Automated Ingest
Jason M. Coposky
@jason_coposky
Executive Director, iRODS Consortium
Automated Ingest

January 14-16 2020
CINES
Montpellier, France

- Packaged and supported solutions
- Require configuration not code
- Derived from the majority of use cases observed in the user community








iRODS Capabilities

Automated Ingest

Provide a flexible and highly scalable mechanism for data ingest
- Directly ingest files
- Capture streaming data
- Register data in place
- Synchronize a file system with the catalog
- Extract and apply metadata
Architecture Overview

- Implemented with Python iRODS Client
- Based on Redis and Redis-Queue
- Any number of workers distributed across servers
- Policy defined through event callbacks to user-provided python functions
- File system metadata cached in Redis to detect changes
- iRODS API is invoked only to update the catalog
Getting Started

sudo apt-get install -y redis-server python-pip
sudo service redis-server start
As the ubuntu user - start Redis server
pip install virtualenv --user python -m virtualenv -p python3 rodssync source rodssync/bin/activate pip install irods_capability_automated_ingest
As the irods user - install automated ingest via pip
Getting Started

export CELERY_BROKER_URL=redis://127.0.0.1:6379/0 export PYTHONPATH=`pwd`
celery -A irods_capability_automated_ingest.sync_task worker -l error -Q restart,path,file
Open a new terminal, activate rodssync as the irods user, and set environment variables for the scanner
As the irods user - start Celery workers
source rodssync/bin/activate export CELERY_BROKER_URL=redis://127.0.0.1:6379/0 export PYTHONPATH=`pwd`
Getting Started

mkdir /tmp/test_dir cp irods_training/stickers.jpg /tmp/test_dir/img0.jpg cp irods_training/stickers.jpg /tmp/test_dir/img1.jpg cp irods_training/stickers.jpg /tmp/test_dir/img2.jpg cp irods_training/stickers.jpg /tmp/test_dir/img3.jpg cp irods_training/stickers.jpg /tmp/test_dir/img4.jpg
Generate some source data
mkdir ./src_dir
cp -r /tmp/test_dir ./src_dir/dir0
Stage source data
git clone https://github.com/irods/irods_training/
Fetch test data
Default Ingest Behavior

By default the framework will register the data in place against the default resource into the given collection
imkdir reg_coll python -m irods_capability_automated_ingest.irods_sync start ./src_dir /tempZone/home/rods/reg_coll
Check the redis monitor terminal for the restart, file and path queues
Default Ingest Behavior

Check that our results are registered in place
$ ils -L reg_coll/dir0 /tempZone/home/rods/reg_coll/dir0: rods 0 demoResc 2157087 2019-05-21.05:54 & img0.jpg generic /var/lib/irods/src_dir/dir0/img0.jpg rods 0 demoResc 2157087 2019-05-21.05:54 & img1.jpg generic /var/lib/irods/src_dir/dir0/img1.jpg rods 0 demoResc 2157087 2019-05-21.05:54 & img2.jpg generic /var/lib/irods/src_dir/dir0/img2.jpg rods 0 demoResc 2157087 2019-05-21.05:54 & img3.jpg generic /var/lib/irods/src_dir/dir0/img3.jpg rods 0 demoResc 2157087 2019-05-21.05:54 & img4.jpg generic /var/lib/irods/src_dir/dir0/img4.jpg
Default Ingest Behavior

Stage a different set of data and re-run the ingest
cp -r /tmp/test_dir ./src_dir/dir1 python -m irods_capability_automated_ingest.irods_sync start ./src_dir /tempZone/home/rods/reg_coll
$ ils -L reg_coll/dir1 /tempZone/home/rods/reg_coll/dir1: rods 0 demoResc 2157087 2019-05-21.06:11 & img0.jpg generic /var/lib/irods/src_dir/dir1/img0.jpg rods 0 demoResc 2157087 2019-05-21.06:11 & img1.jpg generic /var/lib/irods/src_dir/dir1/img1.jpg rods 0 demoResc 2157087 2019-05-21.06:11 & img2.jpg generic /var/lib/irods/src_dir/dir1/img2.jpg rods 0 demoResc 2157087 2019-05-21.06:11 & img3.jpg generic /var/lib/irods/src_dir/dir1/img3.jpg rods 0 demoResc 2157087 2019-05-21.06:11 & img4.jpg generic /var/lib/irods/src_dir/dir1/img4.jpg
Check our results
Customizing the Ingest Behavior

The ingest tool is a callback based system in which the system invokes methods within the custom event handler module provided by the administrator.
These events may then take action such as setting ACLs, providing additional context such as selection of storage resources, or extracting and applying metadata.
Customizing the Ingest Behavior

Method |
Effect |
Default |
|---|---|---|
pre_data_obj_create |
user-defined python |
none |
post_data_obj_create |
user-defined python |
none |
pre_data_obj_modify |
user-defined python |
none |
post_data_obj_modify |
user-defined python | none |
| pre_coll_create | user-defined python | none |
post_coll_create |
user-defined python | none |
| as_user | takes action as this iRODS user | authenticated user |
| target_path | set mount path on the irods server which can be different from client mount path | client mount path |
to_resource |
defines target resource request of operation | as provided by client environment |
operation |
defines the mode of operation | Operation.REGISTER_SYNC |
Available --event_handler methods
Customizing the Ingest Behavior

The operation mode is returned during the 'operation' method which informs the ingest tool as to which behavior is desired for a given ingest job.
To override the default behavior, one of these operations must be selected and returned. These operations are hard coded into the tool, and cover the typical use cases of data ingest.
Customizing the Ingest Behavior

| Operation | New Files | Updated Files |
|---|---|---|
Operation.REGISTER_SYNC (default) |
registers in catalog | updates size in catalog |
| Operation.REGISTER_AS_REPLICA_SYNC | registers first or additional replica |
updates size in catalog |
| Operation.PUT | copies file to target vault, and registers in catalog |
no action |
| Operation.PUT_SYNC | copies file to target vault, and registers in catalog |
copies entire file again, and updates catalog |
| Operation.PUT_APPEND | copies file to target vault, and registers in catalog |
copies only appended part of file, and updates catalog |
Available Operations
Example Ingest Modules

./rodssync/lib/python3.5/site-packages/irods_capability_automated_ingest/examples/put_with_resc_name.py
from irods_capability_automated_ingest.core import Core from irods_capability_automated_ingest.utils import Operation class event_handler(Core): <-- expected interface @staticmethod def to_resource(session, target, path, **options): <-- method return "regiResc2a" <-- expected side effect @staticmethod def operation(session, target, path, **options): <-- method return Operation.PUT <-- operation
For this example we would want to change 'regiResc2a' to our target resource
Example Ingest Modules

./rodssync/lib/python3.5/site-packages/irods_capability_automated_ingest/examples/sync_root_with_resc_name.py
from irods_capability_automated_ingest.core import Core from irods_capability_automated_ingest.utils import Operation class event_handler(Core): @staticmethod def to_resource(session, target, path, **options): return "regiResc2Root" @staticmethod def operation(session, target, path, **options): return Operation.PUT_SYNC
Note that this uses a different operation which will not just ingest new data but synchronize previously ingested data
Custom Ingest with Metadata Extraction

Install the exifread python library
pip install exifread
./rodssync/lib/python3.5/site-packages/irods_capability_automated_ingest/examples/put_with_resc_name_image_metadata.py
Create a new ingest collection
imkdir put_coll
With the editor of your choice, create and edit:
Custom Ingest with Metadata Extraction

Implement the usual event handler and operation
We will implement two additional event handlers:
-
post_data_obj_create
-
post_data_obj_modify
Using the python iRODS client we will apply metadata extracted by exifread
Should the metadata key already exist we overwrite it with the new value
Custom Ingest with Metadata Extraction

import exifread from irods_capability_automated_ingest.core import Core from irods_capability_automated_ingest.utils import Operation from irods.meta import iRODSMeta def add_exif_metadata(session, target, path): with open(path, 'rb') as f: obj = session.data_objects.get(target) tags = exifread.process_file(f, details=False) for (k, v) in tags.items(): if k not in ('JPEGThumbnail','TIFFThumbnail','Filename','EXIF MakerNote'): if k in obj.metadata.keys(): obj.metadata[k] = iRODSMeta(k, v) else: obj.metadata.add(str(k), str(v), '') class event_handler(Core): @staticmethod def to_resource(session, meta, **options): return "demoResc" @staticmethod def operation(session, meta, **options): return Operation.PUT @staticmethod def post_data_obj_create(hdlr_mod, logger, session, meta, **options): add_exif_metadata(session, meta['target'], meta['path']) @staticmethod def post_data_obj_modify(hdlr_mod, logger, session, meta, **options): add_exif_metadata(session, meta['target'], meta['path'])
Custom Ingest with Metadata Extraction

Should we want this module to register in place, we can simply change the operation returned to REGISTER_SYNC or REGISTER_AS_REPLICA_SYNC
cp -r /tmp/test_dir ./src_dir/dir2
Stage new test data
python -m irods_capability_automated_ingest.irods_sync start ./src_dir /tempZone/home/rods/put_coll --event_handler irods_capability_automated_ingest.examples.put_with_resc_name_image_metadata
Launch the ingest job (all one line)
Custom Ingest with Metadata Extraction

$ ils -l put_coll/dir2 /tempZone/home/rods/put_coll/dir2: rods 0 demoResc 2157087 2019-05-22.11:16 & img0.jpg rods 0 demoResc 2157087 2019-05-22.11:16 & img1.jpg rods 0 demoResc 2157087 2019-05-22.11:16 & img2.jpg rods 0 demoResc 2157087 2019-05-22.11:16 & img3.jpg rods 0 demoResc 2157087 2019-05-22.11:16 & img4.jpg
Check our results
Custom Ingest with Metadata Extraction

$ imeta ls -d put_coll/dir2/img0.jpg AVUs defined for dataObj sync_coll/dir4/img0.jpg: attribute: EXIF ApertureValue value: 7983/3509 units: ---- attribute: EXIF BrightnessValue value: 2632/897 units: ---- <snip> ---- attribute: Thumbnail YResolution value: 72 units:
Inspect our newly harvested metadata
Any python library can now be leveraged to extract and apply metadata. Mime type can be detected and mapped to the appropriate metadata extraction.
The Landing Zone

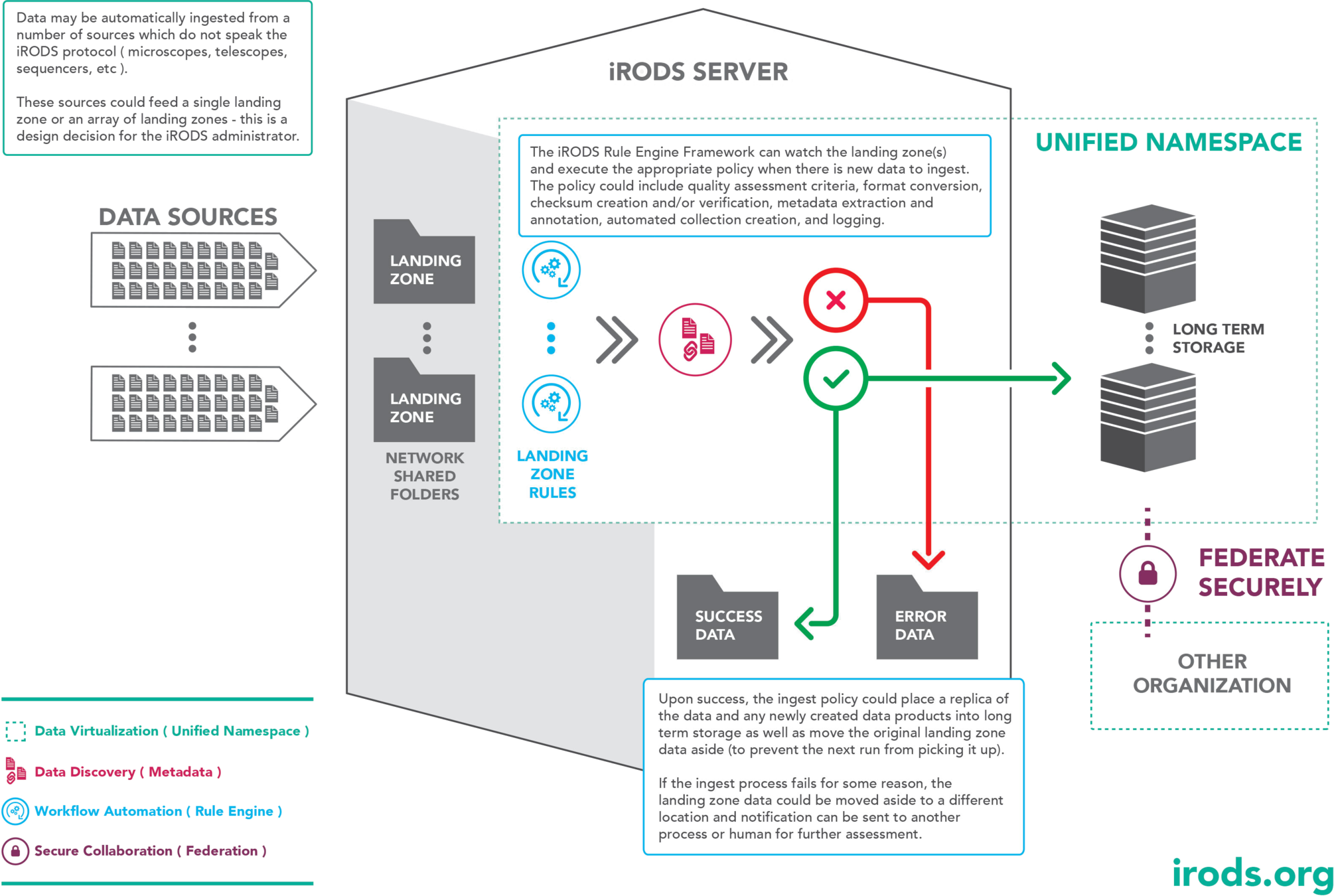
The Landing Zone

In this use case, data is written to disk by an instrument or another source we can run an ingest job on that directory. Once data is ingested it is moved out of the way in order to improve ingest performance. These ingested files can be removed later as a matter of local administrative policy.
The critical difference between a pure file system scan and a landing zone is that the LZ is not considered the single point of truth, it is a staging area for ingest (moving files out of the way).
In a file system scan, the file system is the canonical replica and the catalog and other replicas are kept in sync.
The Landing Zone

Create a new ingest collection
imkdir lz_coll
Create the landing zone and ingested directories
mkdir /tmp/landing_zone
mkdir /tmp/ingested
Stage data for ingest
cp -r /tmp/test_dir /tmp/landing_zone/dir0
Preparing a Landing Zone
With the editor of your choice, create and edit:
./rodssync/lib/python3.5/site-packages/irods_capability_automated_ingest/examples/lz_put_with_resc_name_image_metadata.py
The Landing Zone

import exifread
import os
from irods_capability_automated_ingest.core import Core
from irods_capability_automated_ingest.utils import Operation
def add_exif_metadata(session, target, path):
with open(path, 'rb') as f:
obj = session.data_objects.get(target)
try:
tags = exifread.process_file(f, details=False)
for (k, v) in tags.items():
if k not in ('JPEGThumbnail','TIFFThumbnail','Filename','EXIF MakerNote'):
if k in obj.metadata.keys():
obj.metadata[k] = iRODSMeta(k, v)
else:
obj.metadata.add(str(k), str(v), '')
except:
pass
class event_handler(Core):
@staticmethod
def to_resource(session, meta, **options):
return "demoResc"
@staticmethod
def operation(session, meta, **options):
return Operation.PUT
@staticmethod
def post_data_obj_create(hdlr_mod, logger, session, meta, **options):
path = meta['path']
add_exif_metadata(session, meta['target'], meta['path'])
new_path = path.replace('/tmp/landing_zone', '/tmp/ingested')
try:
dir_name = os.path.dirname(new_path)
os.makedirs(dir_name, exist_ok=True)
os.rename(path, new_path)
except:
logger.info('FAILED to move ['+path+'] to ['+new_path+']')
The Landing Zone

python -m irods_capability_automated_ingest.irods_sync start /tmp/landing_zone /tempZone/home/rods/lz_coll --event_handler irods_capability_automated_ingest.examples.lz_put_with_resc_name_image_metadata
Launch the ingest job
$ ls -l /tmp/ingested/dir0/ total 10540 -rw-rw-r-- 1 irods irods 2157087 May 23 05:24 img0.jpg -rw-rw-r-- 1 irods irods 2157087 May 23 05:24 img1.jpg -rw-rw-r-- 1 irods irods 2157087 May 23 05:24 img2.jpg -rw-rw-r-- 1 irods irods 2157087 May 23 05:24 img3.jpg -rw-rw-r-- 1 irods irods 2157087 May 23 05:24 img4.jpg
Check the ingested directory for the files
The Landing Zone

$ ils -l lz_coll/dir0 /tempZone/home/rods/lz_coll/dir0: rods 0 demoResc 2157087 2019-05-22.11:16 & img0.jpg rods 0 demoResc 2157087 2019-05-22.11:16 & img1.jpg rods 0 demoResc 2157087 2019-05-22.11:16 & img2.jpg rods 0 demoResc 2157087 2019-05-22.11:16 & img3.jpg rods 0 demoResc 2157087 2019-05-22.11:16 & img4.jpg
Check the ingested collection lz_coll
The Landing Zone

Check the ingested metadata
$ imeta ls -d lz_coll/dir0/img0.jpg AVUs defined for dataObj lz_coll/dir0/img0.jpg: attribute: EXIF ApertureValue value: 7983/3509 units: ---- attribute: EXIF BrightnessValue value: 2632/897 units: ---- <snip> ---- attribute: Thumbnail YResolution value: 72 units:
Streaming Ingest

For this use case some instruments periodically append output to a file or set of files.
We will configure an ingest job for a file to which a process will periodically append some data.
#!/bin/bash file=$1 touch $file for i in {1..1000}; do echo "do more science!!!!" >> $file sleep 2 done
Edit append_data.sh (make sure it's executable)
Streaming Ingest

Modify:
./rodssync/lib/python3.5/site-packages/irods_capability_automated_ingest/examples/append_with_resc_name.py
from irods_capability_automated_ingest.core import Core from irods_capability_automated_ingest.utils import Operation class event_handler(Core): @staticmethod def to_resource(session, target, path, **options): return "regiResc2a" @staticmethod def operation(session, target, path, **options): return Operation.PUT_APPEND
"regiResc2a" should become "demoResc"
Streaming Ingest

Prepare the source directory
rm -r ./src_dir/*
In another terminal start the data creation process
./append_data.sh ./src_dir/science.txt
Create a new ingest collection
imkdir stream_coll
Streaming Ingest

Check the results --- science.txt is growing in size
$ ils -l stream_coll/science.txt rods 0 demoResc 374 2019-05-23.10:13 & science.txt $ ils -l stream_coll/science.txt rods 0 demoResc 418 2019-05-23.10:13 & science.txt $ ils -l stream_coll/science.txt rods 0 demoResc 440 2019-05-23.10:13 & science.txt <SNIP>
Start the ingest process with a restart interval of 1s
python -m irods_capability_automated_ingest.irods_sync start \
./src_dir /tempZone/home/rods/stream_coll \
--event_handler \
irods_capability_automated_ingest.examples.append_with_resc_name -i 1
Streaming Ingest

Listing a periodic ingest job
$ python -m irods_capability_automated_ingest.irods_sync list
{"singlepass": [], "periodic": ["3defdd48-943c-11e9-aba6-123bf4b544e2"]}
Halting a running periodic ingest job
$ python -m irods_capability_automated_ingest.irods_sync \
stop 3defdd48-943c-11e9-aba6-123bf4b544e2
File System Scanning

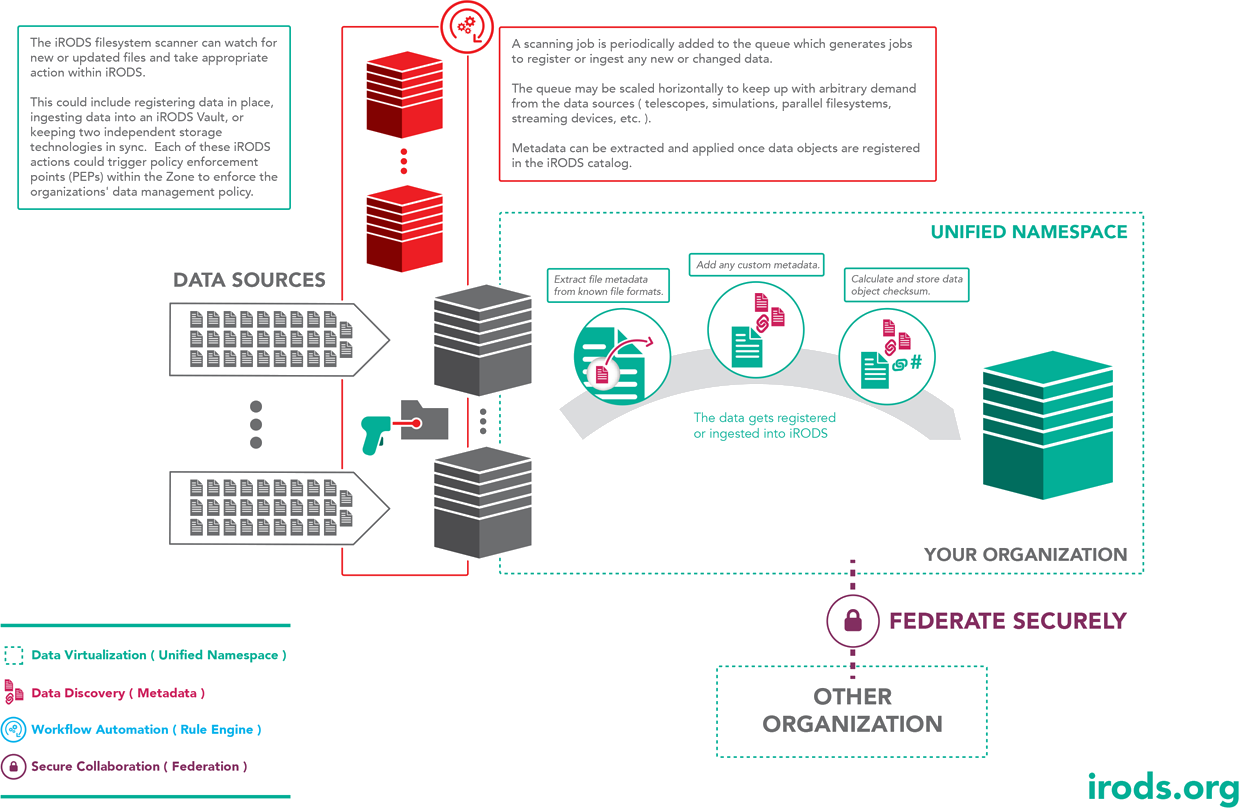
File System Scanning

This implementation will periodically scan a source directory, register the data in place, or update system metadata for changed files.
In this use case, the file system is considered the single point of truth for ingest. Changes are detected during the scan, and the system metadata is updated within the catalog.
File System Scanning

Clean up the source directory and stage data
rm -r ./src_dir/*
cp -r /tmp/test_dir ./src_dir/dir0
Create a new target collection
imkdir scan_coll
Launch the scanner with a period of 1 second
python -m irods_capability_automated_ingest.irods_sync start ./src_dir /tempZone/home/rods/scan_coll --event_handler irods_capability_automated_ingest.examples.register -i 1
File System Scanning

Investigate the results
$ ils -l scan_coll/dir0 /tempZone/home/rods/scan_coll/dir0: rods 0 demoResc 2157087 2019-05-24.07:28 & img0.jpg rods 0 demoResc 2157087 2019-05-24.07:28 & img1.jpg rods 0 demoResc 2157087 2019-05-24.07:28 & img2.jpg rods 0 demoResc 2157087 2019-05-24.07:28 & img3.jpg rods 0 demoResc 2157087 2019-05-24.07:28 & img4.jpg
Stage new data and investigate data registration
$ cp -r /tmp/test_dir ./src_dir/dir1
$ ils -l scan_coll/
/tempZone/home/rods/scan_coll:
C- /tempZone/home/rods/scan_coll/dir0
C- /tempZone/home/rods/scan_coll/dir1
Wait for it...
File System Scanning

Investigate the results
$ ils -l scan_coll/dir1 /tempZone/home/rods/scan_coll/dir1: rods 0 demoResc 2157087 2019-05-24.07:28 & img0.jpg rods 0 demoResc 2157087 2019-05-24.07:28 & img1.jpg rods 0 demoResc 2157087 2019-05-24.07:28 & img2.jpg rods 0 demoResc 2157087 2019-05-24.07:28 & img3.jpg rods 0 demoResc 2157087 2019-05-24.07:28 & img4.jpg
If it is not running, in another terminal
$ ./append_data.sh ./src_dir/science.txt
Launch the append script to test streaming writes
File System Scanning

Investigate the results
$ ils -l scan_coll /tempZone/home/rods/scan_coll: rods 0 demoResc 154 2019-05-24.07:45 & science.txt C- /tempZone/home/rods/scan_coll/dir0 C- /tempZone/home/rods/scan_coll/dir1 $ ils -l scan_coll/science.txt rods 0 demoResc 264 2019-05-24.07:45 & science.txt $ ils -l scan_coll/science.txt rods 0 demoResc 308 2019-05-24.07:45 & science.txt $ ils -l scan_coll/science.txt rods 0 demoResc 308 2019-05-24.07:45 & science.txt
File System Scanning

Managing a periodic ingest job
$ python -m irods_capability_automated_ingest.irods_sync list
{"periodic": ["c07e84bc-943c-11e9-aba6-123bf4b544e2"], "singlepass": []}
Halting a periodic ingest job
$ python -m irods_capability_automated_ingest.irods_sync \ stop b0f2d6fe-5f47-11e8-b142-080027e8658d
Questions?

CINES 2020 - Automated Ingest
By jason coposky
CINES 2020 - Automated Ingest
CINES 2020 Training Module
- 1,416



