Cloud of Reproducible Records



Faical Yannick P. Congo
faical.congo@nist.gov
MRaDS
Wednesday, September 27, 2017
Institute for Bioscience and Biotechnology Research
https://slides.com/faicalyannickcongo/corr/live
Buckle up for this Journey
BUT BEFORE...
LET'S START WITH SOME FACTS
version control?
execution management?
research pedigree?
code#execution versioning!
execution tools!
coRR!
The scientist's nightmares
IN...
3 Scenarios


Early RESEARCH BACK and Forth SWING


I have to
run my study again and
again on...
different datasets
different code
different parameters
How do i keep track of all that in a meaningful way to me and others?
Research pipeline rationale



week
day
month
year
semerter
rational 1
rational 2
rational 3
rational 4
rational 5
rational 6
How do i keep track of all the rationale i had during my study timeline?
scale
during a study timeline, a scientist will take actions for different reasons!
using existing publications materials



publication
extract these
reconstruct
execute
corroborate
research involves others using your study in theirs. it needs a lot more!
How do i capture everything needed for a pair to run my study easily?

Humm...
What I need is
sort of
- A provenance tracking Tool?
- A work-flow management tool?
- A pedigree capture tool?
what about version control?
BUT
Which one?
Source Code
EXECUTION
YES: They are two very different things!
Source Code
EXECUTION
CODE MODIFICTAIONS
RUN CHANGES
Cleaner with Branches
LIBRARIES TRACKING
easy backups
SYSTEM Changes
why not use source codeversion control to do all the job?
Let's SEE...
The things that can affect your executions but not your code:
- A change in hardware may affect your run. Your code less likely.
- An OS update may affect your run. Change your code less likely.
- Using different data may affect your run. Your code less likely.
- Changing parameters may affect your run. Your code less likely.
seriously! Do not use source code version control for execution version control
- Runs as different commits in the same branch?
- Runs as different branches in the same repository?
- Runs as different repositories?
each of these are not appropriate as is
RUNS as different commits in the same branch?
- All runs will have to produce a file treated as source code
- A commit becomes an nondeterminate state
- Does not scale well for huge amounts of runs
BASICALLY treat a run as a code change?
RUNS as different branches in the same repository?
- A branch becomes an nondeterminate state
BASICALLY treat a run as a new branch?
- The notion of merging two branches has to be redefined.
- Scaling is still a big problem.
RUNS as different repositories?
- A repository becomes an nondeterminate state
- The notion of a repository as a project loses all its sense
BASICALLY treat a run as a new repo?
- Scaling is still a big problem.
so It is settled then!
CODE VERSION CONTROL IS NOT APPROPRIATE FOR EXECUTION
YET!
What is worse than not using version control for your code today?
Let's ASSUME we have the solution
Cloud Storage
without it, all your work is seating on a single point of failure: loss, corruption, deletion, ...
cloud storage and all its subsequent features are a must have!
WHY? The two best success Stories:
Github
bitbucket
BACKUP, COLLABORATION, exposure, dissemination, open-source, custom features

scientists need source code version control
scientists need a cloud storage option: Github|Bitbucket
BUT for executions
scientists need execution version control tools
scientists need a cloud storage alternative with them
luckily: There are quite a few!
executions management systems
NOTEBOOKS
WORKFLOWS
PROVENANCE
SYSTEM
HERE ARE 40 of them
Sumatra
reprozip
cde
panda
noworkflow
maestrowf
hubzero
galaxy
taverna
aiida
kepler
fireworks
ergatis
anduril
askalon
airavata
biobkie
autosubmit
bioclipse
hyperflow
cknowledge
cuneiform
nextflow
knime
nipype
openmole
ogrange
pegasus
scicumulus
vistrails
yabi
tavaxy
jupyter
zeppelin
beaker
nteract
kajero
ucalc
eve
hyperdeck
Great then: We just have to use these!
we are here because it is not that simple
YES, but wait...
with source code version control tools
we have the following features
git
Mercurial
subversion
bazaar
cvs
fossil

migration
cloud services

sadly out of these, only half are
Sumatra
reprozip
cde
panda
noworkflow
maestrowf
hubzero
galaxy
taverna
aiida
kepler
fireworks
ergatis
anduril
askalon
airavata
biobkie
autosubmit
bioclipse
hyperflow
cknowledge
cuneiform
nextflow
knime
nipype
openmole
ogrange
pegasus
scicumulus
vistrails
yabi
tavaxy
jupyter
zeppelin
beaker
nteract
kajero
ucalc
eve
hyperdeck
connected and none has a
migration capability






















this gave birth to
Cloud of Reproducible Records



migration capability between the tools
common cloud services features
corr is a nist-mgi funded project

usnistgov/corr
OPEN-SOURCE
DEMO live soon
collaborations



docker-hub
corr [microservices] architecture

rest
file system | s3
rest
js | css | html5
mongodb
HOME TOP
HOME menu
HOME bottom

HOME docs

HOME search
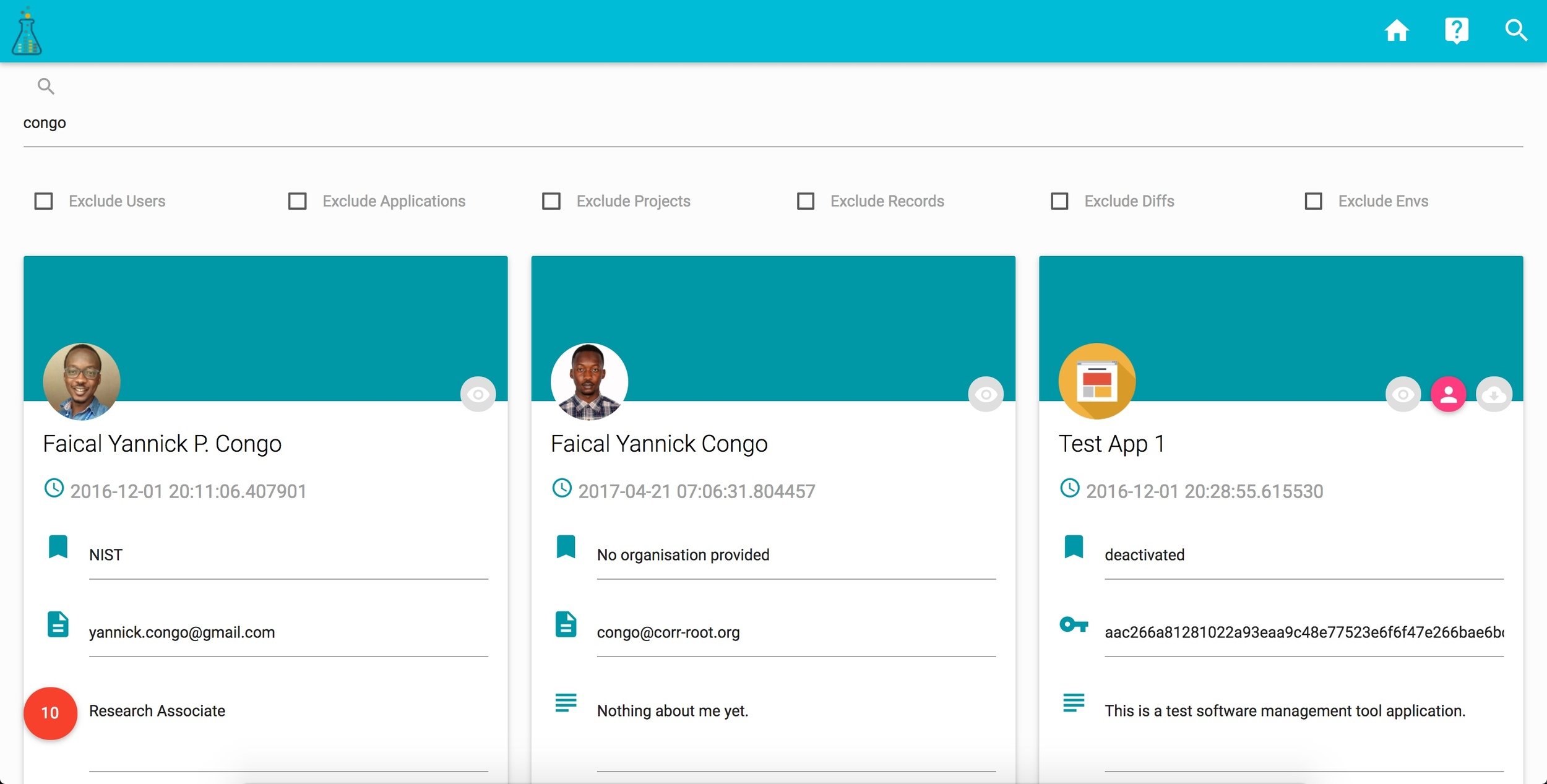
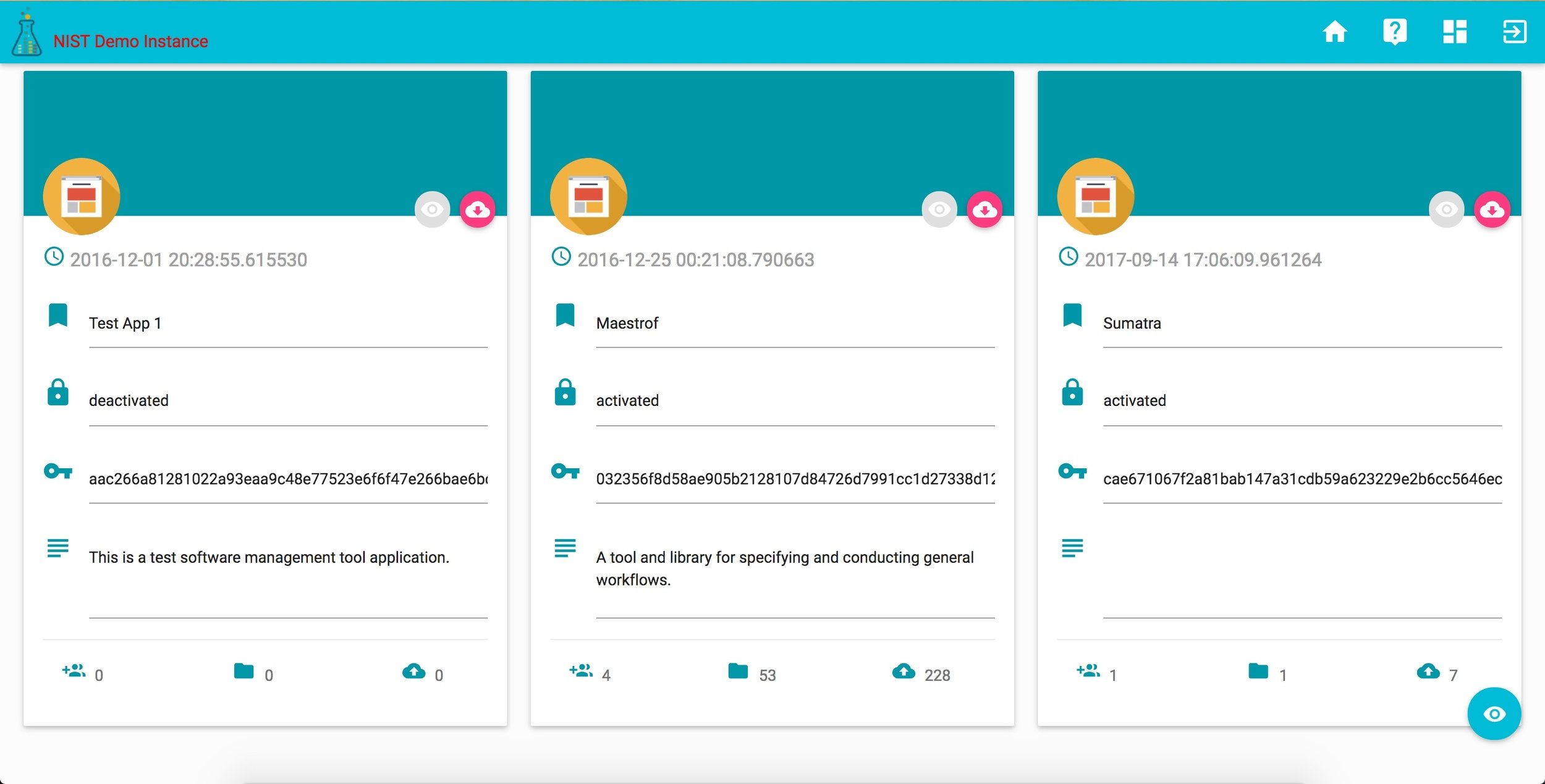
corr ACCOUNT

corr DASHBOARD
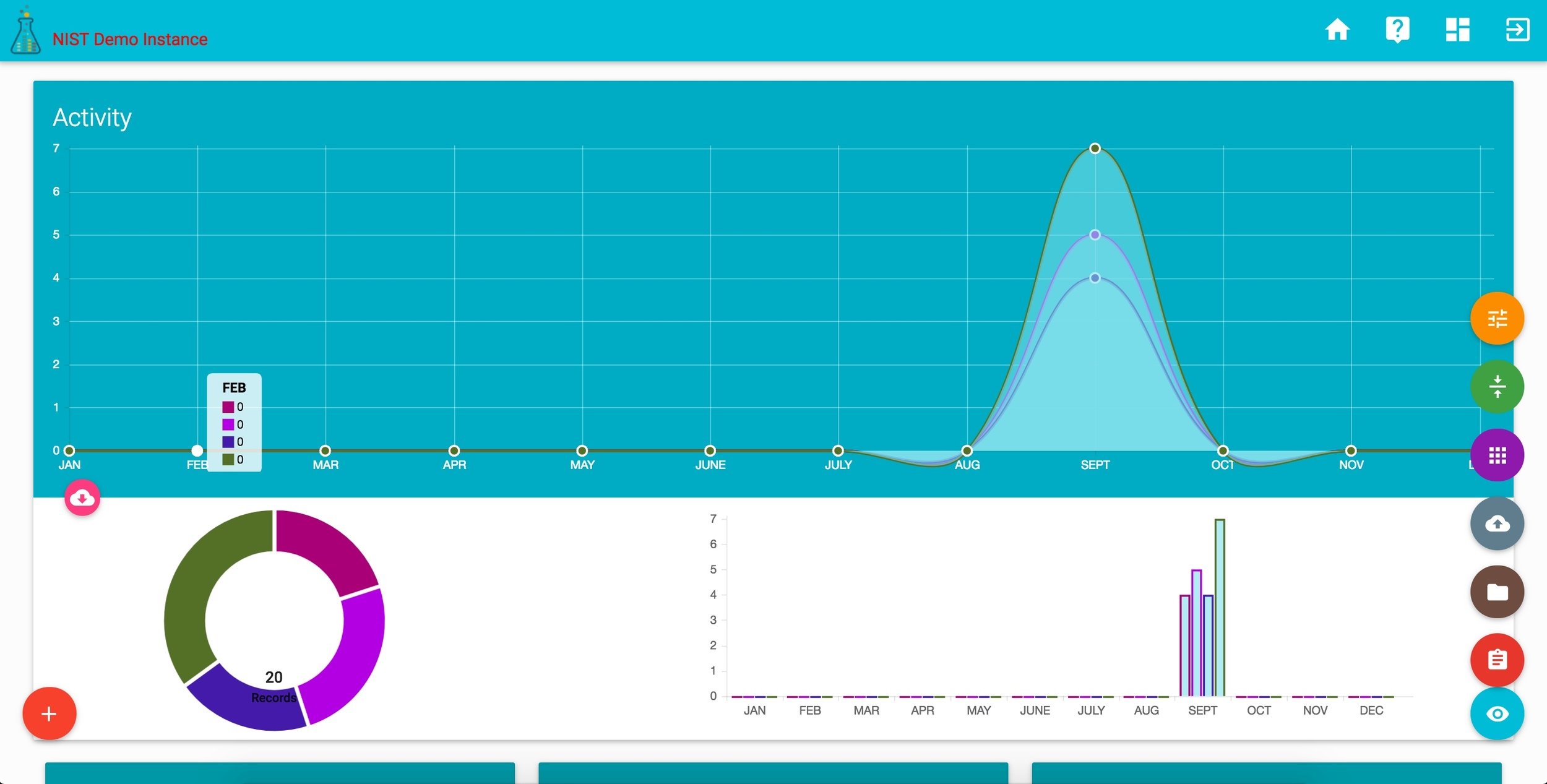
corr projects
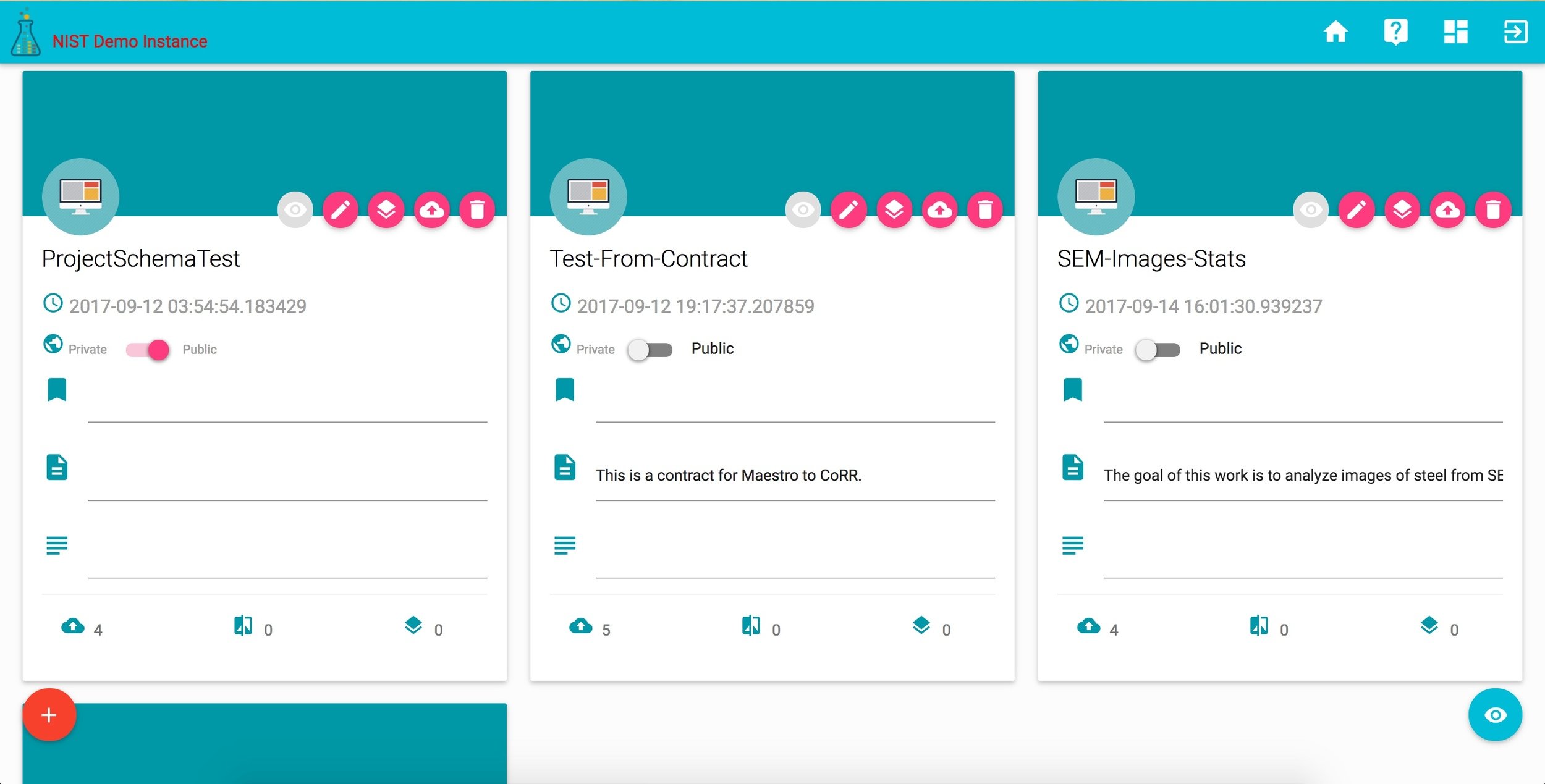
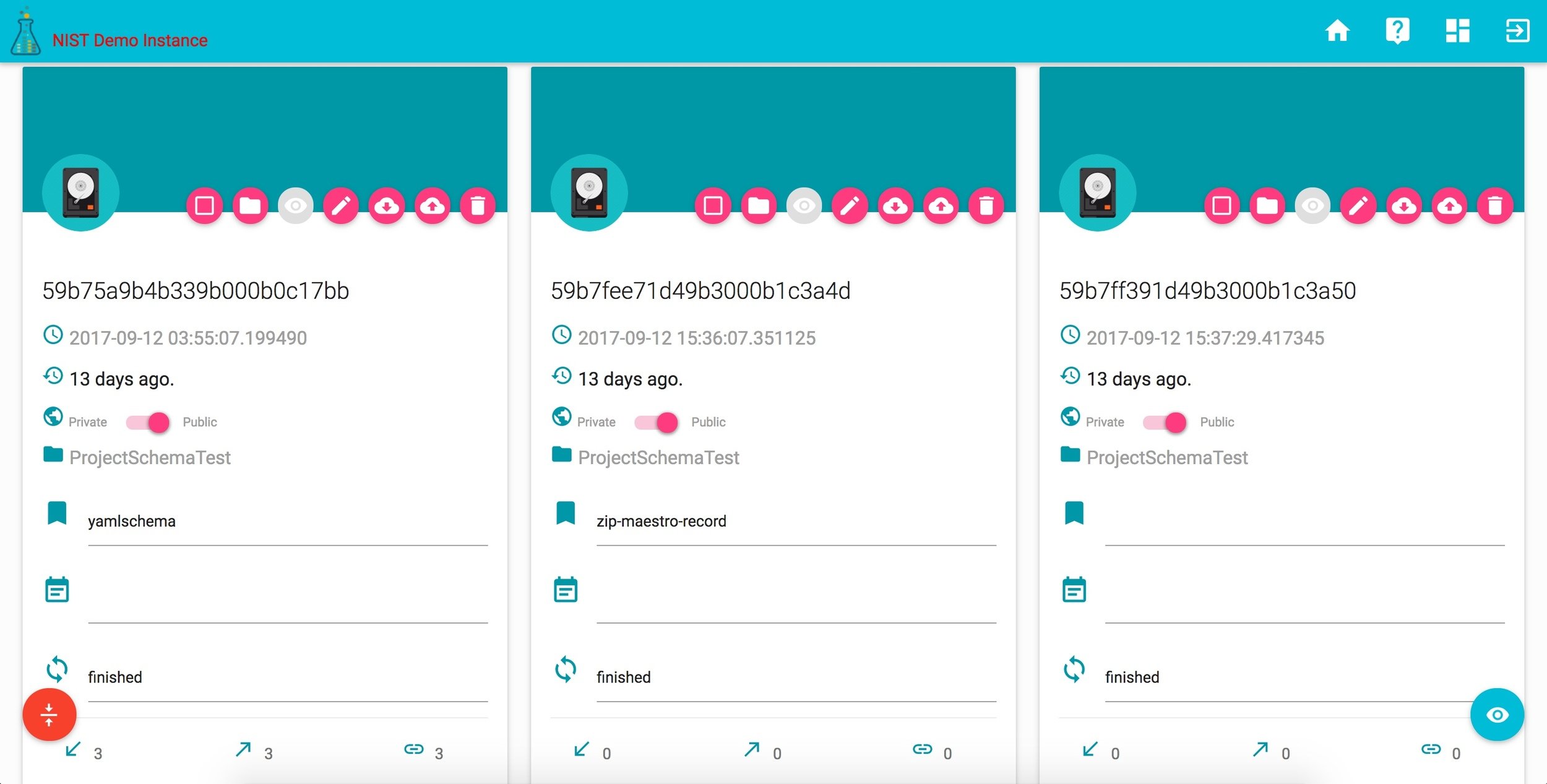
corr records
corr tools
research project demo
Credit: ShenG Yen Li, Daniel Wheeler
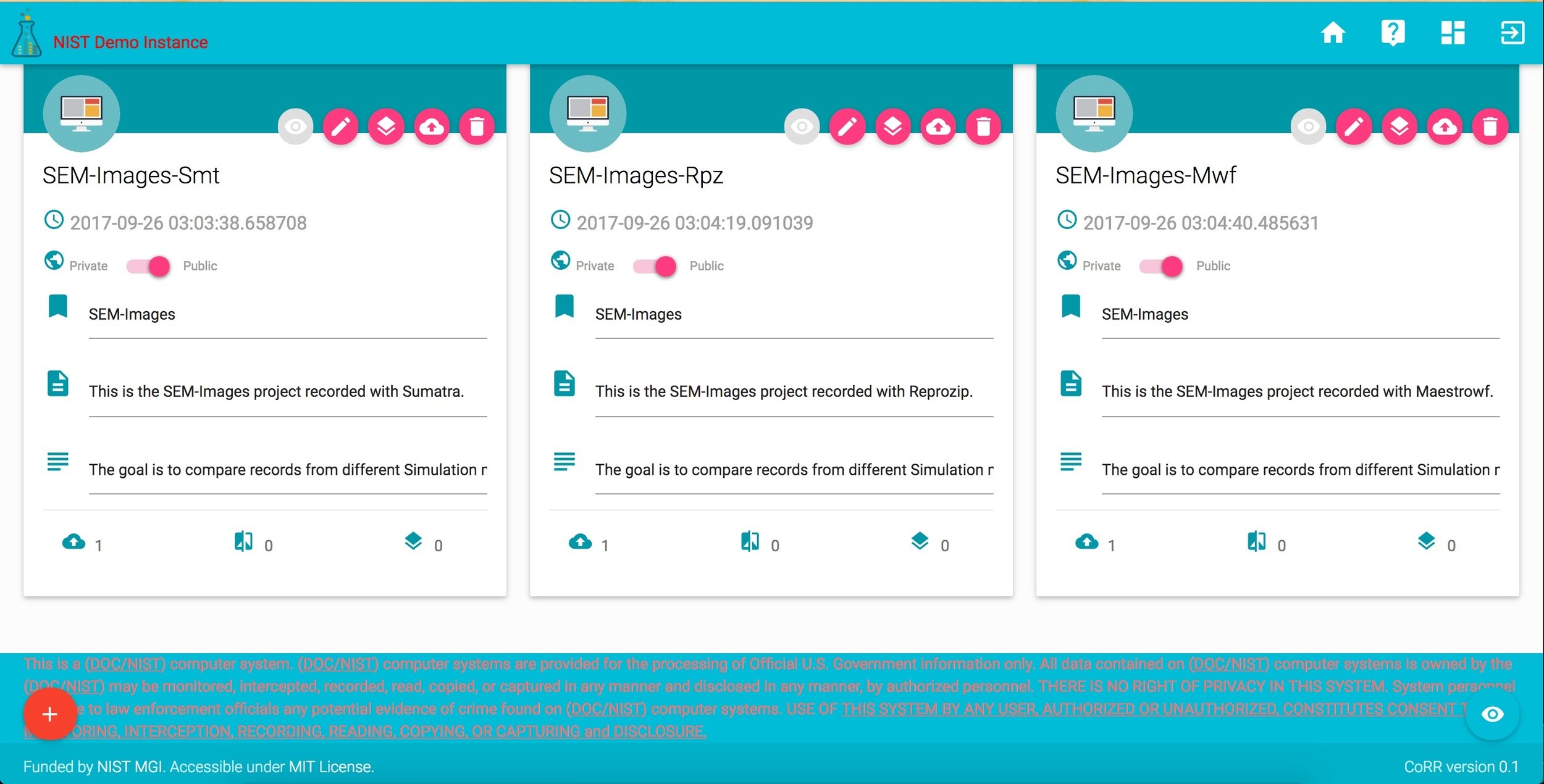
sem images
threshold
min-size
clean
reveal
pearlite
ferrite
cemmentite
save
json fractions
study
demo
1
2
3
4
5
8
7
6
without corr
with corr


study
sumatra


study
reprozip


study
maestrowf


study
sumatra


study
reprozip


study
maestrowf



study
def threshold(filename):
result = dict(filename=filename,
threshold_image=threshold_image(filename),
**extract_metadata(filename))
return resultdef min_size(data):
data['min_size'] =
f_min_size(data['scale_microns'],
data['scale_pixels'])
return datadef clean(data):
data['clean_image'] =
~remove_small_holes(~data['threshold_image'],
data['min_size'])
return datadef reveal(data):
data['pearlite_image'] =
reveal_pearlite(data['clean_image'])
return datadef cemmentite(data):
data['cemmentite_fraction'] =
frac1(data['clean_image'])
return datadef ferrite(data):
data['ferrite_fraction'] =
frac0(data['clean_image'])
return datadef pearlite(data):
data['pearlite_fraction'] =
frac1(data['pearlite_image'])
return datadef save(data):
clean_name = data['filename'].
split("/")[-1].
split(".")[0]
file_path = "{0}.json".format(clean_name) filtered_data = {}
filtered_data['filename'] = clean_name
filtered_data['pearlite_fraction'] = data['pearlite_fraction']
filtered_data['ferrite_fraction'] = data['ferrite_fraction']
filtered_data['cemmentite_fraction'] = data['cemmentite_fraction']
with open(file_path, "w") as save_file:
save_file.write(json.dumps(filtered_data, sort_keys=True,
indent=4, separators=(',', ': ')))demo
sumatra
## Setup Version Control
$ git init
$ git add --all
$ git commit -m "Setting up the repo."## Setup Sumatra without CoRR
$ smt init SEM-Images-Smt .
## Setup Sumatra with CoRR
$ smt init -s=config.json SEM-Images-Smt .## Run Study without Sumatra
$ python study.py## Run Study with Sumatra
$ smt run --executable=python --main=study.py## Produces:
## - A folder: .git## Produces:
## - A folder: .smt
## - A file: .smt/project
## - A file: .smt/records## Produces:
## - A list of json files.## Produces:
## - A row in .smt/records
## - A list of json files.demo
reprozip
## Trace the study run
$ reprozip trace SEM-Image-Rpz python study.py
## Trace with CoRR
$ reprozip trace -s=config_file SEM-Image-Rpz python study.py## Produces:
## - A folder: .reprozip
## - A file: bundle.rpzdemo
maestrowf
## Run the study
$ maestro -s -d 1 -y -c -t 2 sem-study.yaml
## Archive the run with CoRR
$ archive -f config.json record_path/sem-study.yaml -d 1## Produces:
## - A folder: sample_output
## - A folder: */sem-images-maestrowf
## - A folder: */*/*_recordIDprojects in CoRR
contracts module
thank you!
Cloud of Reproducible Records

Faical Yannick P. Congo
faical.congo@nist.gov
Wednesday, September 27, 2017
Institute for Bioscience and Biotechnology Research
corr stickers coming up!
federation capability!
demo users registrations
questions?
more time? Hands on!
Cloud of Reproducible Records

Faical Yannick P. Congo
faical.congo@nist.gov
Wednesday, September 27, 2017
Institute for Bioscience and Biotechnology Research
how many laptops?
who has done this before?
instance available
follow me :-)
http://10.5.100.207:5000/
https://github.com/usnistgov/MRaDS-2017-Demo-Study
corr
By Faical Yannick Congo
corr
- 905



