From Sequencing to Awareness

Bioinformatic Open Days 2020
C I Mendes • Mário Ramirez Lab
Twitter: @ines_cim
The dengue virus




| Transmission & Disease
The dengue virus
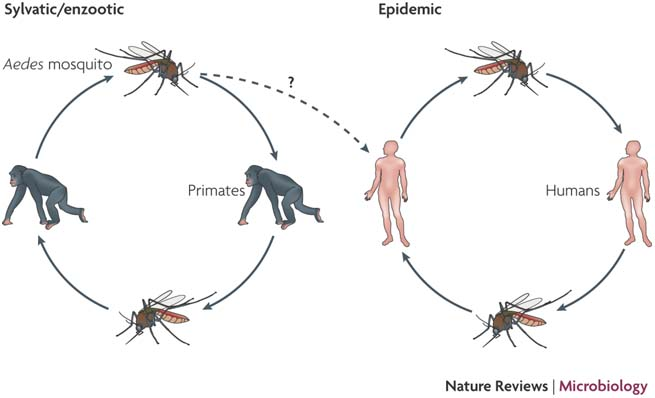
doi:10.1038/nrmicro1690
| Transmission & Disease
Sequential infection increases the risk of a severe form of the infection - dengue hemorrhagic fever.
The dengue virus
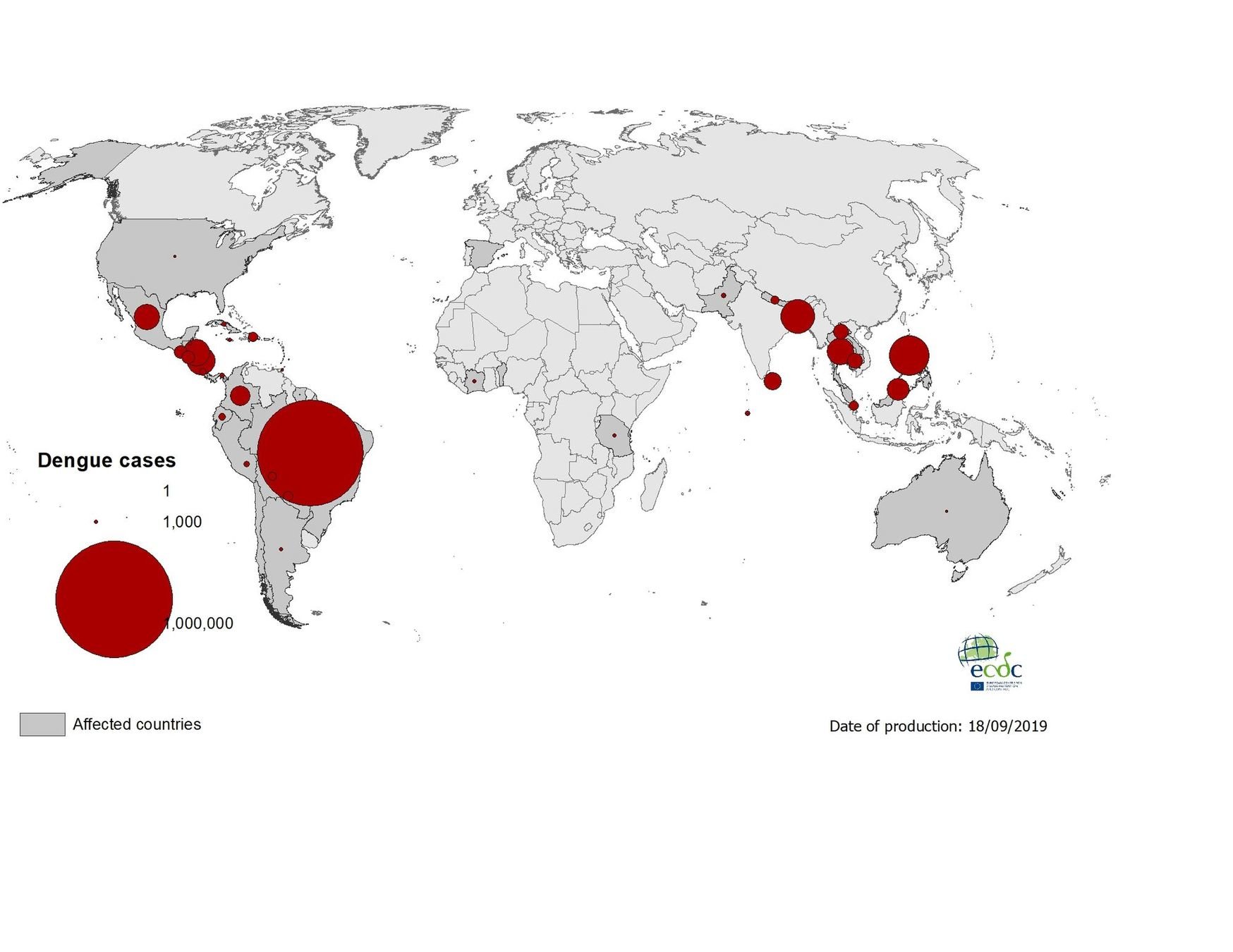

Geographical distribution of dengue cases reported worldwide, July to September 2019
17 September 2019 - laboratory-confirmed autochthonous case of dengue in Barcelona, Spain
| Transmission & Disease
The dengue virus
| The virus
Dengue hemorrhagic fever:
- The virus presents 4 different serotypes, sub-classified in many genotypes
- After "primary" infection with one serotype, "secondary" infections by one or more of the other serotypes can precipitate ‘antibody dependant enhancement’ (ADE)
- Infection with one type gives little immune protection against the other types
The dengue virus
| The virus
- DENV-1 - genotypes I-V
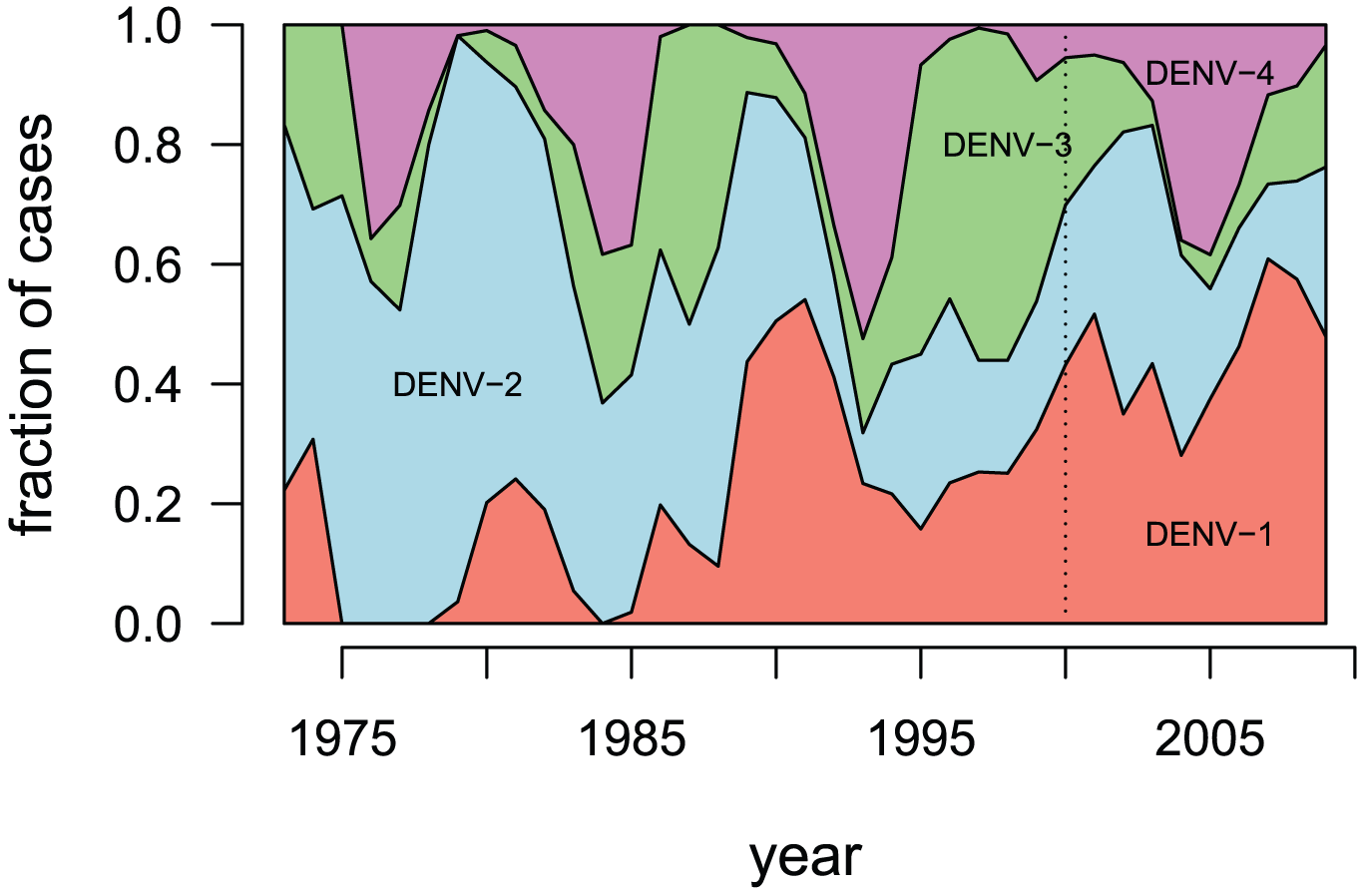
- DENV-2 - genotypes Asian I, Asian II, Cosmopolitan, American, Asian/American & Sylvatic
- DENV-3 - genotypes I-V
- DENV-4 - genotypes I - III & Sylvatic
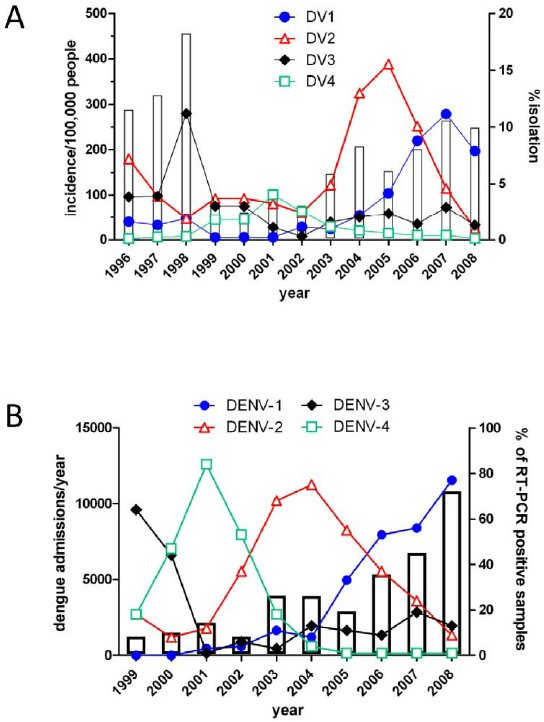
https://doi.org/10.1371/journal.pntd.0001876.g002
https://doi.org/10.1371/journal.pntd.0000757
Thailand
Viet Nam
The dengue virus
DENV: (+)ssRNA (~11Kb; 1 ORF)
The single polyprotein encodes:
-
Structural Proteins:
-
C – capsid
-
prM – pre-membrane
-
M - membrane
-
E - envelope
-
-
Non-Structural Proteins:
-
NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5
-

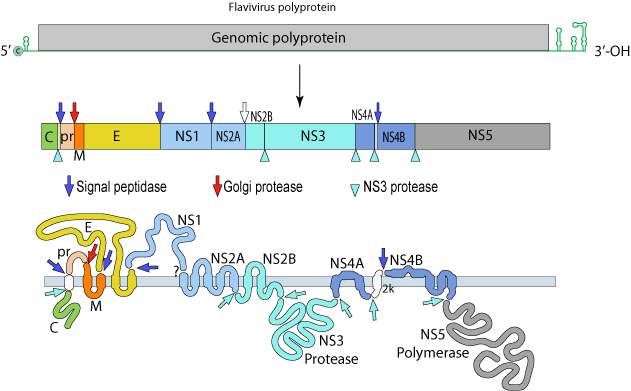
| The virus
The dengue virus
| The virus
Empower the use of HTS to monitor the dissemination of the disease
RNA Extraction
PCR
Amplification
HTS Sequencing
➔
➔
How do the different genotypes model transmission and infection?

| The Workflow
- Ready to use but customizable
- Scalable
- Reproducible
- Stand-alone (confidential data)
- Easy to explore and share results
Requirements


A solution




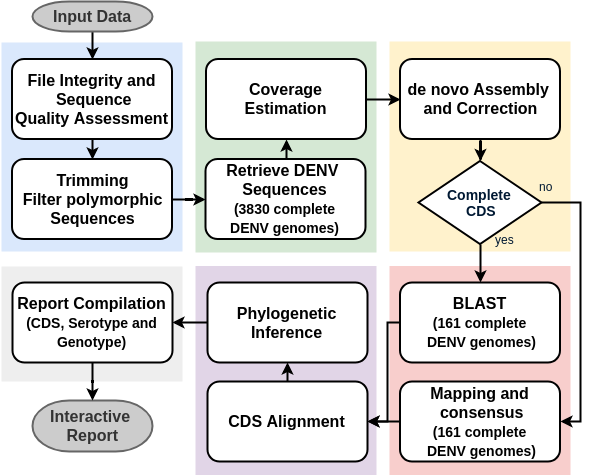
A ready-to-use, reproducible, scalable workflow for the identification, serotyping, genotyping and phylogenetic analysis of DENV from HTS data
| The Workflow




DENV Identification
- 3830 complete DENV genomes from the NIAID Virus Pathogen Database and Analysis Resource (ViPR)
- complete genome sequence
- human host (exception of DENV-1 III, monkey)
- collection year (1950-2018)
In Silico Typing:
- 161 representative sequences of all sero and genotypes.
| The Workflow

| Test datasets

Shotgun Metagenomics dataset:
- 9 plasma samples
- 13 serum samples
- 1 spiked sample with the 4 serotypes
- Positive and Negative controls


nextflow run DEN-IM.nf -profile slurm_shifter --fastq="fastq/*_{1,2}.*"- HPC with 300 Cores/Processing Power and 3 TB RAM
- On average, each sample took 7 minutes to analyse. A total of 75 CPU hours were used to analyse the 25 samples, with a total of 17Gb in size. This analysis resulted in 69Gb of data.

| Test datasets
| Where to find it?

https://doi.org/10.1101/628073




Git, Nextflow (java) and a container engine (Docker, singularity, shifter...).
apt-get install gitcurl -s https://get.nextflow.io | bash
apt-install docker-ce
Clone (or run remotely)
git clone https://github.com/B-UMMI/DEN-IM.git
This work was funded by: FCT - "Fundação para a Ciência e a Tecnologia" (SFRH/BD/129483/2017) and the Abel Tasman Talent Program grant from the UMCG, University of Groningen, Groningen, The Netherland
Special thanks to E Lizarazo, M P Machado, D N Silva, A Tami, M Ramirez, N Couto, J W A Rossen, J A Carriço







Copy of DEN-IM
By Pedro Cerqueira
Copy of DEN-IM
Slide presentation for BOD'2020
- 368



