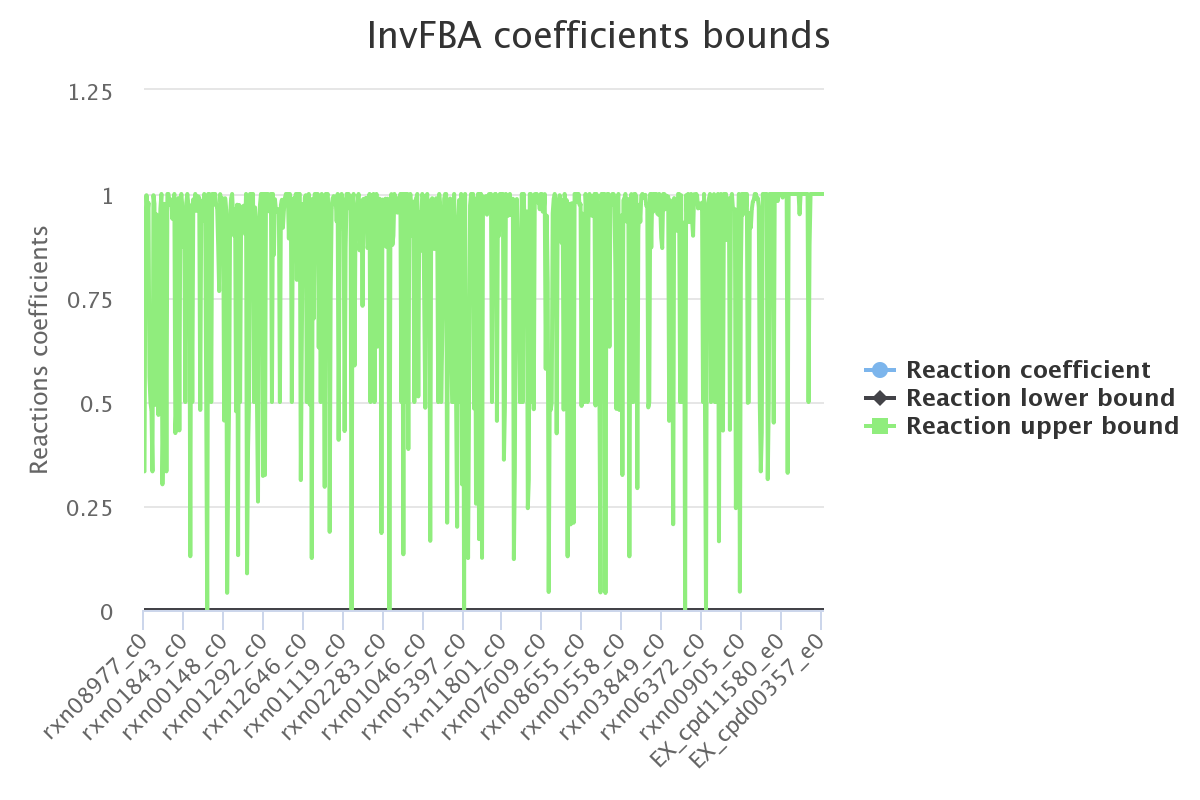
Decomposition
SPOT max value
Ran on all non null coefficients
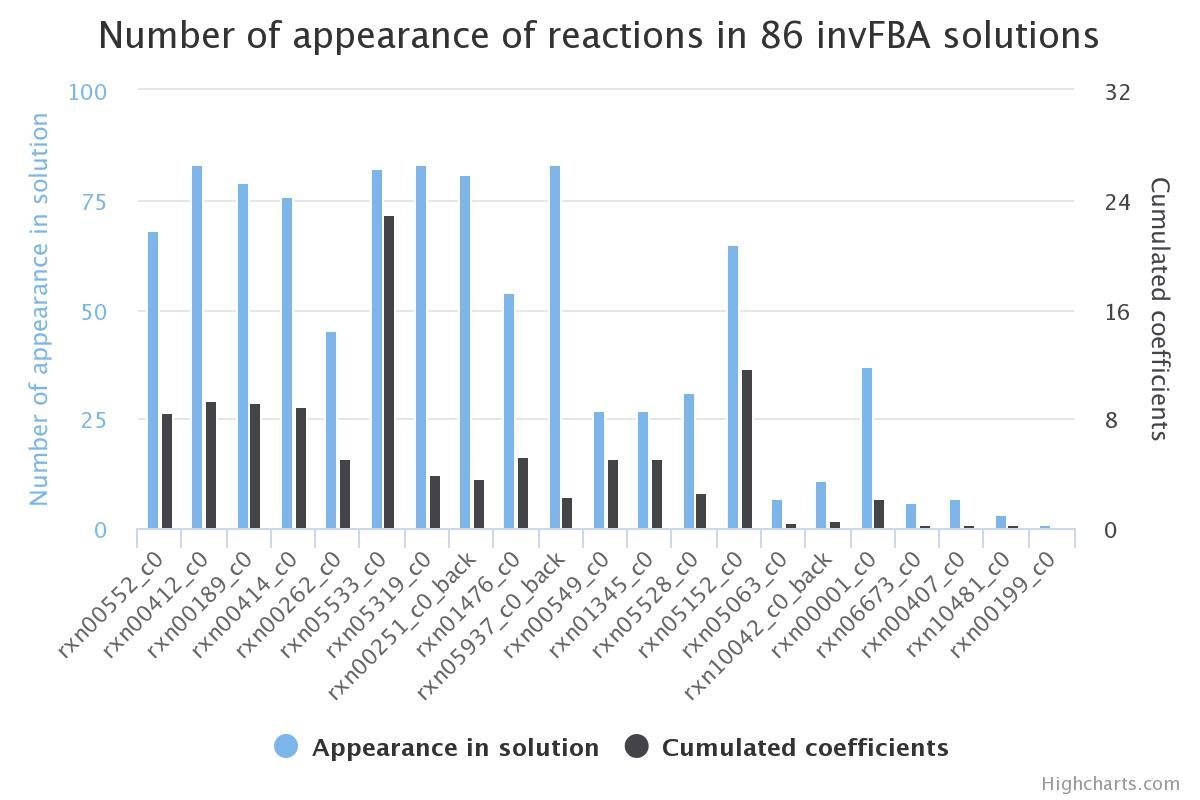
Test on all conditions
Questions
Fluxes obtained with SPOT ranges between 0 and 1
-> What about the reaction bounds in invFBA ?
Decomposition of reversible reactions
-> Adapt invFBA model ?
-> Take max(for,back) and run with classical invFBA model ?
Filter 10e-6 values (gurobi warning about large range matrix)
Validation ??
Set non exchange reaction to 0 ?
Biomass reaction ?
InvFBA-pointJeremie
By edelage
InvFBA-pointJeremie
- 489


