Reproducible data analysis with Snakemake
Johannes Köster
2019
https://koesterlab.github.io
150k downloads since 2015

















497 citations (+150 in 2019)
Snakemake is popular
https://snakemake.readthedocs.io
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
shell:
"some-tool {input} > {output}"Concise DSL
https://snakemake.readthedocs.io
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
script:
"scripts/mytask.py"Python scripts
https://snakemake.readthedocs.io
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
script:
"scripts/mytask.R"R scripts
https://snakemake.readthedocs.io
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
script:
"scripts/mytask.jl"Julia scripts
https://snakemake.readthedocs.io
import matplotlib.pyplot as plt
import pandas as pd
d = pd.read_table(snakemake.input[0])
d.hist(bins=snakemake.config["hist-bins"])
plt.savefig(snakemake.output[0])No boilerplate
rule mytask:
input:
"data/{sample}.txt"
output:
"result/{sample}.txt"
script:
"scripts/mytask.py"https://snakemake.readthedocs.io

rule mytask:
input:
"path/to/{dataset}.txt"
output:
"result/{dataset}.txt"
script:
"scripts/myscript.R"
rule myfiltration:
input:
"result/{dataset}.txt"
output:
"result/{dataset}.filtered.txt"
shell:
"mycommand {input} > {output}"
rule aggregate:
input:
"results/dataset1.filtered.txt",
"results/dataset2.filtered.txt"
output:
"plots/myplot.pdf"
script:
"scripts/myplot.R"Implicit dependencies
https://snakemake.readthedocs.io



workstation
compute server
cluster






grid computing
cloud computing



Scalability
https://snakemake.readthedocs.io
rule mytask:
input:
"path/to/{dataset}.txt"
output:
"result/{dataset}.txt"
conda:
"envs/mycommand.yaml"
shell:
"mycommand {input} > {output}"channels:
- bioconda
- conda-forge
dependencies:
-mycommand =2.3.1Conda integration
https://snakemake.readthedocs.io
rule mytask:
input:
"path/to/{dataset}.txt"
output:
"result/{dataset}.txt"
singularity:
"docker://some/container"
shell:
"mycommand {input} > {output}"Singularity integration
https://snakemake.readthedocs.io
rule mytask:
input:
"path/to/{dataset}.txt"
output:
"result/{dataset}.txt"
conda:
"envs/mycommand.yaml"
singularity:
"docker://some/os"
shell:
"mycommand {input} > {output}"Singularity + Conda
https://snakemake.readthedocs.io
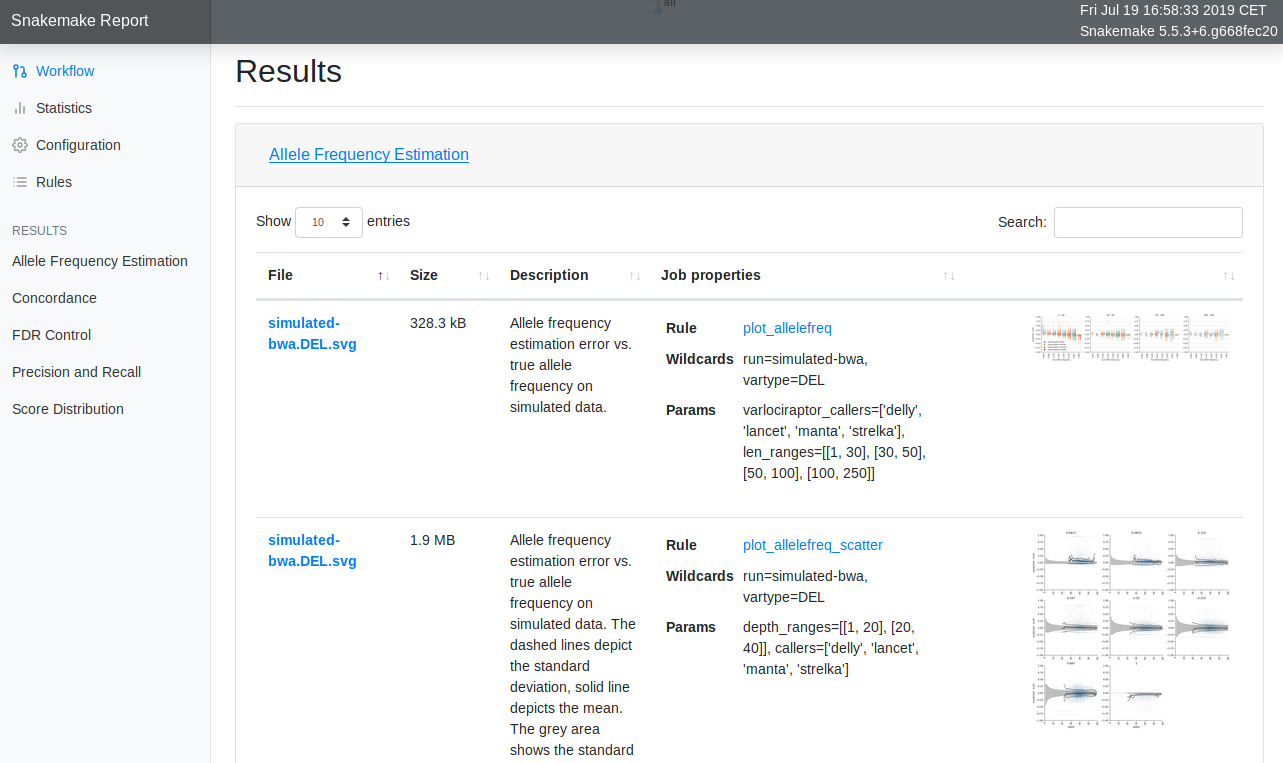
Snakemake
https://snakemake.readthedocs.io

Snakemake
https://snakemake.readthedocs.io
dataset
results
dataset
dataset
dataset
dataset
dataset
portability
scalability
automation/ documentation
https://snakemake.readthedocs.io
Snakemake DoDSC
By Johannes Köster
Snakemake DoDSC
DoDSC 2019
- 1,939



