Options for producing OpenMC h5 cross sections
Jonathan Shimwell
IAEA Consultancy Meeting on Model Code Output
&
Application Nuclear Data Form Structure
Obtaining h5 cross section files
- Download individual isotopes using the openmc-data-downloader Python package https://github.com/openmc-data-storage/openmc_data_downloader
- Download compressed (tar) libraries from OpenMC.org
-
Process h5 files using the data repository https://github.com/openmc-dev/data
- Process ACE files using convert*.py scripts
- Process ENDF files using generate*.py scripts
Obtaining h5 cross section files
| Library | Release | Processed by | Download from openmc.org | Download from openmc-data-downloader | Download ACE files and convert HDF5 | Download ENDF files and generate HDF5 | Convert local ACE files |
|---|---|---|---|---|---|---|---|
| CENDL | 3.1 3.2 |
generate_cendl.py | |||||
| ENDF/B | VII.0 | LANL | ✓ | convert_mcnp70.py | |||
| ENDF/B | VII.1 | LANL |
✓ | convert_mcnp71.py | |||
| ENDF/B | VII.1 | NNDC | ✓ | ✓ | convert_nndc71.py | generate_endf.py | |
| ENDF/B | VIII.0 | LANL | ✓ | convert_lib80x.py | |||
| ENDF/B | VIII.0 | NNDC | ✓ | ||||
| FENDL | 2.1, 3.0, 3.1a, 3.1d |
✓ (3.1d) | convert_fendl.py | ||||
| JENDL | 4.0 | generate_jendl.py | |||||
| JEFF | 3.2 | ✓ | convert_jeff32.py | ||||
| JEFF | 3.3 | ✓ | convert_jeff33.py | ||||
| TENDL | 2015, 2017, 2019 | ✓ (2019) | convert_tendl.py |
There are several ways of obtaining the cross section data in the h5 file format required for neutronics simulations.
OpenMC data repository
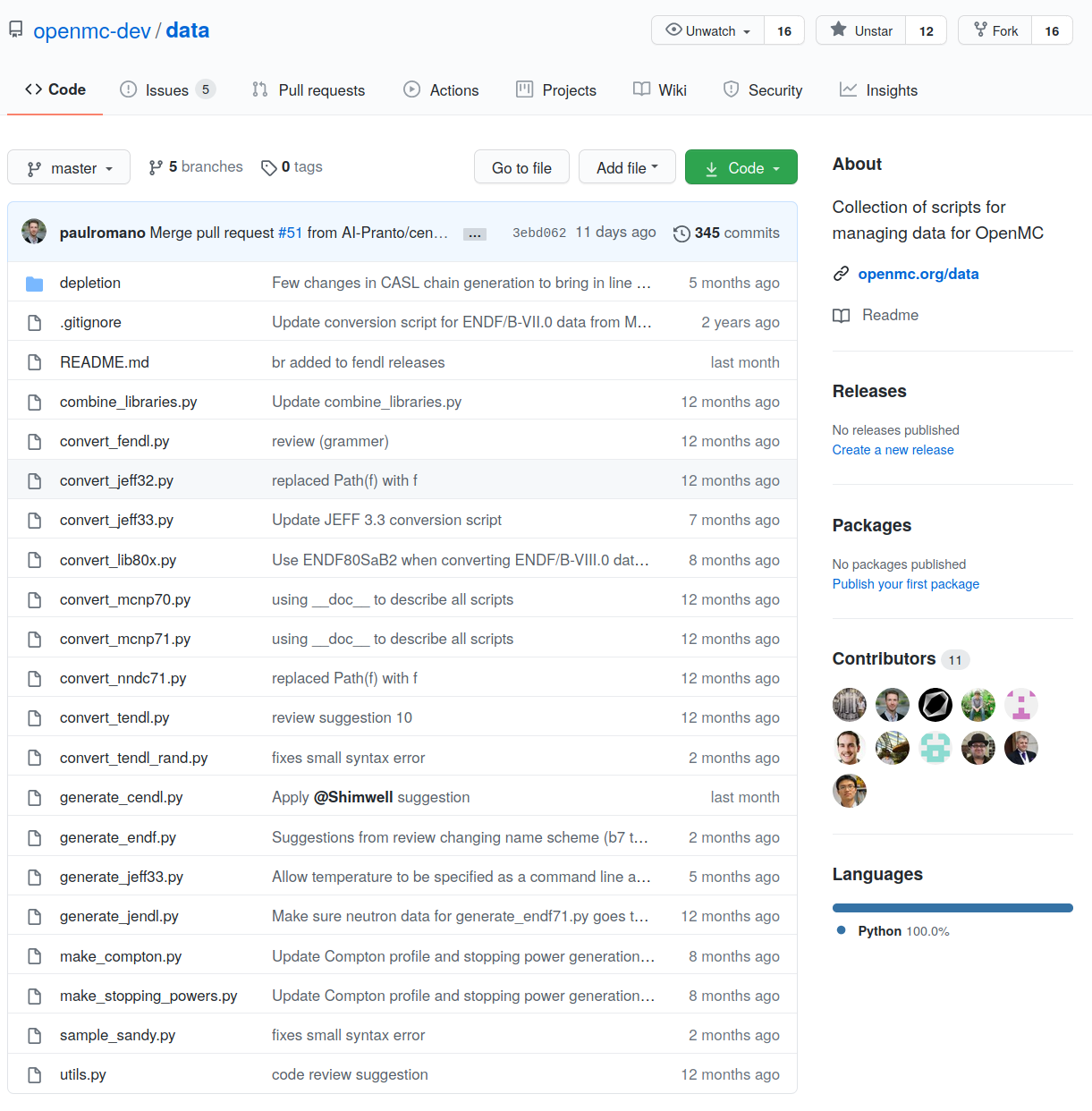
- Python scripts
- Permissive open source license (MIT)
- 11 contributors
- Storage and sharing opportunity to avoid reproducing the same scripts
- Useful for developers
- Generate scripts require NJOY install
- PyPi package with pip install on the horizon
git clone https://github.com/openmc-dev/data.git
cd data
python convert_fendl.py --release 3.1d
>> downloading, extracting and processing ...
ls
>> fendl-3.1d-ace/
>> fendl-3.1d-endf/
>> fendl-3.1d-download/
>> fendl-3.1d-hdf5/
ls fendl-3.1d-hdf5/
>> cross_sections.xml
>> neutron/
>> photon/
ls fendl-3.1d-hdf5/neutron
>> Ag107.h5
>> Ag109.h5
>> Al27.h5
>> Ar36.h5
>> ...
export OPENMC_CROSS_SECTIONS=fendl-3.1d-hdf5/cross_sections.xml
- Example usage of one of the Python scripts
- This particular script downloads 429MB of compressed ACE and ENDF files
- Extracts the files
- Processes the files into h5 file for use in OpenMC
Obtaining h5 cross sections - data repo
OpenMC.org
-
https://openmc.org includes a data section with downloads
- Official Libraries
- LANL-Based Data Libraries
- Other Libraries
- Depletion chains


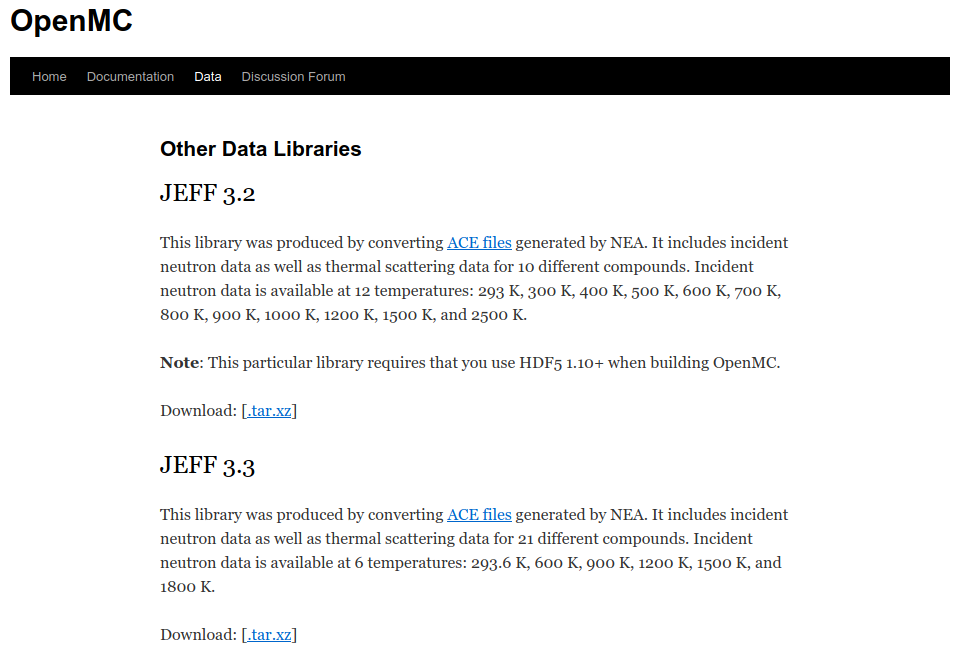
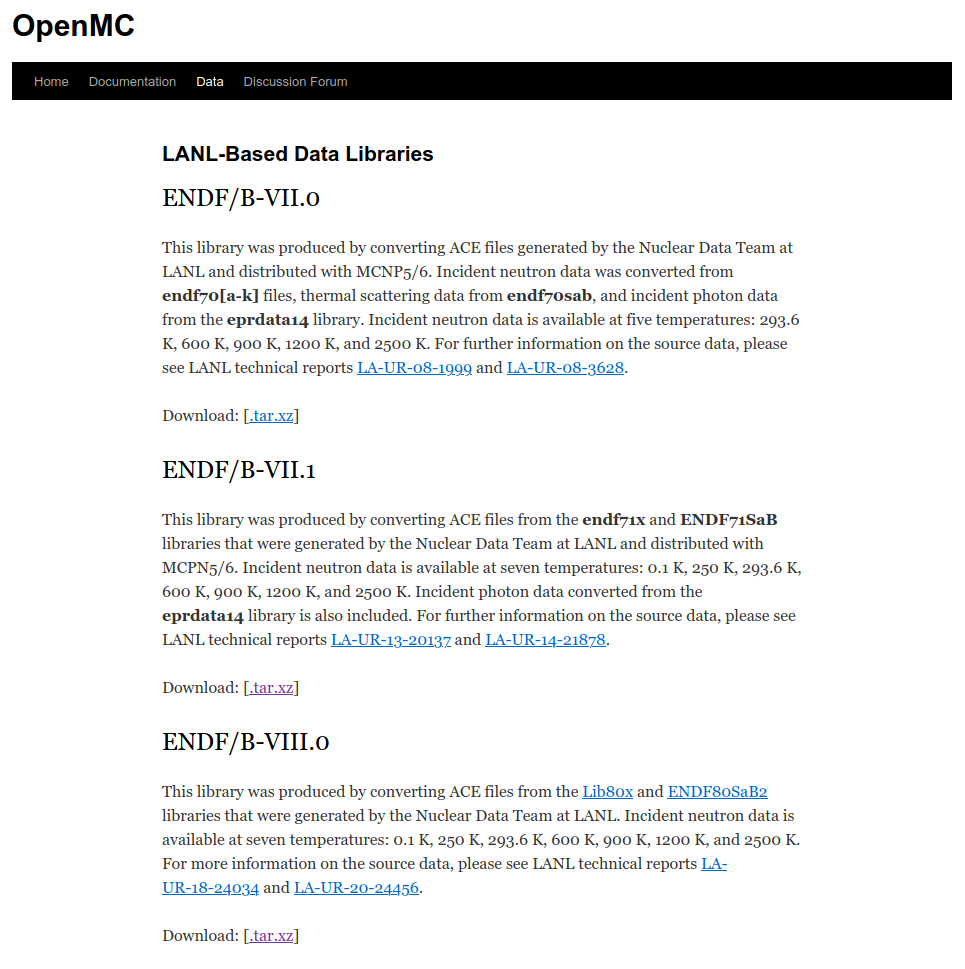
- OpenMC documentation pages includes information on the nuclear data interface https://docs.openmc.org
Obtaining h5 cross sections - OpenMC.org
- The h5 files can be used in simulations once the OPENMC_CROSS_SECTION environmental variable has been set
wget -O nndc-b7.1.tar.xz https://anl.box.com/shared/static/9igk353zpy8fn9ttvtrqgzvw1vtejoz6.xz
>> dowloading compressed file ...
mkdir nndc-b7.1-hdf5
tar -xf nndc-b7.1.tar.xz -C nndc-b7.1-hdf5
>> extracting compressed file ...
ll nndc-b7.1-hdf5
>>cross_sections.xml
>>neutron/
>>photon/
ll nndc-b7.1-hdf5/neutron
>>Ac225.h5
>>Ac226.h5
>>Ac227.h5
>>Ag107.h5
>>Ag109.h5
>>...
export OPENMC_CROSS_SECTIONS=nndc-b7.1-hdf5/cross_sections.xml
- Downloads can be scripted using wget
Obtaining h5 cross sections - OpenMC.org
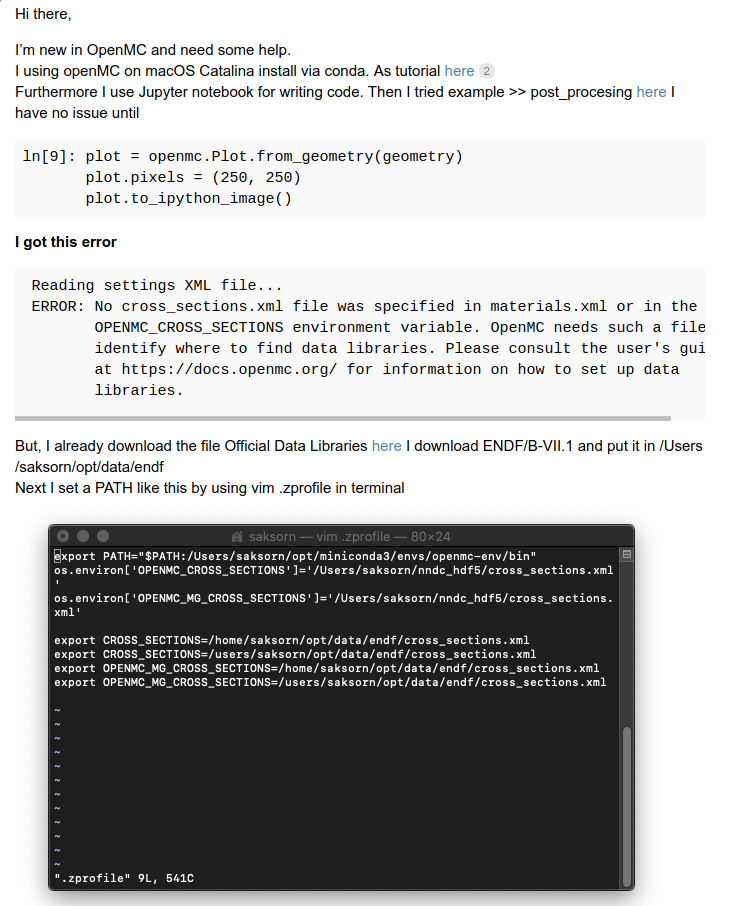
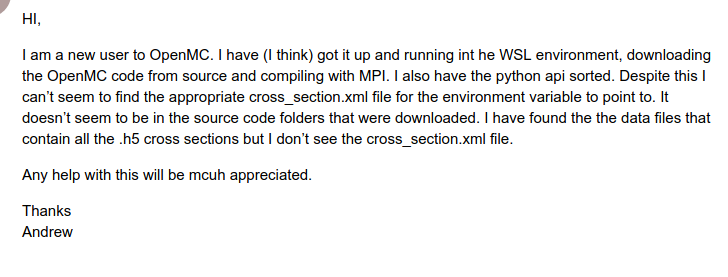

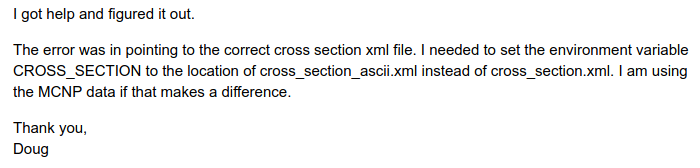
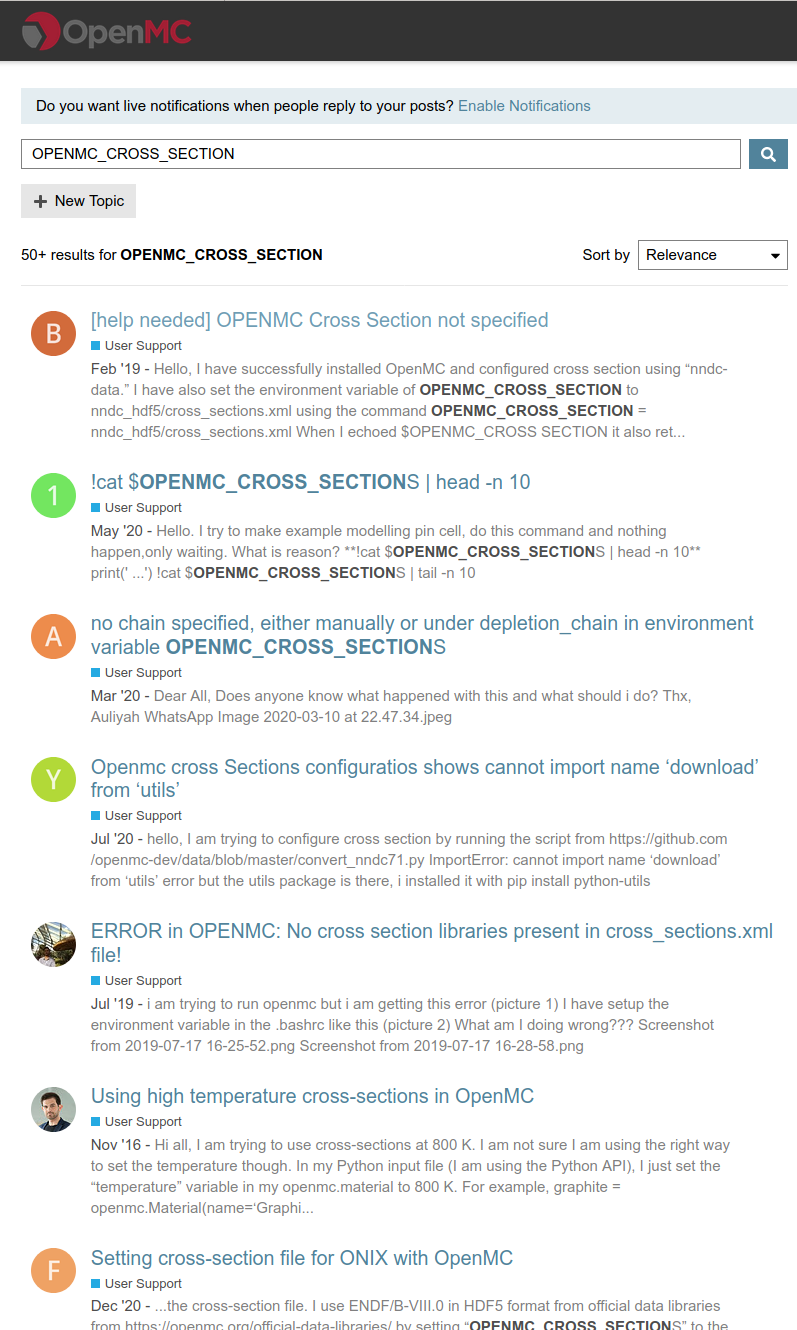

OpenMC Docker image production


- OpenMC automatically produces a new docker image with every code merge
- Github Actions is used to build the docker image and upload it to Dockerhub
https://hub.docker.com/r/openmc/openmc
https://github.com/openmc-dev/openmc/actions
Data storage repo - processing
Contains repository for the openmc-data-downloader Python package.
Contains separate repositories for each library release
Repositories can be templated
Open source, permissively licensed (MIT) https://github.com/openmc-data-storage
Data storage repo - enviroment
Use of the openmc/openmc-dev image this contains NJOY as well
Addition of the openmc-data repo to the base image
FROM openmc/openmc:develop
RUN git clone https://github.com/openmc-dev/data.git
COPY tests tests/
ENV OPENMC_CROSS_SECTIONS=/share/cross_sections.xmlCopying test suite
See environmental variable



4 Lines
+ + =
Data storage repo - processing
Github action (CI) then commits the new h5 files from the share folder to the repository
Github actions (CI) build and run the new docker image
name: process_test_h5_files
on:
push:
branches:
- main
env:
PROCESS_CMD: 'python data/convert_*****.py -d /share/h5_files -r *****'
jobs:
build:
runs-on: ubuntu-latest
steps:
- uses: actions/checkout@master
with:
persist-credentials: false # otherwise, the token used is the GITHUB_TOKEN, instead of your personal token
fetch-depth: 0 # otherwise, you will failed to push refs to dest repo
- name: Create custom readme
run: |
python readme_writer.py
cat README.md
- name: Build and test with Docker
run: |
docker build --tag nuc_data_maker .
rm -rf h5_files
docker run -v /home/runner/work/${{ github.event.repository.name }}/${{ github.event.repository.name }}:/share nuc_data_maker /bin/bash -c "${{ env.PROCESS_CMD }} && pytest -rP tests/test_xs_in_simulations.py"
- name: Commit files
run: |
git config --local user.email "action@github.com"
git config --local user.name "GitHub Action"
git add h5_files/*.h5
git add h5_files/cross_sections.xml
git add README.md
git commit -m "Added readme with repo name (bot commit)" -a
- name: Push changes
uses: ad-m/github-push-action@master
with:
github_token: ${{ secrets.GITHUB_TOKEN }}
branch: ${{ github.ref }}
The new docker image is then run externally with a shared volume mount



40 Lines
4 Lines
+ + =
Data storage repo - testing


Each h5 cross section file is then used in an OpenMC simulation
Testing the h5 file produced has converted successfully and does not cause errors during usage

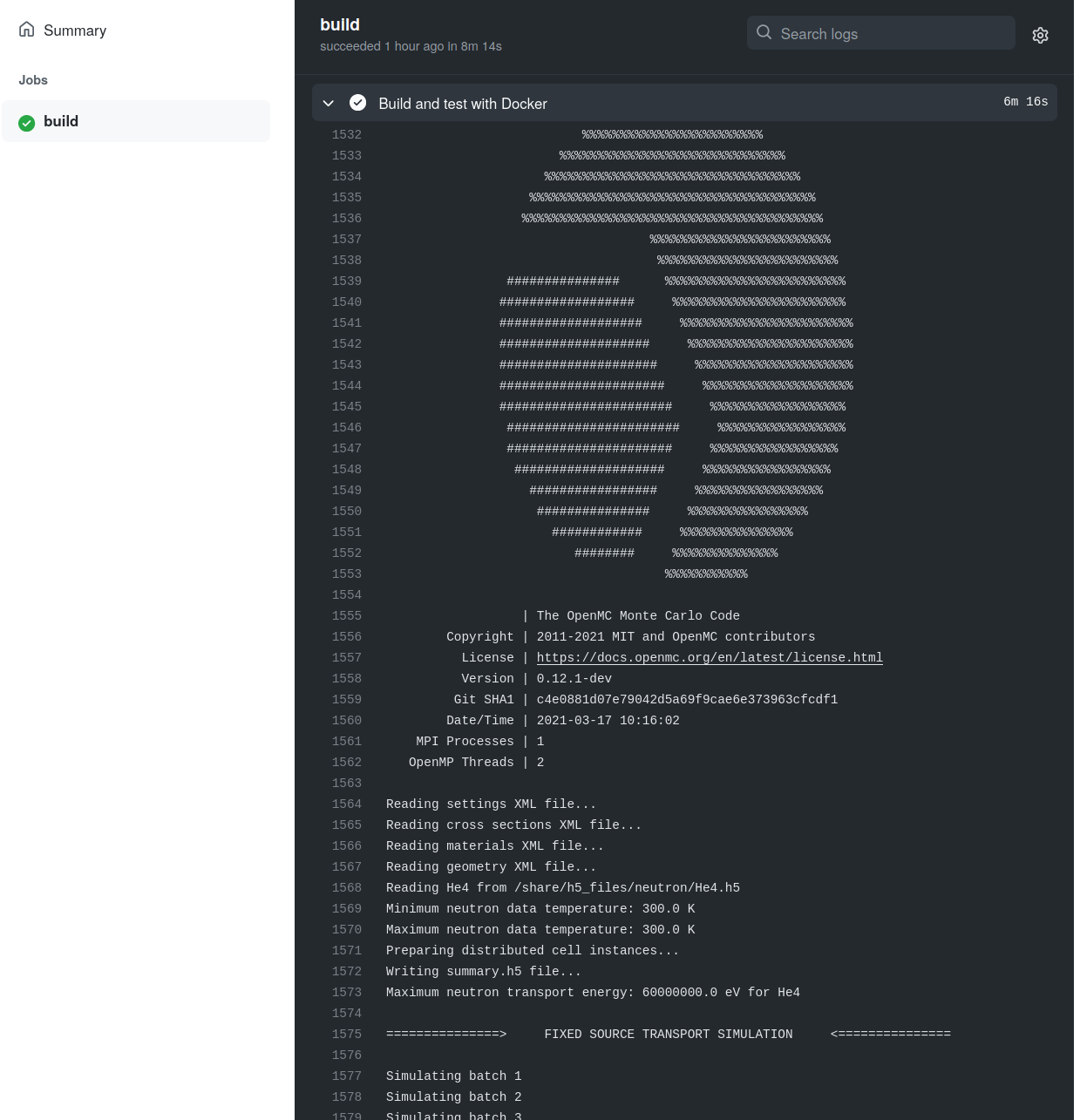
import os
import unittest
import xml.etree.ElementTree as ET
import openmc
os.environ["OPENMC_CROSS_SECTIONS"] = '/share/h5_files/cross_sections.xml'
class TestH5FilesInSimulations(unittest.TestCase):
def test_simulation_runs_with_isotopes(self):
tree = ET.parse('/share/h5_files/cross_sections.xml')
root = tree.getroot()
for elem in root:
if elem.attrib['type'] == 'neutron':
isotope_name = elem.attrib['materials']
sett = openmc.Settings()
sett.batches = 100
sett.inactive = 0
sett.particles = 500
sett.run_mode = 'fixed source'
mat1 = openmc.Material(1, "pure_isotope_material")
mat1.add_nuclide(isotope_name, 1)
mat1.set_density('g/cm3', 1)
mats = openmc.Materials([mat1])
outer_surface = openmc.Sphere(r=10, boundary_type='vacuum')
cell1 = openmc.Cell(region=-outer_surface)
cell1.fill = mat1
universe = openmc.Universe(cells=[cell1])
geom = openmc.Geometry(universe)
model = openmc.model.Model(geom, mats, sett)
model.run()40 Lines
4 Lines
39 Lines
+ + =
Data storage repo - storage
Gigabytes of h5 files each with their own URL endpoint
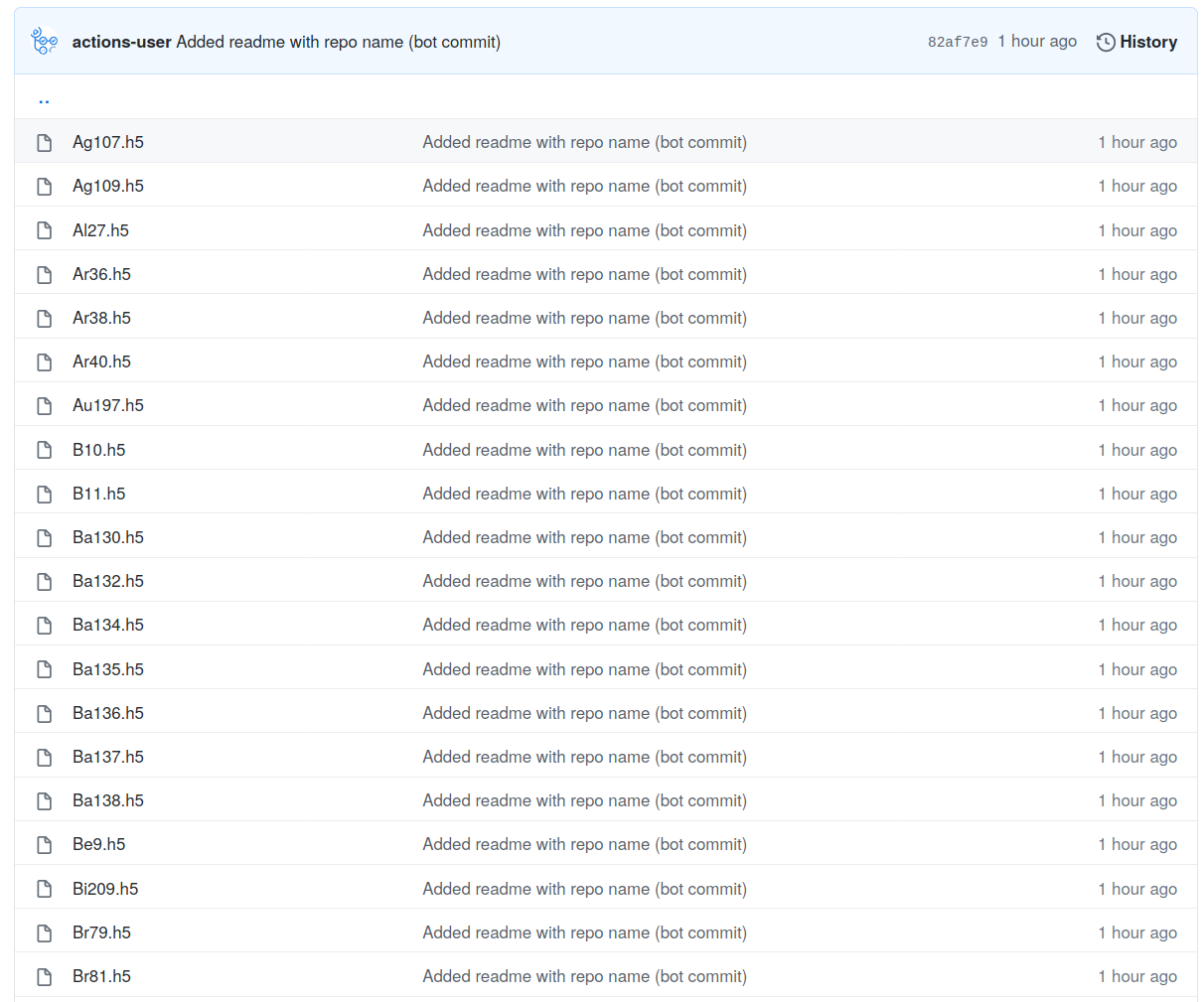
https://github.com/openmc-data-storage/FENDL-3.1d/blob/main/h5_files/neutron/Ag107.h5?raw=true



+ + =
40 Lines
4 Lines
39 Lines
Data storage repo - openmc-data-downloader
Easy to install Python package
pip install openmc_data_downloaderhttps://github.com/openmc-data-storage/openmc_data_downloader
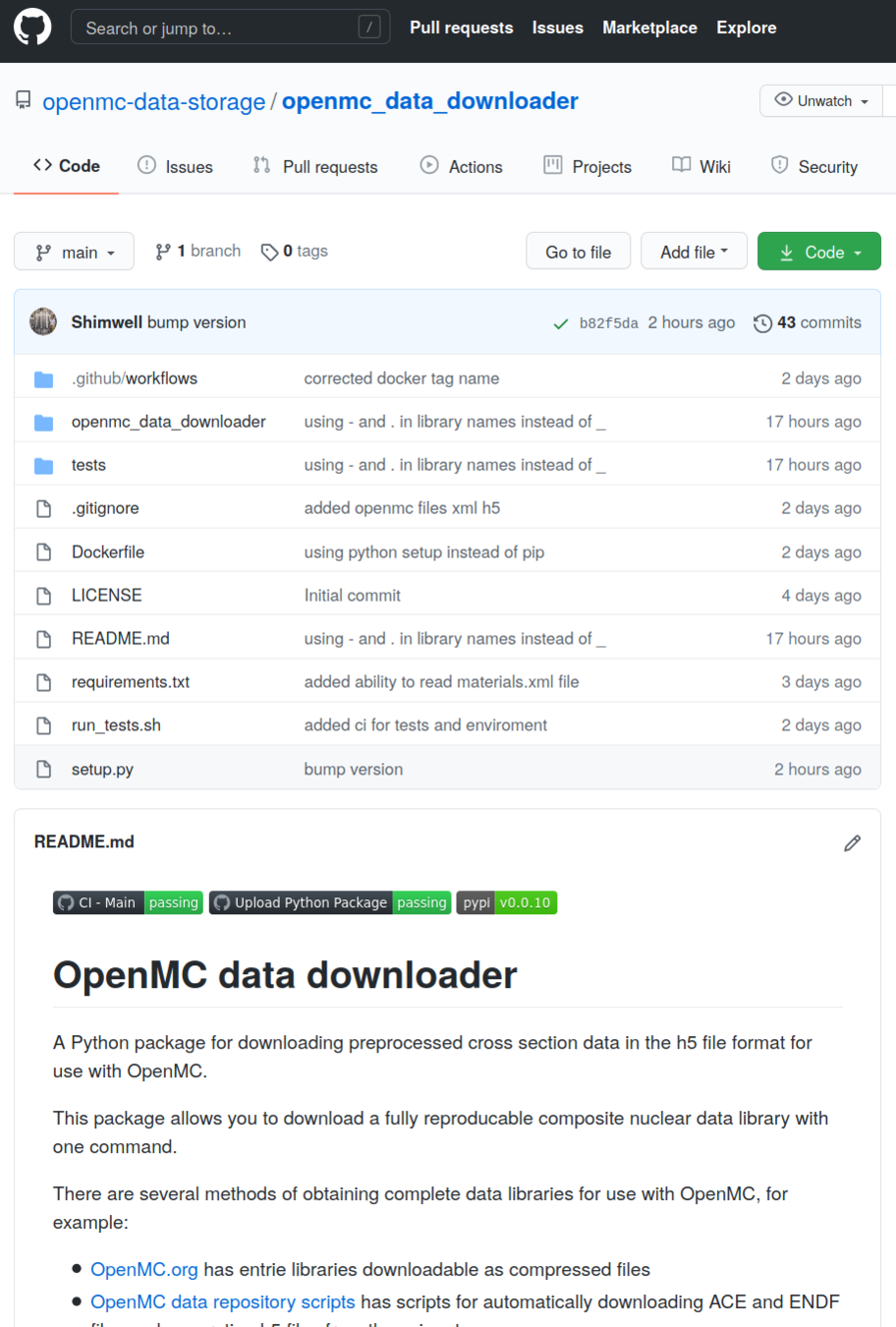
Knowledge of all the URLs of all the h5 files available.
Translation from user instructions to a list of h5 files to download

Internal data based that gets filtered by library / isotopes etc
{
'element': 'Zn',
'isotope': 'Zn66',
'library': 'FENDL-3.1d',
'local_file': 'FENDL-3.1d_Zn66.h5',
'remote_file': 'Zn66.h5',
'url': 'https://github.com/openmc-data-storage/FENDL-3.1d/raw/main/h5_files/neutron/Zn66.h5'
},
...Data storage repo - openmc-data-downloader
Command line usage
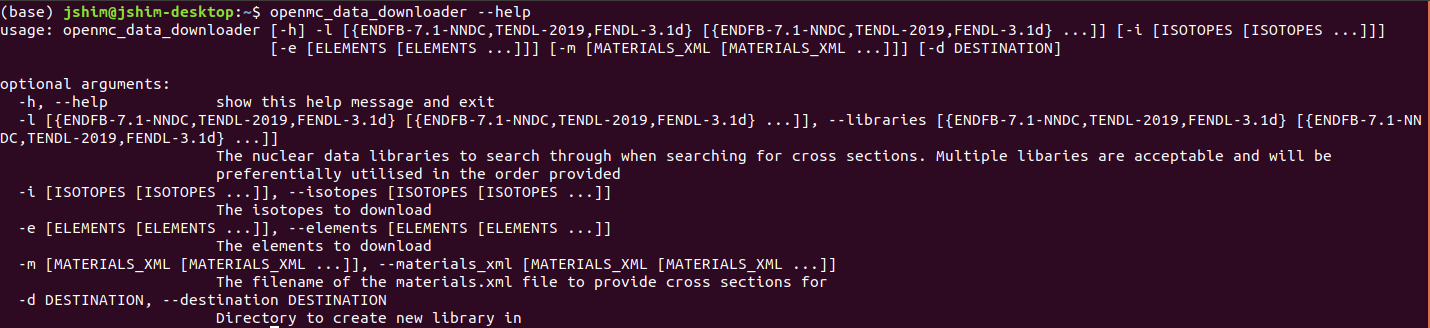
openmc_data_downloader --helpData storage repo - openmc-data-downloader
openmc_data_downloader -l FENDL-3.1d -i Li6Downloads one isotope
openmc_data_downloader -l TENDL-2019 -i Li6 Li7Downloads two isotopes
openmc_data_downloader -l TENDL-2019 -e LiDownloads one element
openmc_data_downloader -l TENDL-2019 -e Li Si NaDownloads two elements
openmc_data_downloader -l ENDFB-7.1-NNDC -i Be9 -d my_h5_filesDownloads to a directory
openmc_data_downloader -l TENDL-2019 -e Li Si Na -i Fe56 U235Downloads a combination of elements and isotopes
openmc_data_downloader -l TENDL-2019 -m materials.xmlDownloads all isotopes in a materials.xml file
openmc_data_downloader -l FENDL-3.1d TENDL-2019 -i Li6 U235Downloads isotopes with library preference
Just in time library generator
import openmc
import openmc_data_downloader as odd
mat1 = openmc.Material(1, "breeder_mat")
mat1.add_element('Pb', 84.2, percent_type='ao')
mat1.add_element('Li', 15.8, percent_type='ao')
mat1.set_density('g/cm3', 11.3)
mats = openmc.Materials([mat1])
odd.just_in_time_library_generator(
libraries='FENDL-3.1d',
materials=mat1
)
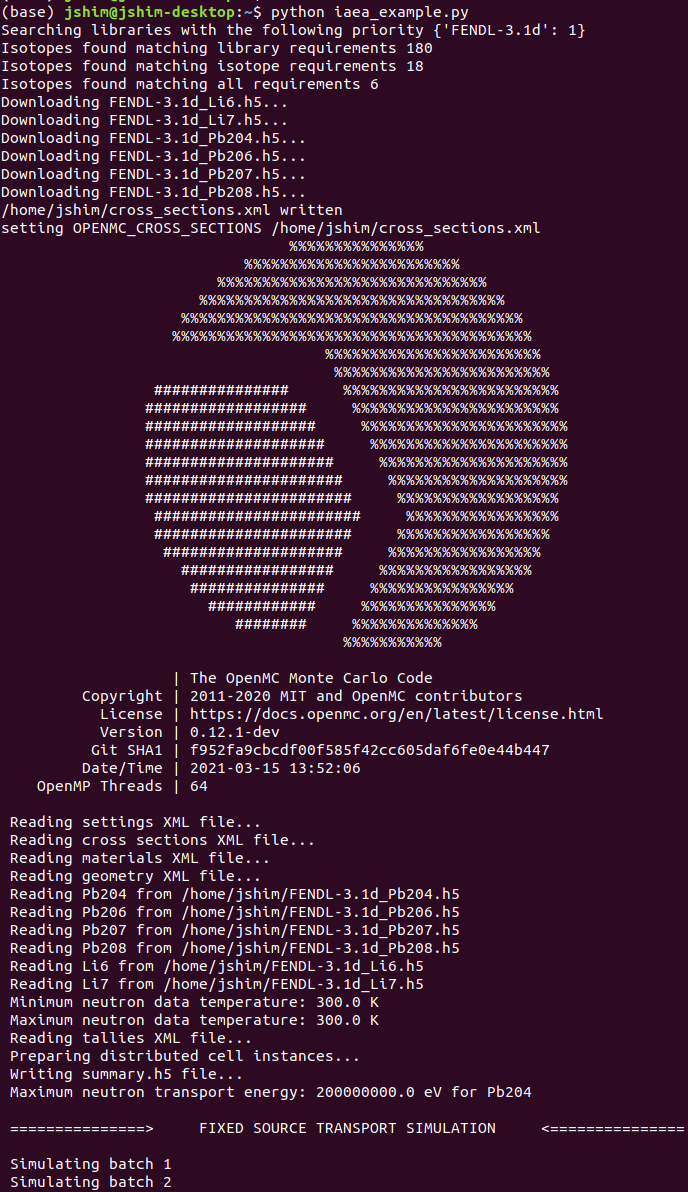
Extra import
Material
Download h5 cross section files for the materials, accepts lists or individual items
Resulting download of h5 files and setting of OPENMC_CROSS_SECTIONS environmental varible
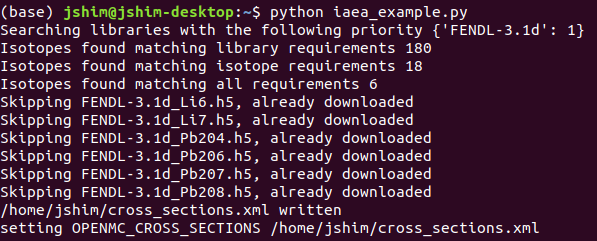
Re-running the same Python script and h5 downloading can be skipped as they already exist
Just in time library generator - in simulation
import openmc
import openmc_data_downloader as odd
mat1 = openmc.Material(1, "breeder_mat")
mat1.add_element('Pb', 84.2, percent_type='ao')
mat1.add_element('Li', 15.8, percent_type='ao')
mat1.set_density('g/cm3', 11.3)
mats = openmc.Materials([mat1])
outer_surface = openmc.Sphere(r=500, boundary_type='vacuum')
cell1 = openmc.Cell(region=-outer_surface)
cell1.fill = mat1
universe = openmc.Universe(cells=[cell1])
geom = openmc.Geometry(universe)
sett = openmc.Settings()
sett.batches = 100
sett.particles = 500
sett.run_mode = 'fixed source'
source = openmc.Source()
source.space = openmc.stats.Point((0, 0, 0))
source.angle = openmc.stats.Isotropic()
source.energy = openmc.stats.Discrete([14e6], [1])
sett.source = source
odd.just_in_time_library_generator(
libraries='FENDL-3.1d',
materials=mat1
)
model = openmc.model.Model(geom, mats, sett)
model.run()

Extra import
Materials
CSG
Settings
Point source
Download h5 cross section files for the materials
Run the simulation
Scale using repository templates
Less time spent making a repo for new nuclear data
Architecture + customization = new nuclear data repo
Live demo
~ 1 min human time
Building blocks
OpenMC Python API
Openmc-dev data repository
Automated processing and testing via CI
Downloading of reproducible collections of isotopes
+URL endpoints for ACE and ENDF libraries
+ Run commands and containerized enviroment
+URL end points for h5 files and libraries
IncidentNeutron, from_endf and from_ace
convert*.py and generate*.py
docker build, docker run and pytest
Summary
There are several methods of obtaining the h5 cross section files needed to run OpenMC simulations.
Each method has a specific use case.
- OpenMC.org - GUI, script, entire libraries, h5 data
- OpenMC-dev data repo - scripts for processing libraries
- openmc-data-downloader - reproducible mixed micro libraries
Future work
- Ability to specify particle type to allow photons
- Ability to specify temperature
Reproducible nuclear data enviroment
By neutronics_workshop
Reproducible nuclear data enviroment
- 538



