Flat Buffers
https://google.github.io/flatbuffers/
Flashback to structs in c
struct contact
{
char name[50];
char phone[10];
int age;
};Data is stored as bytes, co-located in memory. It's very small (compared to serialized formats like json), it's fast to read, but it's hard to transport, why?
* If I gave someone else a memory address to read, they'd need to know how long each field is (ex, first 50 bytes are the name field)
* They'd need to know the order of the bytes (big vs little endian)
* What if consumer is java and not c? what's a struct?
Typical deserialization (not just Java)
* Given a byte array
* (for non-schema formats like json) scan the array, look for tokens, parse out structure (i.e. look for {}, "", :, etc)
* Create a new object in memory
* Copy the data into the new memory object
[ A1, 00, B7, 23 ]
class Foo {
int bar; // 161
int baz; // 9143
}needed to know byte order, had to copy data, data in memory twice, extra GC
Flat Buffer approach (simplified)
* Given a byte array
* Given a schema
* data "on demand" read out of the byte array according to schema
[ A1, 00, B7, 23 ]
class Foo(val byteArray) {
fun getBar(): Int {
// jump to offset 0, read 2 bytes as little endian int
}
fun getBaz(): Int {
// jump to offset 2, read 2 bytes as little endian int
}
}Flat Buffer use (tl;dr)
* Write Schema file
* Generate Java classes (or c++/python/etc) from schema
* Use FlatBufferBuilder to build FBO
* Traverse your byte buffer in place
Flat Buffer (physIQ)
* Write Schema file
contracts/flatbuffers/schema/com.physiq.vitalink.sdk.flatbuffers.series.fbs
table Int8Channel {
data:[byte];
}
union ChannelDataUnion {
Int8Channel,
...
StringChannel
}
table ChannelData {
readings:ChannelDataUnion;
}
table SamplingSetData {
channels:[ChannelData];
}
Flat Buffer (physIQ)
* Generate Java Classes
cloud/code/sdk/java-sdk/build/generated/source/
flatbuffers-generator/main/java/com/physiq/
vitalink/sdk/flatbuffers/series/Int8Channel.java
public final class Int8Channel extends Table {
public static Int8Channel getRootAsInt8Channel(ByteBuffer _bb) { return getRootAsInt8Channel(_bb, new Int8Channel()); }
...
public static int createInt8Channel(FlatBufferBuilder builder,
int dataOffset) {
builder.startObject(1);
Int8Channel.addData(builder, dataOffset);
return Int8Channel.endInt8Channel(builder);
}
Flat Buffer (physIQ)
* Build FBO
sampling_sets:
- alias: min-avg
channels:
- alias: hr
desc: Minute average of heart rate.
name: Heart Rate
type: INT16
classification: HR
units: BPMtable SeriesFrame {
frameId:long;
samplingSets:[SamplingSetData];
}
table SamplingSetData {
id:byte;
channels:[ChannelData];
}
cloud/code/flink/processor/sbm/src/test/kotlin/com/physiq/vitalink/timeseries/sbm/DataHelpers.kt
fun buildVitalHRSeriesFrame(frameNumber: Int, x: ShortArray): ByteArray {
return FlatBufferBuilder().apply {
finish(SeriesFrame.createSeriesFrame(
this, // builder
17460 + frameNumber.toLong(), // frame id
-1, // ingested at micros
SeriesFrame.createSamplingSetsVector(
this, // builder
intArrayOf(SamplingSetData.createSamplingSetData(
this, // builder
0.toByte(), // sampling set number
0, // start offset
SamplingSetData.createChannelsVector(
this, // builder
intArrayOf(ChannelData.createChannelData(
this, // builder
0.toByte(), // vector number
ReadingType.INT16,
Int16Channel.createInt16Channel(
this, // builder
Int16Channel.createDataVector(
this, // builder
x // shortArray
)
)
))
)
))),
-1,
0
))
}.sizedByteArray()
} val result = Int16Channel().apply {
flatBufferObject.obj.dataAsSeriesFrame().samplingSets(0)
.channels(0).readings(this)
}
val hr = result.data(0)
Read the data without copies
short hp = monster.hp();
Vec3 pos = monster.pos();Compare to example code from google
Benchmarks and Benefits
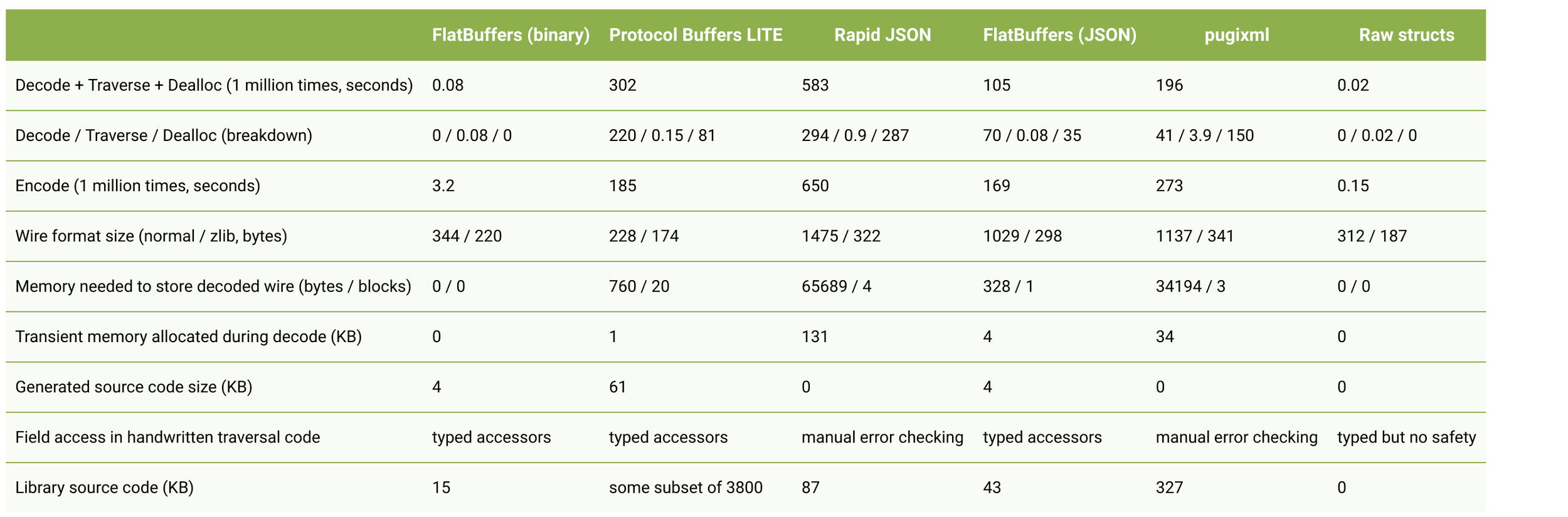
also less GC = better performance for us jvm'ers
Further Reading
Flat buffers https://google.github.io/flatbuffers/
C structs https://www.geeksforgeeks.org/structures-c/
Flat Buffers
By Philip Doctor
Flat Buffers
- 1,812



