Aquatic microbial ecology at DPES
DPES, University of Toronto-Scarborough (Feb 8th, 2022)
Jesse McNichol (he/him) : PhD, Biological Oceanography
Postdoctoral Scholar, University of Southern California

How do aquatic microbial communities* influence the Earth System**, and vice-versa***?

*whole community methods

**primary productivity, elemental cycling

***effect of anthropogenic pressure on aquatic ecosystems
Microbe art: @claudia_traboni
Core research question
Microbiology
Environmental science
Bioinformatics
Evolution, population genetics
Ecosystem modelling, global change biology
Microbial ecology, biogeochemistry
3 "pillars"
Microbiology
Bioinformatics
How do aquatic microbial communities influence the Earth System, and vice-versa?

Pillars support core research question
Environmental science
In silico
Laboratory
Field work
Holistic ecosystem knowledge
Reductionist (multiple entry points)
Interdisciplinary opportunities
Entry points, interconnections
Outline

1. Current work





2. HK/Taiwan work




3. PhD work


1. Current work

Outline
Ecology & environmental science
Bioinformatics


Research questions
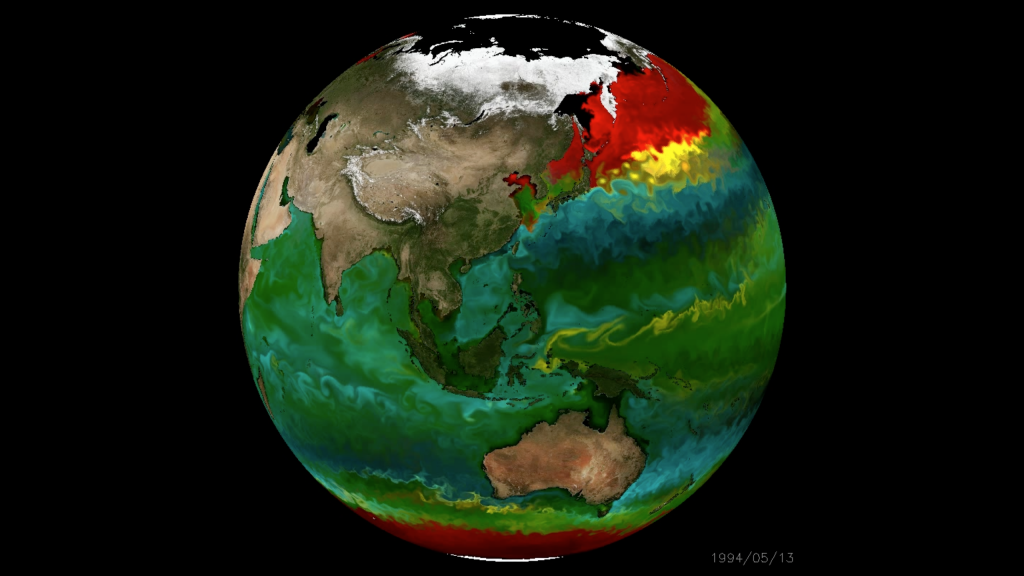
An in silico global phytoplankton model (DARWIN project, MIT)
-
How robust are model predictions?
- Need "ground truth" biogeographic distribution data
- What is the function of microbial "black box"?
- Can models and data help predict consequences of climate change?
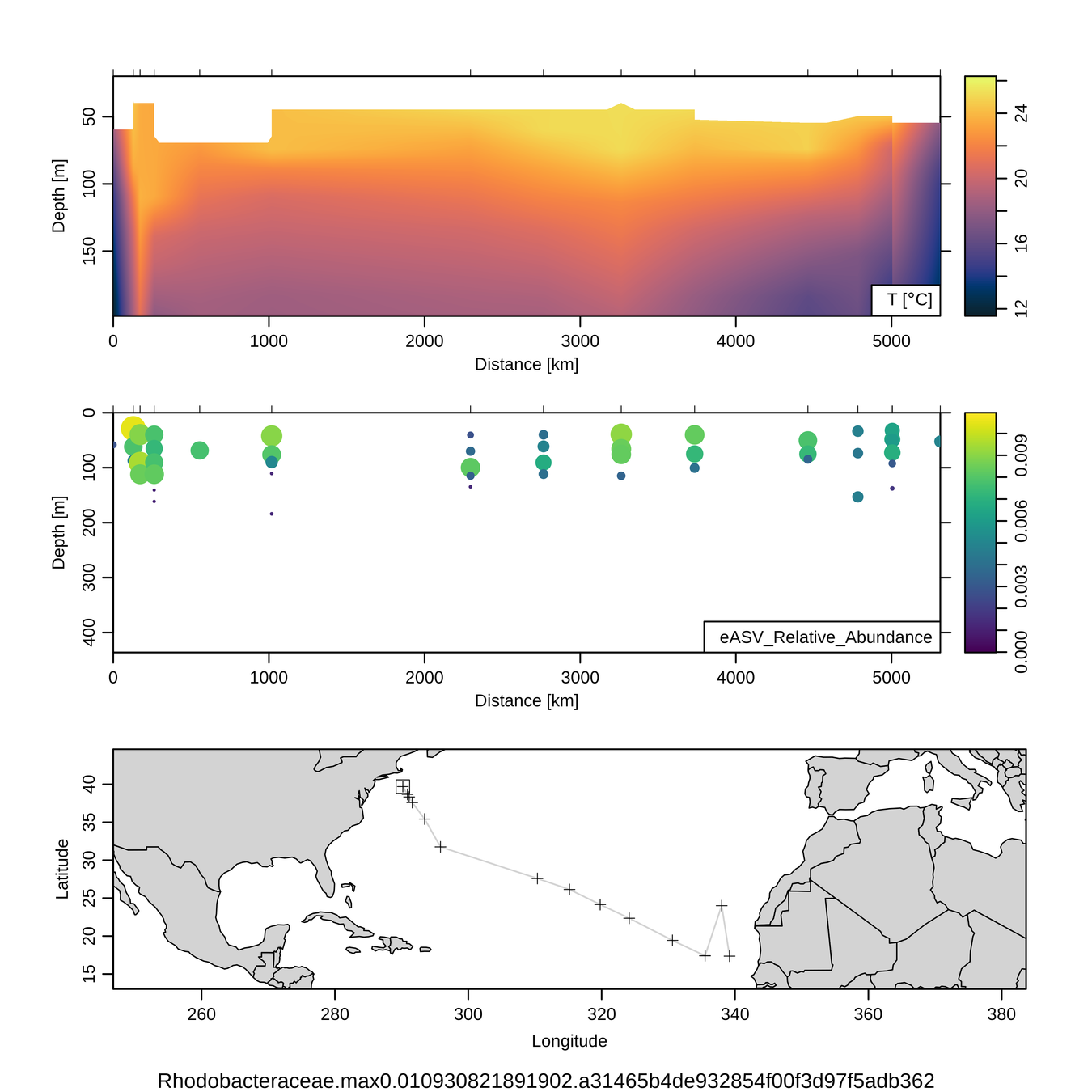
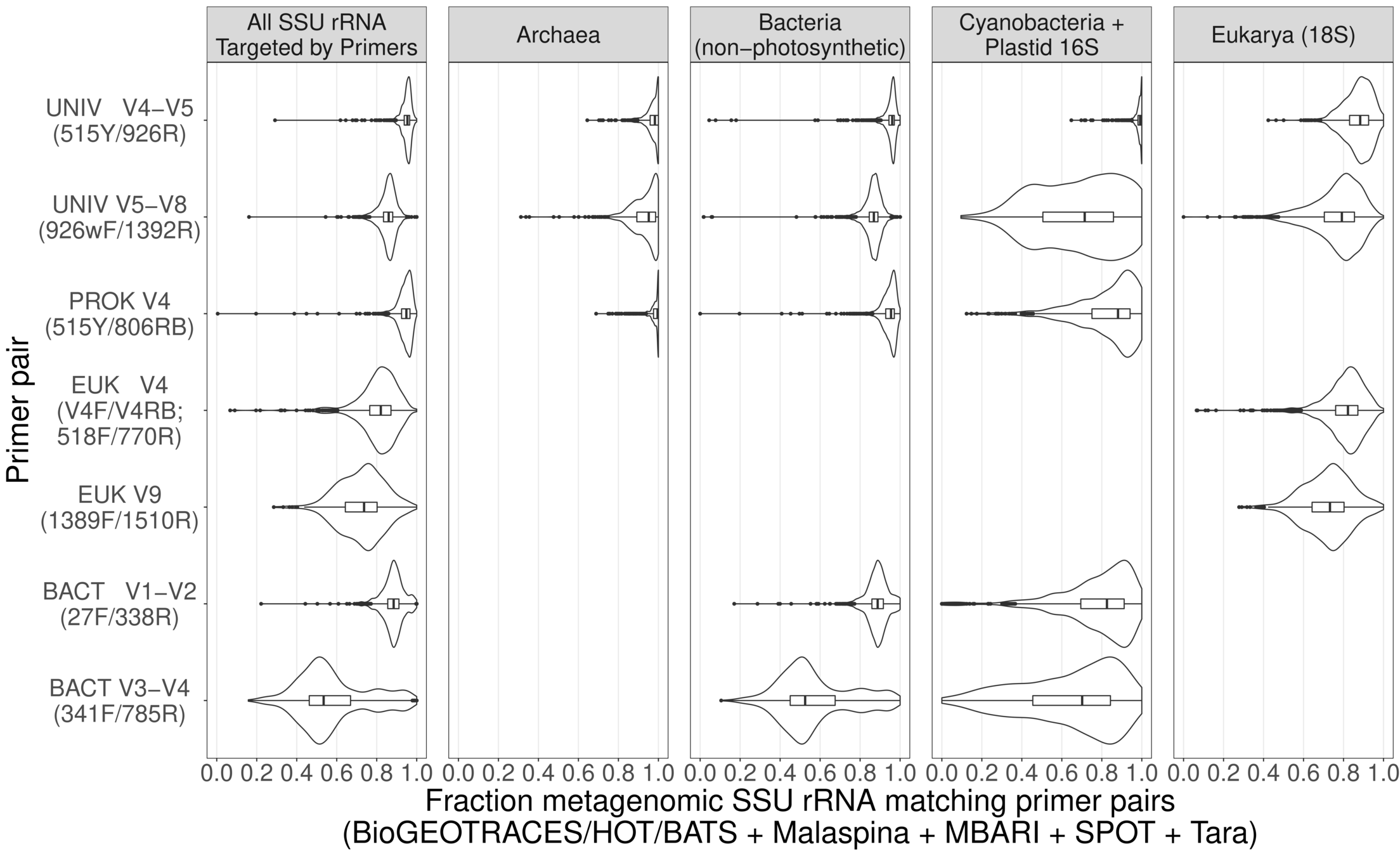
- Goal: measure abundance of microbial life across space / time in global oceans using PCR "barcodes"
- Comprehensive (515Y / 926R primers target all cellular life)
- Sensitive with deep sequencing
- Specific with "denoising" algorithms




Microbe art: @claudia_traboni
Assembling "ground truth" data


Accomplishments & novelty
1. Led in silico method optimization
Universal primers for barcoding work almost* perfectly across global oceans
2. Developed collaborations and generated data




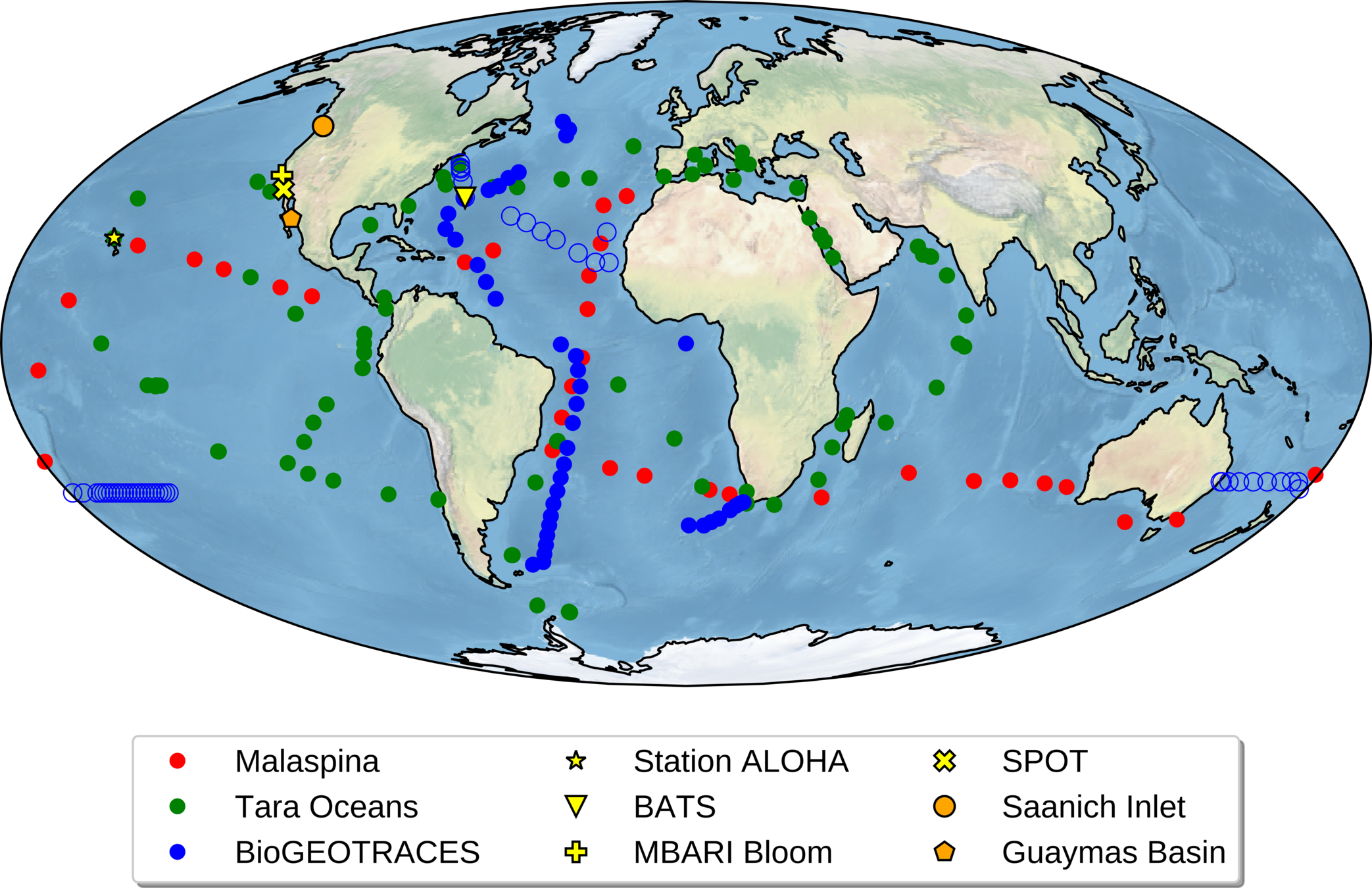
Global metagenomes
Over 800 globally-distributed barcode samples allow model-data intercomparison


Craig Carlson, UCSB
Current & future collaborations





Ecological niche differentiation
Taxon abundance across depth
Current & future collaborations





Model-data intercomparison


Taxon abundance across Pacific Ocean transect
Yubin Raut, USC
Current & future collaborations




Mathematical framework for comparing ASVs to other data types

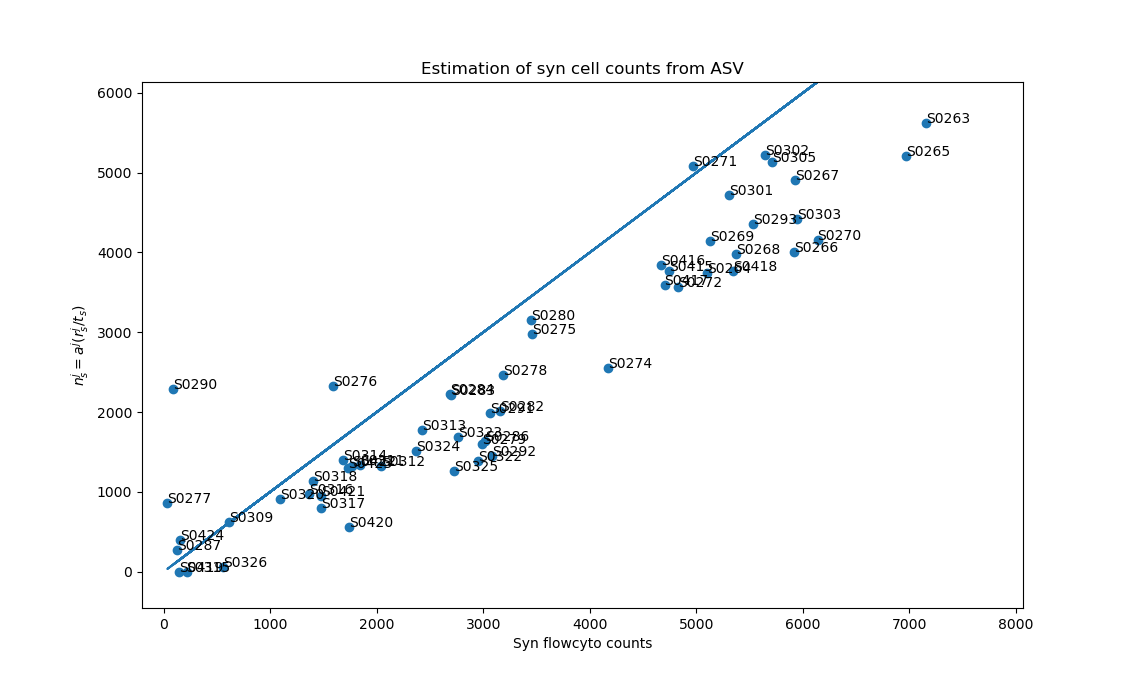
x axis = Synechococcus flow cytometry counts (cells/mL)
y axis = Calculated Synechococcus cell density (cells/mL)
Enrico Ser-Giacomi, MIT
Current & future collaborations





Building models of marine microheterotroph function
Bacterial / archaeal abundance and diversity across Pacific Ocean transect

Emily Zakem, Carnegie/USC
Microbial biogeography & modelling at DPES
-
Short term: Set up bioinformatic infrastructure, develop "bite-size" biogeographic projects:
- Analysis and interpretation to support CBIOMES
-
Med. term - apply for grants:
- NSERC Discovery Grant
- CBIOMES /Simons Foundation Young Investigator awards
- CFI to set up microbiome sequencing facility
-
Long term:
- Develop local / pan-Canadian collaborations to apply these techniques to diverse ecosystems (+ "eDNA"?)



In silico
- Collaboration with modellers
Laboratory
- Method optimization / intercalibration
Field work
- New data collection
Pathways for mentees







1. Current work

Outline
Ecology & environmental science
Bioinformatics




2. HK/Taiwan work
Outline
Bioinformatics
Microbiology
Research questions
- What bio(geo)chemical processes do microbes catalyze?
- How does speciation/evolution affect biogeochemistry?
- What are the specific proteins/pathways responsible and their properties?
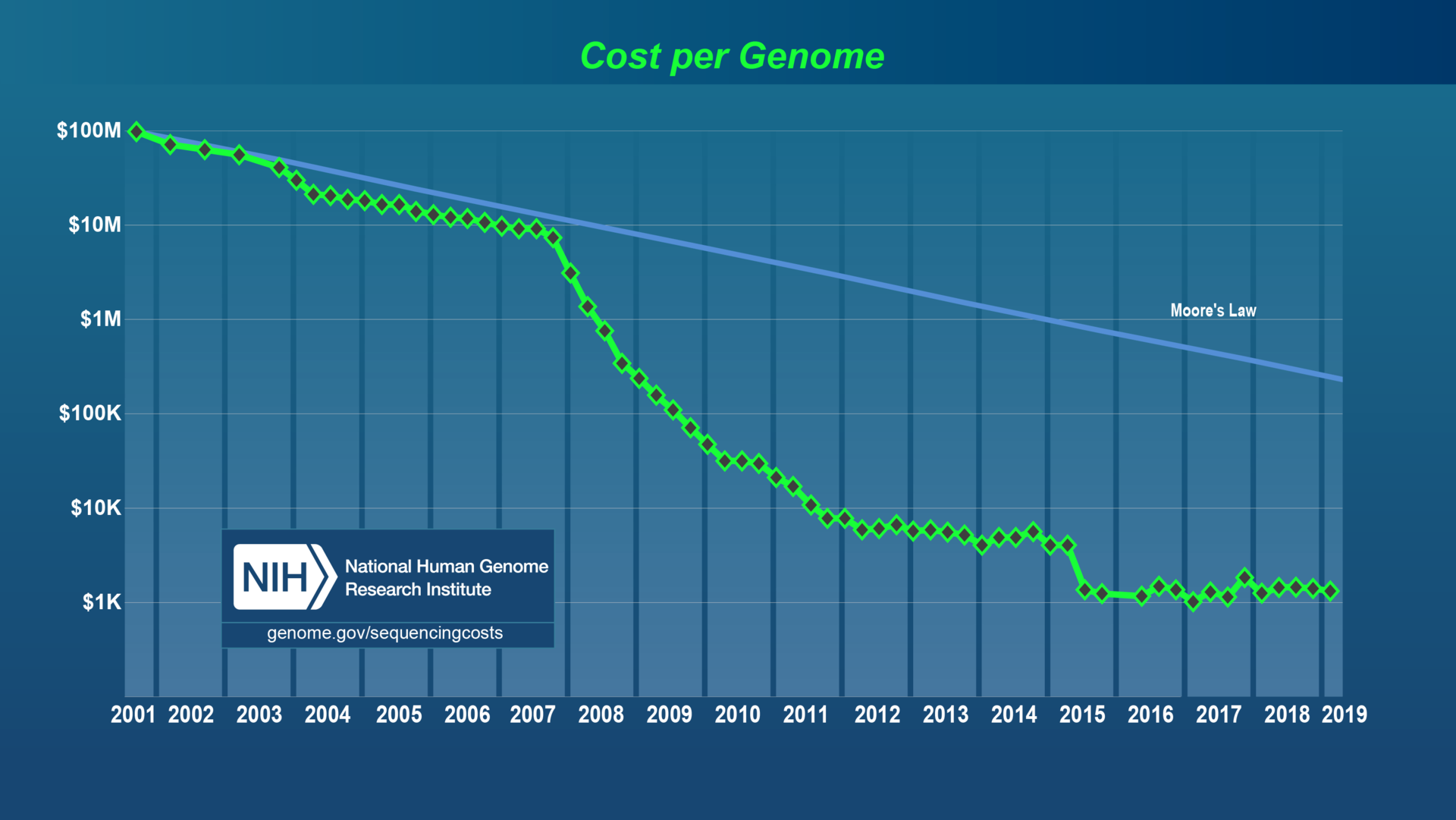
DNA sequencing is now low-cost...

...allowing us to sequence environmental "microbiomes"


Methods

1. Cultures or environmental genomes
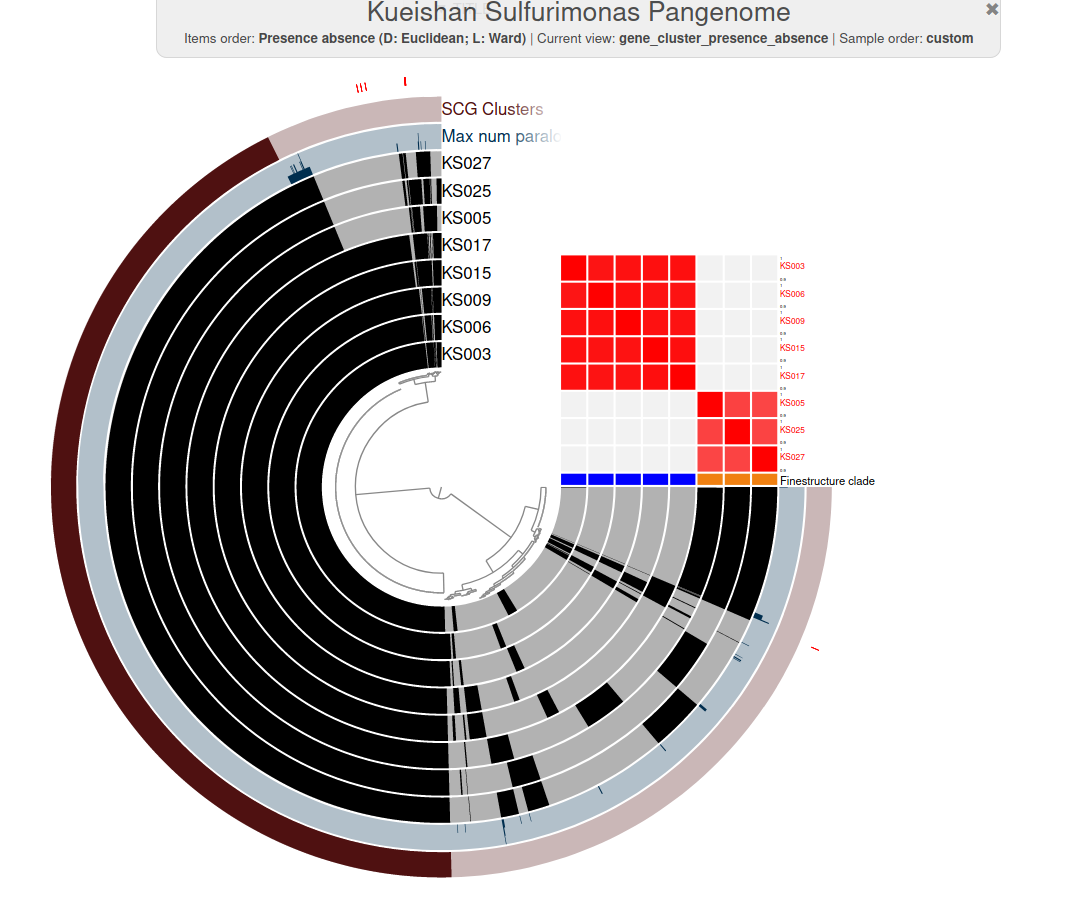
Plot made with anvi'o
2. (Pan)genomic analysis with anvi'o

3. Diversity, evolution, biogeochemistry






Accomplishments
- Developed novel, high-throughput, isolation methods
- Assembled collection of isolates with full genomes
Heterotrophs (marine sediments)

Chemoautotrophs (shallow-water vents)
New genus
Xiaoyuan Feng, CUHK
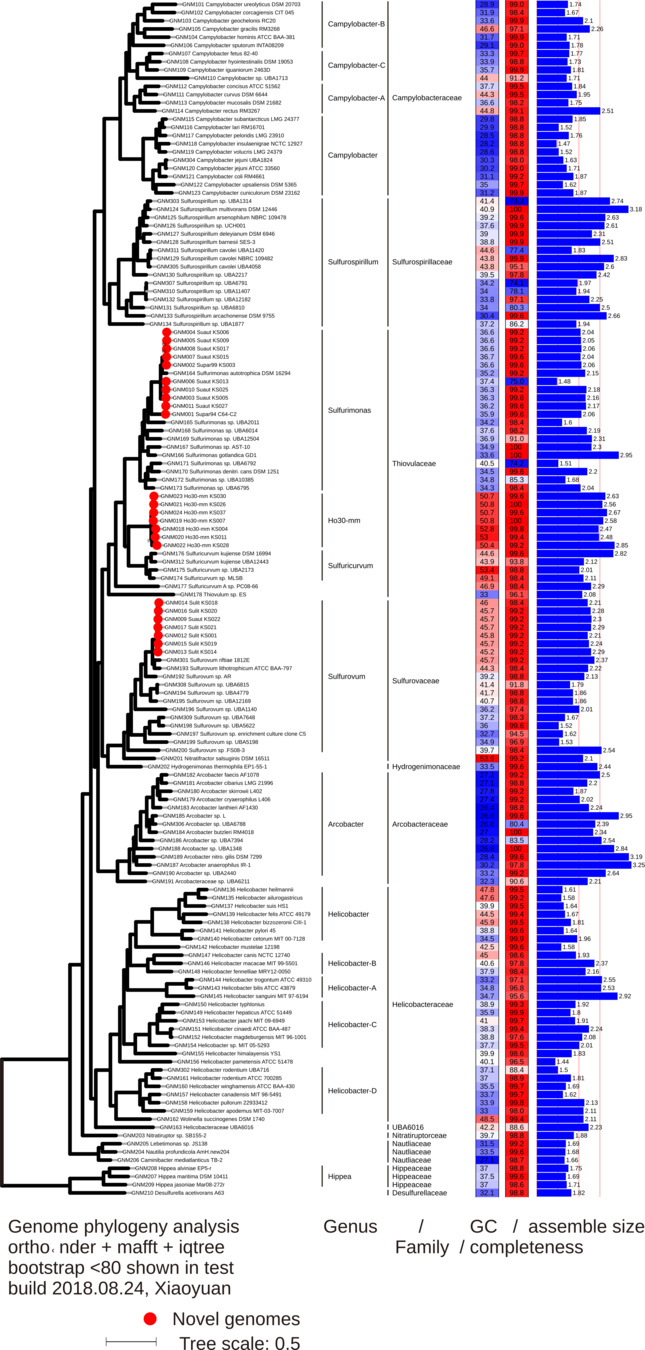
Genomics to biogeochemical insights
Plot made with anvi'o
Clade 1:
+ denit, + N2'ase
N source & sink
Clade 2:
- denit, + N2'ase
N source



Diagram = Li et al., 2018.

N2
In silico
- Study evolution and explore new genomes
Laboratory
- Describe novel taxa, study their physiology
Field work
- Explore new ecosystems
Pathways for mentees





Annie Wing-Yi Lo, CUHK
- Set up equipment for O2-sensitive microbes
- Retrieve cultures / samples / data
- Train students in cultivation, genomic analysis
- Medium term: Develop methods to identify essential genes (Tn-Seq)
- Long term: Conduct isolation campaigns in model systems, do biochemical assays


Cultivation and genomics at DPES




Microbial evolution at DPES
- Campylobacteria likely originated in Earth's "middle ages", but timing uncertain
-
Genomic analysis can place their evolution in Earth Systems context
-
A long-term, collaborative project (proposal ready)
-
A long-term, collaborative project (proposal ready)


?




2. HK/Taiwan work
Outline
Bioinformatics
Microbiology




3. PhD work
Outline
Ecology & environmental science
Microbiology
Bioinformatics
Research questions
- What is the effect of subsurface productivity on the deep sea?
- How do physicochemical factors affect ecology and productivity?

Deep-sea hydrothermal vents: Oases fueled by geochemical energy


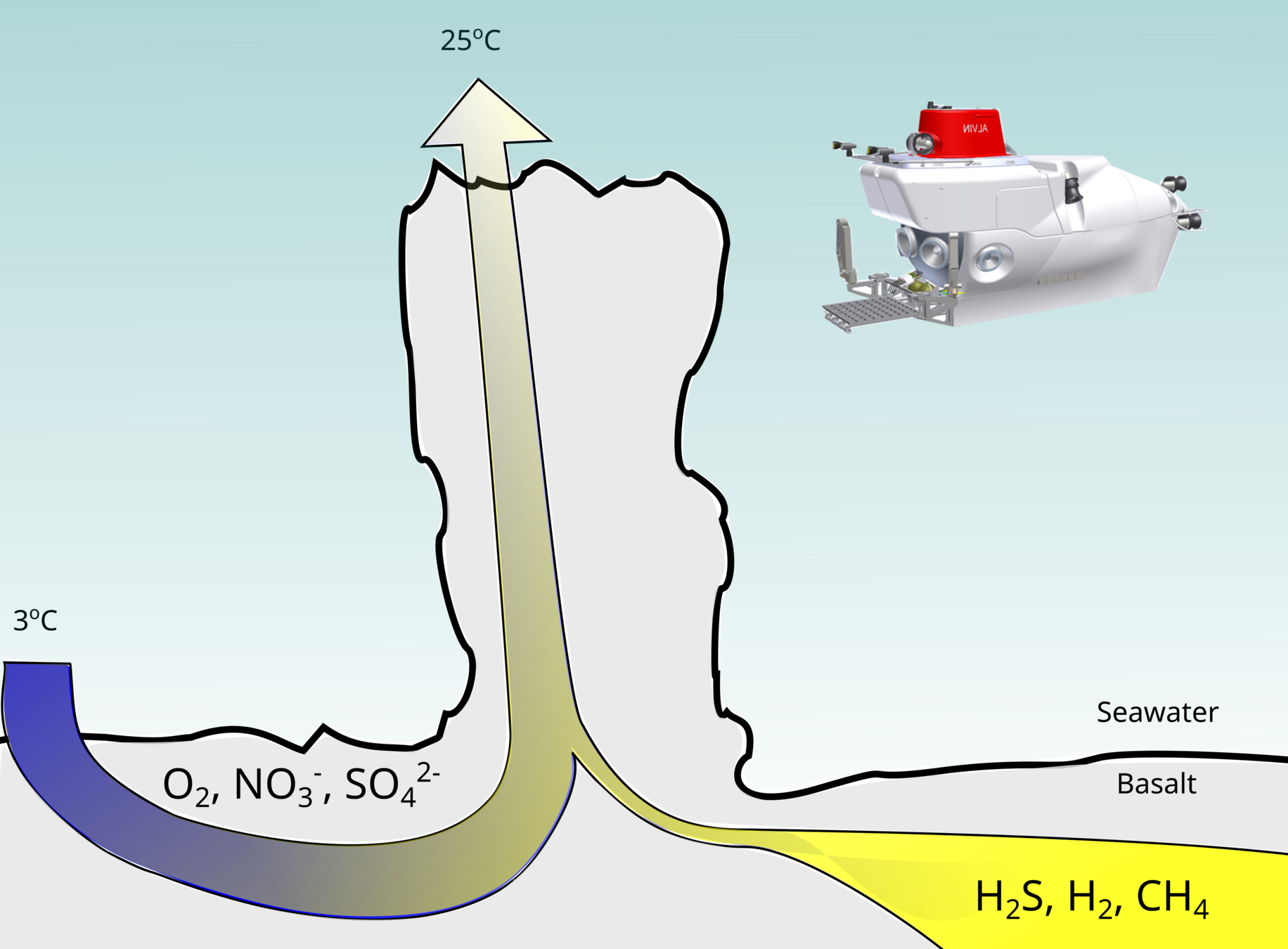
- Developed novel high-pressure incubations
- Quantified bulk and single-cell CO2 fixation (NanoSIMS)
Methods

FISH
SIP




Impact - biogeochemistry

Subsurface C production > 10 - 10,000x photic zone export
Subsurface C production rivals chemosynthetic symbioses



Impact - ecology


O2 caused major shift in community over ~ 24 h

Have single-cell genomes - good student project
Biochemical underpinnings of this response?
O2 reduced the efficiency of carbon fixation

Process studies = holistic research
Holistic, process studies require a "model system"
Ecology & environmental science

Microbiology

Bioinformatics


- Set up lab (FISH, isotope-labeling)
-
Medium term:
- Continue hydrothermal vent work
- Investigate new model systems
-
Long term:
- Study the effect of climate change on global aquatic ecosystems
Process studies at DPES



What I would bring to your department
Microbiology
Ecology & environmental science
Bioinformatics
- > 10 years experience in marine microbial ecology
- Can train mentees in lab, computational, field techniques
- Diverse interests: oceanography, modelling, analytical method development, natural history, evolution
- Passionate about integrating cutting-edge "microbiome" research techniques into my teaching (CURE)
- Track record of successful interdisciplinary collaborations, and excited to build connections at DPES / UofT
Mentoring philosophy & DEI
Instrumental
- Real data, skills
- Student-driven research
- Important questions, diverse collaborations
Psychosocial
Evidence-based approaches
- Building "soft skills"
- Foster emotional belonging
- Student-centered mentoring philosophy
- Modern pedagogy
- Scaffolding approaches
- Culturally-aware mentoring
Microbe art: @claudia_traboni
Core motivation
Earth Systems Science more important than ever
- Would bring unique perspective and passion to Global Environmental Change program




Acknowledgements
WHOI / UFZ: Stefan Sievert, Jeff Seewald, François Thomas, Niculina Musat, Craig Taylor
CUHK / Academia Sinica: Haiwei Luo, Annie Wing-Yi Lo, Benny Chan
USC/MIT/UCSB/Dalhousie/Carnegie: Jed Fuhrman, Yubin Raut, Enrico Ser-Giacomi, Paul Berube, Steven Biller, Mick Follows, Stephanie Dutkiewicz, Craig Carlson, Zoe Finkel, Emily Zakem








Sources
| Images used in presentation were adapted from: |
|---|
|
Line diagram (slide 9): J. A. Fuhrman, J. A. Cram, D. M. Needham, Marine microbial community dynamics and their ecological interpretation. Nature Reviews Microbiology 13, 133–146 (2015). Metagenome image (slide 19): V.A. Iverson et al., Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science, 335(6068): 587-590 (2012). Anvi'o used to make image on slide 20: https://merenlab.org/software/anvio/; https://peerj.com/articles/1319/ Shallow-water vent image (slides 20, 22): Y. Li et al., Coupled Carbon, Sulfur, and Nitrogen Cycles Mediated by Microorganisms in the Water Column of a Shallow-Water Hydrothermal Ecosystem. Frontiers in Microbiology. 9:2718 (2018). Speciation image (slides 20, 22): R. Stepanauskas et al., Gene exchange networks define species-like units in marine prokaryotes. bioRxiv, 2020.09.10.291518 (2020). Oxygen diagram (slide 25): L.R. Kump, The Rise of Atmospheric Oxygen. Nature, 451(7176): 277-8 (2008). Water column image on slide 30 adapted from: M. Hügler, S. M. Sievert, Beyond the Calvin Cycle: Autotrophic Carbon Fixation in the Ocean. Annu. Rev. Marine. Sci. 3, 261–289 (2011). |
Unless otherwise noted, other images are either my own (un)published work, ⓒWHOI, or public domain images from Wikimedia Commons
deck
By jcmcnch
deck
- 64



